Tendon 3D Scaffolds Establish a Tailored Microenvironment Instructing Paracrine Mediated Regenerative Amniotic Epithelial Stem Cells Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Electrospinning and Fabrication of PLGA Fleeces and 3D Scaffolds
2.2.1. Imaging and Morphological Analysis of PLGA Fleeces and 3D Scaffolds
2.2.2. Mechanical Characterization of PLGA Fleeces and 3D Scaffolds
2.3. Biological Evaluation of PLGA Fleeces and 3D Scaffolds on AECs
2.3.1. Ethic Statement
2.3.2. Isolation of Ovine AECs
2.3.3. PLGA Constructs Sterilization and Conditioning
2.3.4. Cell Seeding on PLGA Fleeces and 3D Scaffolds
2.3.5. Immunofluorescence Analyses on Seeded AECs
- -
- Cells morphology by calculating the cellular nuclei aspect ratio.
- -
- Tenogenic differentiation by detecting TNMD, a tendon-related late marker expression.
- -
- Mechano-mediated signal activation of by detecting YAP marker’ cell localization.
2.3.6. Reverse Transcription and Real Time Polymerase Chain Reaction (RT-PCR)
2.3.7. Conditioned Medium (CM) Collection and Their Immunomodulatory Properties Evaluation
2.3.8. Western Blot Analysis
2.3.9. CM Biological Effect on THP-1 Cells
2.4. Statistical Analysis
3. Results
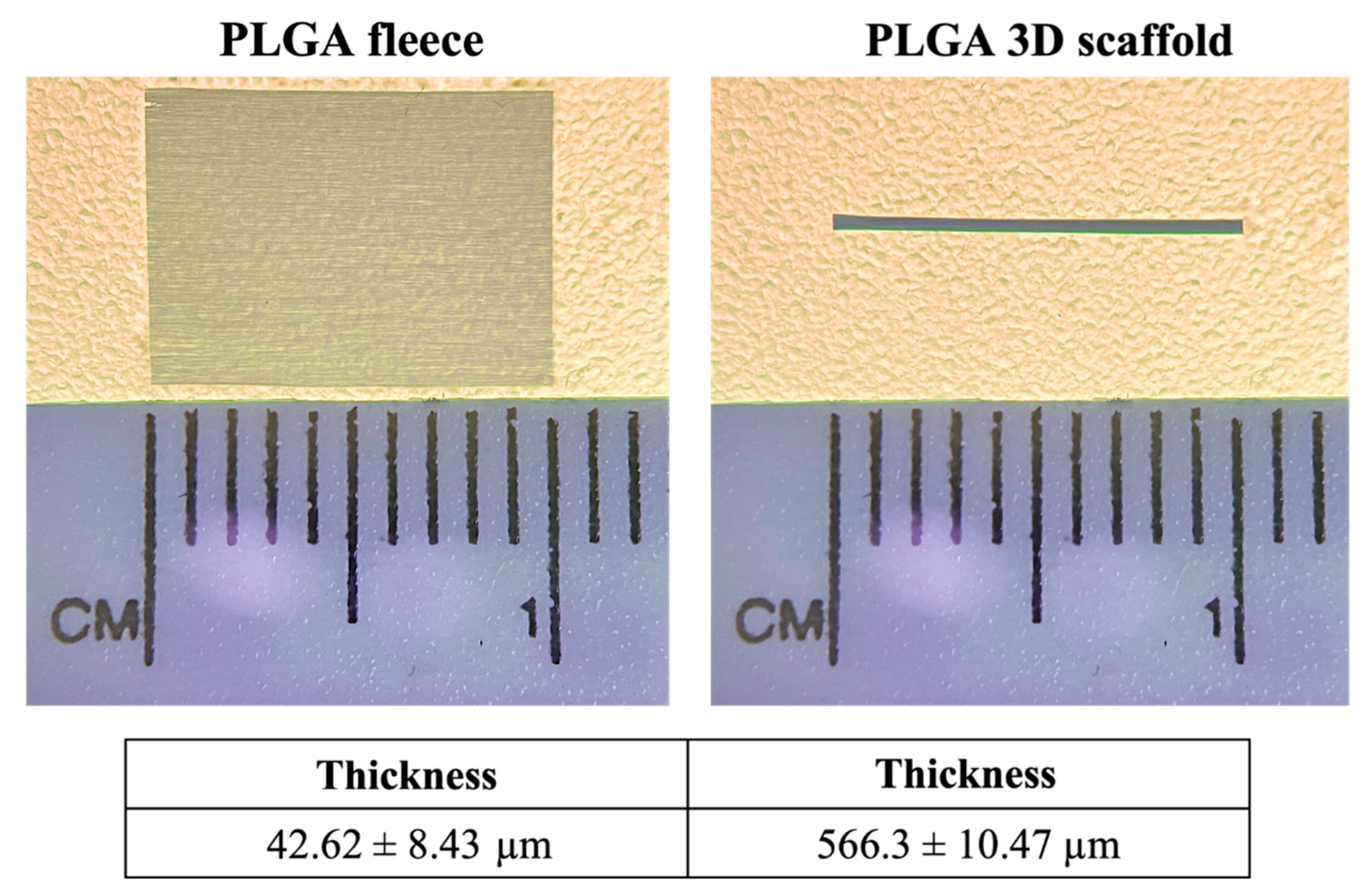
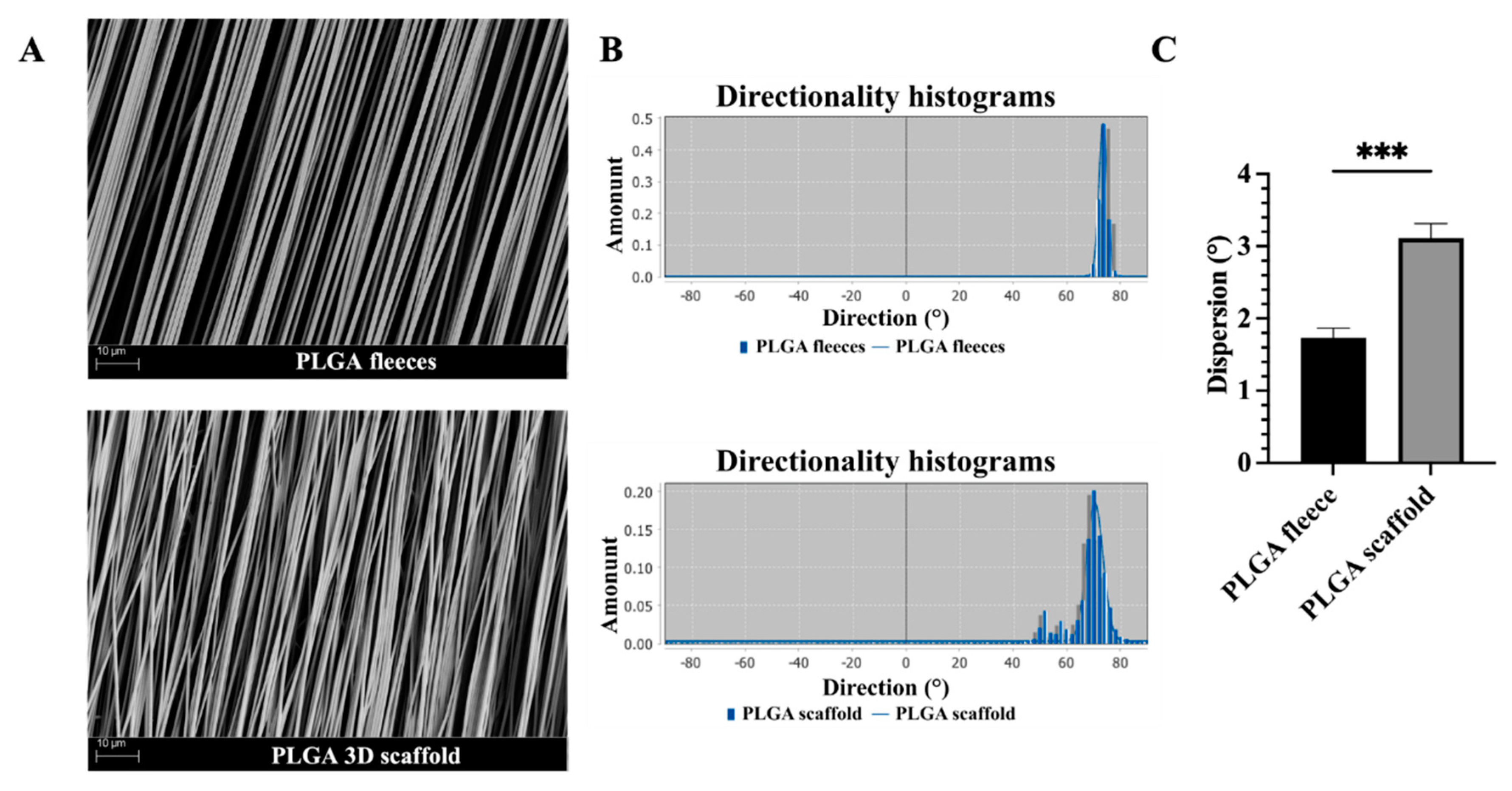
3.1. Topology and Topography of PLGA 3D Scaffolds Resemble Tendon Macroscopy and Microscopy
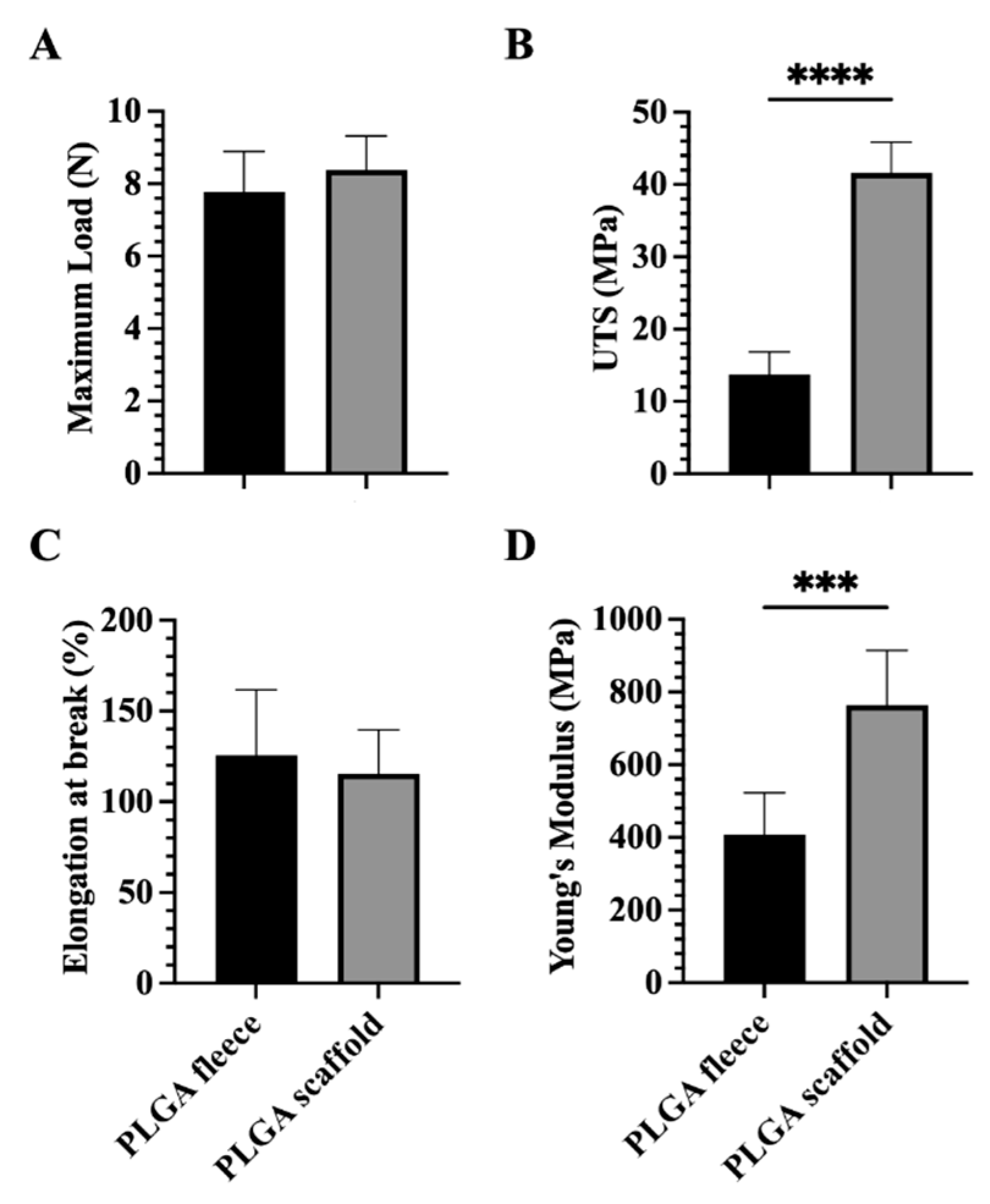
3.2. Mechanical Properties of PLGA 3D Scaffolds Are Enhanced Respect to Fleeces
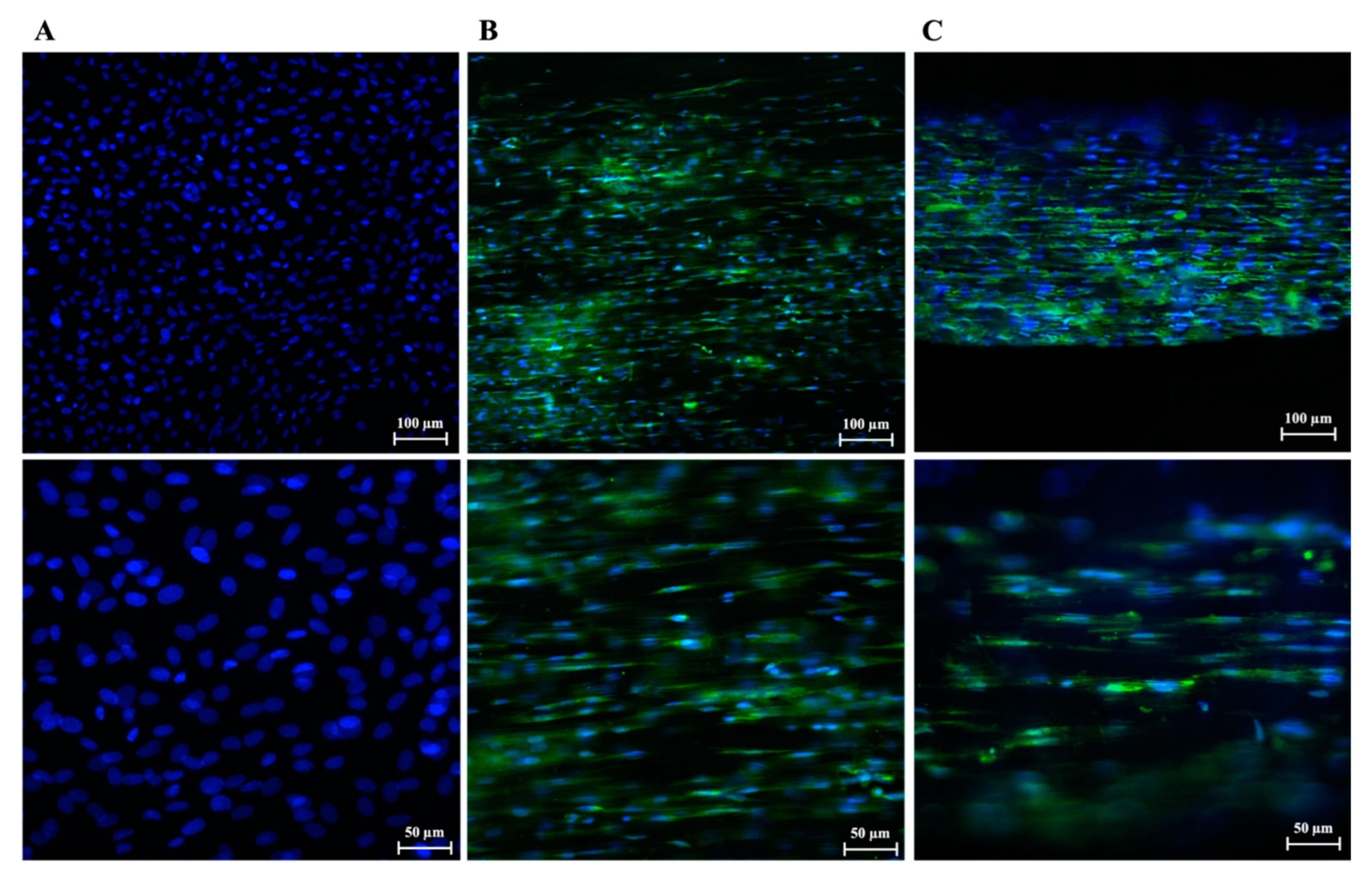
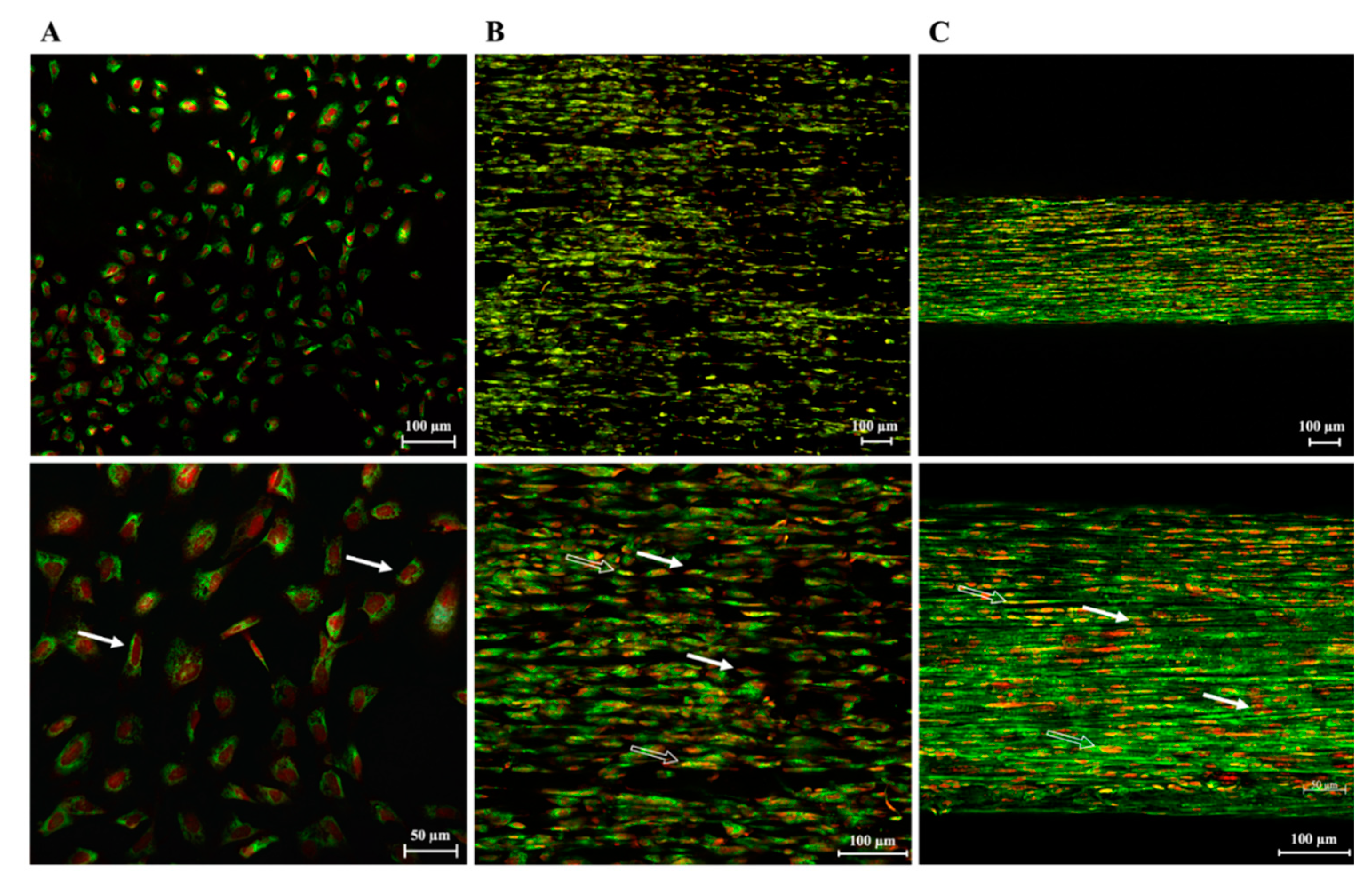
3.3. PLGA Fiber Alignment and 3-Dimensionality of the Construct Influence AECs’ Morphology
3.4. PLGA 3D Scaffolds Possess a Boosted Teno-Inductive Potential on AECs Respect to Fleeces
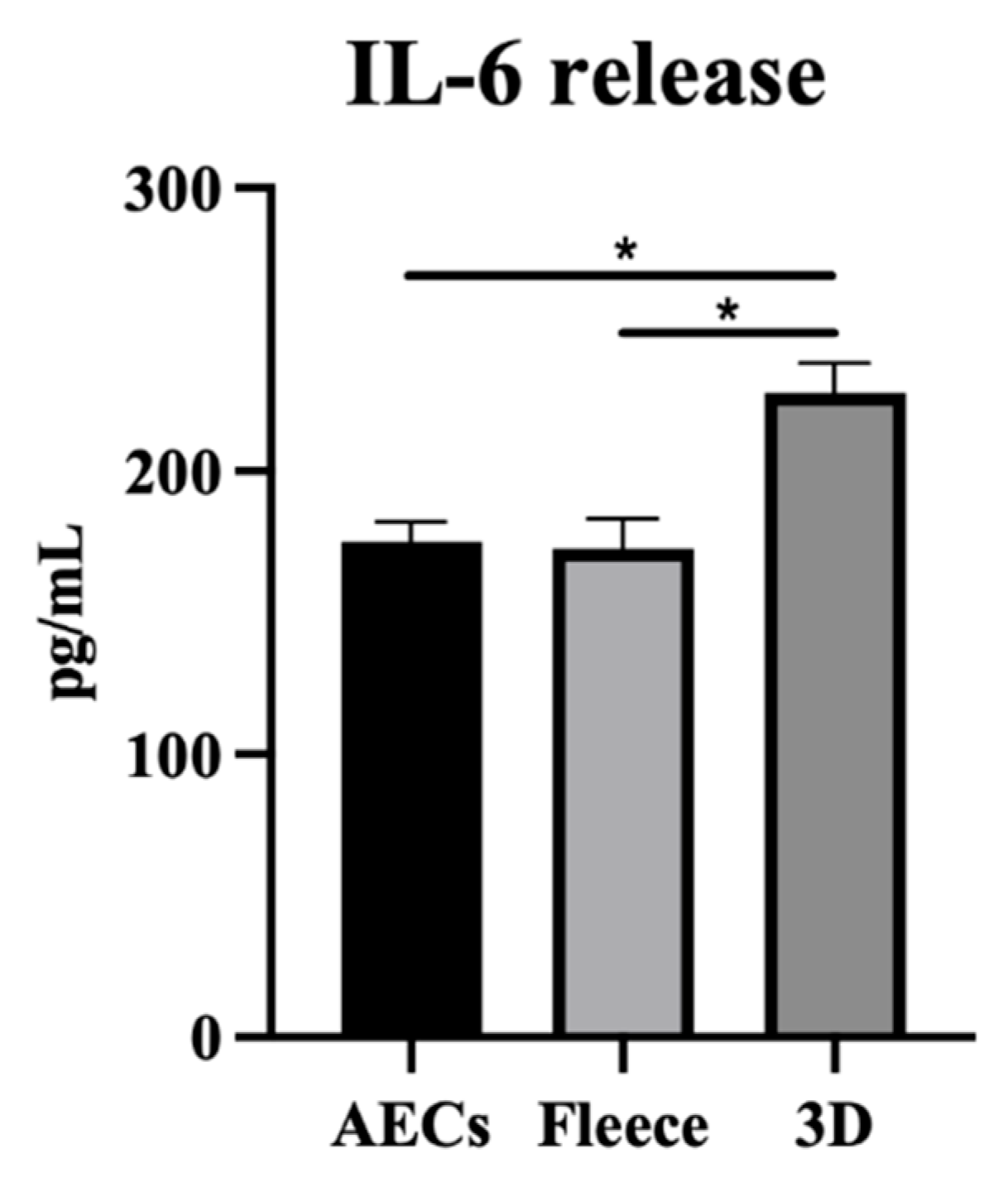
3.5. PLGA 3D Scaffolds Enhance AECs’ Molecular and Paracrine Immune-Modulatory Function
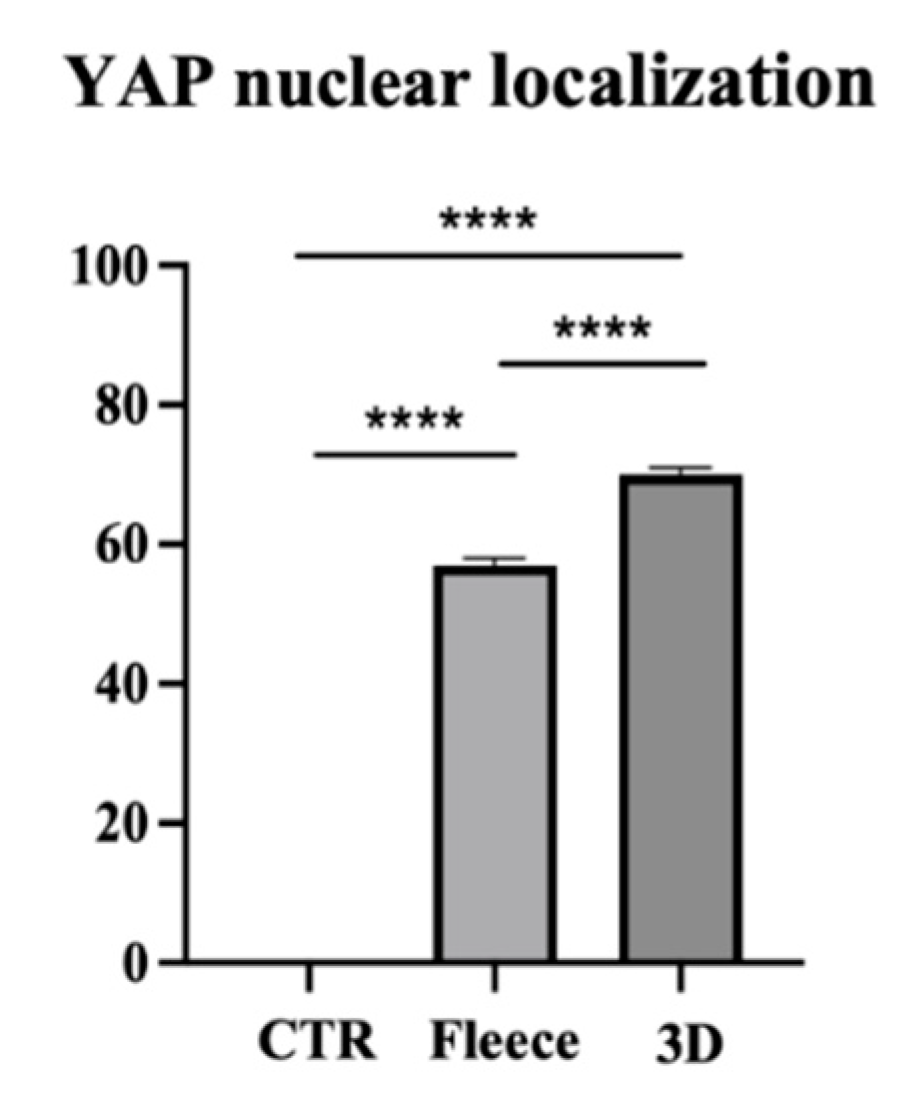
3.6. PLGA 3D Scaffold Greatly Activates AECs’ Mechanosensitive YAP Signaling Pathway over Fleeces
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sayegh, E.T.; Sandy, J.D.; Virk, M.S.; Romeo, A.A.; Wysocki, R.W.; Galante, J.O.; Trella, K.J.; Plaas, A.; Wang, V.M. Recent Scientific Advances Towards the Development of Tendon Healing Strategies. Curr. Tissue Eng. 2015, 4, 128–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomas, A.J.; Ryan, C.N.M.; Sorushanova, A.; Shologu, N.; Sideri, A.I.; Tsioli, V.; Fthenakis, G.C.; Tzora, A.; Skoufos, I.; Quinlan, L.R.; et al. The Past, Present and Future in Scaffold-Based Tendon Treatments. Adv. Drug Deliv. Rev. 2015, 84, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Caddeo, S.; Boffito, M.; Sartori, S. Tissue Engineering Approaches in the Design of Healthy and Pathological in Vitro Tissue Models. Front. Bioeng. Biotechnol. 2017, 5, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carriel, V.; Martínez-Medina, I.; González-Quevedo, D.; Campos, F.; Campos, A. Tissue Engineering Strategies for the Treatment of Tendon Injuries. Bone Jt. Res. 2018, 7, 318–324. [Google Scholar] [CrossRef]
- Longo, U.G.; Lamberti, A.; Petrillo, S.; Maffulli, N.; Denaro, V. Scaffolds in Tendon Tissue Engineering. Stem Cells Int. 2012, 2012, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Su, N.; Gao, P.-L.; Wang, K.; Wang, J.-Y.; Zhong, Y.; Luo, Y. Fibrous Scaffolds Potentiate the Paracrine Function of Mesenchymal Stem Cells: A New Dimension in Cell-Material Interaction. Biomaterials 2017, 141, 74–85. [Google Scholar] [CrossRef]
- Beldjilali-Labro, M.; Garcia, A.G.; Farhat, F.; Bedoui, F.; Grosset, J.-F.F.; Dufresne, M.; Legallais, C.; Garcia Garcia, A.; Farhat, F.; Bedoui, F.; et al. Biomaterials in Tendon and Skeletal Muscle Tissue Engineering: Current Trends and Challenges. Materials 2018, 11, 1116. [Google Scholar] [CrossRef] [Green Version]
- Sensini, A.; Cristofolini, L. Biofabrication of Electrospun Scaffolds for the Regeneration of Tendons and Ligaments. Materials 2018, 11, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.; Ding, J. Poly(Lactide-Co-Glycolide) Porous Scaffolds for Tissue Engineering and Regenerative Medicine. Interface Focus 2012, 2, 366–377. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, M.; John, B. Synthetic Biopolymer Nanocomposites for Tissue Engineering Scaffolds. Prog. Polym. Sci. 2013, 38, 1487–1503. [Google Scholar] [CrossRef]
- Goh, J.C.H.; Sahoo, S. Scaffolds for Tendon and Ligament Tissue Engineering. In Regenerative Medicine and Biomaterials for the Repair of Connective Tissues; Archer, C., Ralphs, J., Eds.; Woodhead Publishing: Cambridge, UK, 2010; pp. 452–463. ISBN 978-1-84569-779-2. [Google Scholar]
- Russo, V.; Tammaro, L.; Di Marcantonio, L.; Sorrentino, A.; Ancora, M.; Valbonetti, L.; Turriani, M.; Martelli, A.; Cammà, C.; Barboni, B. Amniotic Epithelial Stem Cell Biocompatibility for Electrospun Poly(Lactide-Co-Glycolide), Poly(ε-Caprolactone), Poly(Lactic Acid) Scaffolds. Mater. Sci. Eng. C 2016, 69, 321–329. [Google Scholar] [CrossRef]
- Barboni, B.; Curini, V.; Russo, V.; Mauro, A.; Di Giacinto, O.; Marchisio, M.; Alfonsi, M.; Mattioli, M.; Giacinto, O.; Marchisio, M.; et al. Indirect Co-Culture with Tendons or Tenocytes Can Program Amniotic Epithelial Cells towards Stepwise Tenogenic Differentiation. PLoS ONE 2012, 7, e30974–e30987. [Google Scholar] [CrossRef] [Green Version]
- Citeroni, M.R.; Mauro, A.; Ciardulli, M.C.; Di Mattia, M.; El Khatib, M.; Russo, V.; Turriani, M.; Santer, M.; Della Porta, G.; Maffulli, N.; et al. Amnion-Derived Teno-Inductive Secretomes: A Novel Approach to Foster Tendon Differentiation and Regeneration in an Ovine Model. Front. Bioeng. Biotechnol. 2021, 9, 649288. [Google Scholar] [CrossRef]
- Russo, V.; El Khatib, M.; Di Marcantonio, L.; Ancora, M.; Wyrwa, R.; Mauro, A.; Walter, T.; Weisser, J.; Citeroni, M.R.; Lazzaro, F.; et al. Tendon Biomimetic Electrospun PLGA Fleeces Induce an Early Epithelial-Mesenchymal Transition and Tenogenic Differentiation on Amniotic Epithelial Stem Cells. Cells 2020, 9, 303. [Google Scholar] [CrossRef] [Green Version]
- El Khatib, M.; Mauro, A.; Di Mattia, M.; Wyrwa, R.; Schweder, M.; Ancora, M.; Lazzaro, F.; Berardinelli, P.; Valbonetti, L.; Di Giacinto, O.; et al. Electrospun PLGA Fiber Diameter and Alignment of Tendon Biomimetic Fleece Potentiate Tenogenic Differentiation and Immunomodulatory Function of Amniotic Epithelial Stem Cells. Cells 2020, 9, 1207. [Google Scholar] [CrossRef]
- El Khatib, M.; Mauro, A.; Wyrwa, R.; Di Mattia, M.; Turriani, M.; Di Giacinto, O.; Kretzschmar, B.; Seemann, T.; Valbonetti, L.; Berardinelli, P.; et al. Fabrication and Plasma Surface Activation of Aligned Electrospun PLGA Fiber Fleeces with Improved Adhesion and Infiltration of Amniotic Epithelial Stem Cells Maintaining Their Teno-Inductive Potential. Molecules 2020, 25, 3176. [Google Scholar] [CrossRef]
- Barboni, B.; Russo, V.; Curini, V.; Martelli, A.; Berardinelli, P.; Mauro, A.; Mattioli, M.; Marchisio, M.; Bonassi Signoroni, P.; Parolini, O.; et al. Gestational Stage Affects Amniotic Epithelial Cells Phenotype, Methylation Status, Immunomodulatory and Stemness Properties. Stem Cell Rev. Rep. 2014, 10, 725–741. [Google Scholar] [CrossRef] [Green Version]
- Canciello, A.; Russo, V.; Berardinelli, P.; Bernabò, N.; Muttini, A.; Mattioli, M.; Barboni, B. Progesterone Prevents Epithelial-Mesenchymal Transition of Ovine Amniotic Epithelial Cells and Enhances Their Immunomodulatory Properties. Sci. Rep. 2017, 7, 3761. [Google Scholar] [CrossRef] [Green Version]
- Barboni, B.; Russo, V.; Curini, V.; Mauro, A.; Martelli, A.; Muttini, A.; Bernabò, N.; Valbonetti, L.; Marchisio, M.; Di Giacinto, O.; et al. Achilles Tendon Regeneration Can Be Improved by Amniotic Epithelial Cell Allotransplantation. Cell Transplant. 2012, 21, 2377–2395. [Google Scholar] [CrossRef] [Green Version]
- Barboni, B.; Russo, V.; Gatta, V.; Bernabò, N.; Berardinelli, P.; Mauro, A.; Martelli, A.; Valbonetti, L.; Muttini, A.; Di Giacinto, O.; et al. Therapeutic Potential of HAECs for Early Achilles Tendon Defect Repair through Regeneration. J. Tissue Eng. Regen. Med. 2018, 12, e1594–e1608. [Google Scholar] [CrossRef]
- Muttini, A.; Barboni, B.; Valbonetti, L.; Russo, V.; Maffulli, N. Amniotic Epithelial Stem Cells: Salient Features and Possible Therapeutic Role. Sport. Med. Arthrosc. Rev. 2018, 26, 70–74. [Google Scholar] [CrossRef]
- Russo, V.; Paolo, B.; Gatta, V.; Muttini, A.; Stuppia, L.; Parolini, O.; Mattioli, M.; Barboni, B. Cross-Talk between Human Amniotic Derived Cells and Host Tendon Supports Tissue Regeneration. J. Tissue Eng. Regen. Med. 2014, 8, 142. [Google Scholar]
- Tomàs, A.R.; Gonçalves, A.I.; Paz, E.; Freitas, P.; Domingues, R.M.A.; Gomes, M.E. Magneto-Mechanical Actuation of Magnetic Responsive Fibrous Scaffolds Boosts Tenogenesis of Human Adipose Stem Cells. Nanoscale 2019, 11, 18255–18271. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, H.; Liu, H.; Chen, X.; Lu, P.; Zhu, T.; Yang, L.; Yin, Z.; Heng, B.C.; Zhang, Y.; et al. Well-Aligned Chitosan-Based Ultrafine Fibers Committed Teno-Lineage Differentiation of Human Induced Pluripotent Stem Cells for Achilles Tendon Regeneration. Biomaterials 2015, 53, 716–730. [Google Scholar] [CrossRef]
- Russo, V.; El Khatib, M.; Prencipe, G.; Cerveró-Varona, A.; Citeroni, M.R.; Mauro, A.; Berardinelli, P.; Faydaver, M.; Haidar-Montes, A.A.; Turriani, M.; et al. Scaffold-Mediated Immunoengineering as Innovative Strategy for Tendon Regeneration. Cells 2022, 11, 266. [Google Scholar] [CrossRef]
- Wan, S.; Fu, X.; Ji, Y.; Li, M.; Shi, X.; Wang, Y. FAK- and YAP/TAZ Dependent Mechanotransduction Pathways Are Required for Enhanced Immunomodulatory Properties of Adipose-Derived Mesenchymal Stem Cells Induced by Aligned Fibrous Scaffolds. Biomaterials 2018, 171, 107–117. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, Y.; Yildirimer, L.; Xu, T.; Zhao, X. Advanced Technology-Driven Therapeutic Interventions for Prevention of Tendon Adhesion: Design, Intrinsic and Extrinsic Factor Considerations. Acta Biomater. 2021, 124, 15–32. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, C.; Li, R.; Zhong, W.; Xu, G.; Zhang, W. Role of Yes-Associated Protein (YAP) in Regulation of Mesenchymal Stem Cell Tenogenic Differentiation. J. Mol. Histol. 2022. [Google Scholar] [CrossRef]
- Mo, J.-S.; Park, H.W.; Guan, K.-L. The Hippo Signaling Pathway in Stem Cell Biology and Cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef] [Green Version]
- Sensini, A.; Gualandi, C.; Cristofolini, L.; Tozzi, G.; Dicarlo, M.; Teti, G.; Mattioli-Belmonte, M.; Focarete, M.L. Biofabrication of Bundles of Poly(Lactic Acid)-Collagen Blends Mimicking the Fascicles of the Human Achille Tendon—PubMed. Biofabrication 2017, 9, 015025. [Google Scholar] [CrossRef]
- Ali, U.; Zhou, Y.; Wang, X.; Li, T. Electrospinning of Continuous Nanofiber Bundles and Twisted Nanofiber Yarns. In Nanofibers—Production, Properties and Functional Applications; Tong, L., Ed.; InTech: London, UK, 2011; pp. 153–174. ISBN 9789533074207. [Google Scholar]
- Mouthy, P.-A.; Zargar, N.; Hakimi, O.; Lostis, E.; Carr, A. Fabrication of Continuous Electrospun Filaments With Potential for Use as Medical Fibres. Biofabrication 2015, 7, 025006. [Google Scholar] [CrossRef]
- El Khatib, M.; Russo, V.; Prencipe, G.; Mauro, A.; Wyrwa, R.; Grimm, G.; Di Mattia, M.; Berardinelli, P.; Schnabelrauch, M.; Barboni, B. Amniotic Epithelial Stem Cells Counteract Acidic Degradation By-Products of Electrospun PLGA Scaffold by Improving Their Immunomodulatory Profile In Vitro. Cells 2021, 10, 3221. [Google Scholar] [CrossRef]
- Barboni, B.; Russo, V.; Berardinelli, P.; Mauro, A.; Valbonetti, L.; Sanyal, H.; Canciello, A.; Greco, L.; Muttini, A.; Gatta, V.; et al. Placental Stem Cells from Domestic Animals: Translational Potential and Clinical Relevance. Cell Transplant. 2018, 27, 93–116. [Google Scholar] [CrossRef]
- Mauro, A.; Russo, V.; Di Marcantonio, L.; Berardinelli, P.; Martelli, A.; Muttini, A.; Mattioli, M.; Barboni, B. M1 and M2 Macrophage Recruitment during Tendon Regeneration Induced by Amniotic Epithelial Cell Allotransplantation in Ovine. Res. Vet. Sci. 2016, 105, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Chuen, F.S.; Chuk, C.Y.; Ping, W.Y.; Nar, W.W.; Kim, H.L.; Ming, C.K. Immunohistochemical Characterization of Cells in Adult Human Patellar Tendons. J. Histochem. Cytochem. 2004, 52, 1151–1157. [Google Scholar] [CrossRef]
- Di Mattia, M.; Mauro, A.; Delle Monache, S.; Pulcini, F.; Russo, V.; Berardinelli, P.; Citeroni, M.R.; Turriani, M.; Peserico, A.; Barboni, B. Hypoxia-Mimetic CoCl2 Agent Enhances Pro-Angiogenic Activities in Ovine Amniotic Epithelial Cells-Derived Conditioned Medium. Cells 2022, 11, 461. [Google Scholar] [CrossRef]
- Sander, H.; Wallace, S.; Plouse, R.; Tiwari, S.; Gomes, A.V. Ponceau S Waste: Ponceau S Staining for Total Protein Normalization. Anal. Biochem. 2019, 575, 44–53. [Google Scholar] [CrossRef]
- Dufrusine, B.; Di Francesco, A.; Oddi, S.; Scipioni, L.; Angelucci, C.B.; D’Addario, C.; Serafini, M.; Häfner, A.-K.; Steinhilber, D.; Maccarrone, M.; et al. Iron-Dependent Trafficking of 5-Lipoxygenase and Impact on Human Macrophage Activation. Front. Immunol. 2019, 10, 1347. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.M.; Shin, E.K.; Sung, J.H.; Moon, G.J.; Kim, E.H.; Cho, Y.H.; Park, H.D.; Bae, H.; Kim, J.; Bang, O.Y. Efficient Scalable Production of Therapeutic Microvesicles Derived from Human Mesenchymal Stem Cells. Sci. Rep. 2018, 8, 1171. [Google Scholar] [CrossRef] [Green Version]
- Zitnay, J.L.; Weiss, J.A. Load Transfer, Damage, and Failure in Ligaments and Tendons: Damage in Ligaments and Tendons. J. Orthop. Res. 2018, 36, 3093–3104. [Google Scholar] [CrossRef] [Green Version]
- Screen, H.R.C. Hierarchical Approaches to Understanding Tendon Mechanics. J. Biomech. Sci. Eng. 2009, 4, 481–499. [Google Scholar] [CrossRef]
- Erisken, C.; Zhang, X.; Moffat, K.L.; Levine, W.N.; Lu, H.H. Scaffold Fiber Diameter Regulates Human Tendon Fibroblast Growth and Differentiation. Tissue Eng. Part A 2013, 19, 519–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Li, X.; Lipner, J.; Manning, C.N.; Schwartz, A.G.; Thomopoulos, S.; Xia, Y. “Aligned-to-Random” Nanofiber Scaffolds for Mimicking the Structure of the Tendon-to-Bone Insertion Site. Nanoscale 2010, 2, 923–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, S.; Lok Toh, S.; Hong Goh, J.C. PLGA Nanofiber-Coated Silk Microfibrous Scaffold for Connective Tissue Engineering. J. Biomed. Mater. Res. Part B Appl. Biomater. 2010, 95, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Costa-Almeida, R.; Gonçalves, A.I.; Gershovich, P.; Rodrigues, M.T.; Reis, R.L.; Gomes, M.E. Tendon Stem Cell Niche. In Tissue-Specific Stem Cell Niche; Turksen, K., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 221–244. [Google Scholar]
- Kastelic, J.; Galeski, A.; Baer, E. The Multicomposite Structure of Tendon. Connect. Tissue Res. 1978, 6, 11–23. [Google Scholar] [CrossRef]
- Kannus, P. Structure of the Tendon Connective Tissue. Scand. J. Med. Sci. Sport. 2000, 10, 312–320. [Google Scholar] [CrossRef]
- Goh, K.L.; Listrat, A.; Béchet, D. Hierarchical Mechanics of Connective Tissues: Integrating Insights from Nano to Macroscopic Studies. J. Biomed. Nanotechnol. 2014, 10, 2464–2507. [Google Scholar] [CrossRef]
- Driscoll, T.P.; Cosgrove, B.D.; Heo, S.-J.; Shurden, Z.E.; Mauck, R.L. Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophys. J. 2015, 108, 2783–2793. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shu, Z.; Qian, K.; Wang, J.; Zhu, H. Harnessing the Properties of Biomaterial to Enhance the Immunomodulation of Mesenchymal Stem Cells. Tissue Eng. Part B Rev. 2019, 25, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Docheva, D.; Müller, S.A.; Majewski, M.; Evans, C.H. Biologics for Tendon Repair. Adv. Drug Deliv. Rev. 2015, 84, 222–239. [Google Scholar] [CrossRef] [Green Version]
- Russo, V.; El Khatib, M.; Prencipe, G.; Citeroni, M.R.; Faydaver, M.; Mauro, A.; Berardinelli, P.; Cerveró-Varona, A.; Haidar-Montes, A.A.; Turriani, M.; et al. Tendon Immune Regeneration: Insights on the Synergetic Role of Stem and Immune Cells during Tendon Regeneration. Cells 2022, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Orr, S.B.; Chainani, A.; Hippensteel, K.J.; Kishan, A.; Gilchrist, C.; Garrigues, N.W.; Ruch, D.S.; Guilak, F.; Little, D. Aligned Multilayered Electrospun Scaffolds for Rotator Cuff Tendon Tissue Engineering. Acta Biomater. 2015, 24, 117–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.M.; Erisken, C.; Iskratsch, T.; Sheetz, M.; Levine, W.N.; Lu, H.H. Polymer Fiber-Based Models of Connective Tissue Repair and Healing. Biomaterials 2017, 112, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Chen, X.; Chen, J.L.; Shen, W.L.; Hieu Nguyen, T.M.; Gao, L.; Ouyang, H.W. The Regulation of Tendon Stem Cell Differentiation by the Alignment of Nanofibers. Biomaterials 2010, 31, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Gugutkov, D.; Gustavsson, J.; Ginebra, M.P.; Altankov, G. Fibrinogen Nanofibers for Guiding Endothelial Cell Behavior. Biomater. Sci. 2013, 1, 1065. [Google Scholar] [CrossRef]
- Wanjare, M.; Hou, L.; Nakayama, K.H.; Kim, J.J.; Mezak, N.P.; Abilez, O.J.; Tzatzalos, E.; Wu, J.C.; Huang, N.F. Anisotropic Microfibrous Scaffolds Enhance the Organization and Function of Cardiomyocytes Derived from Induced Pluripotent Stem Cells. Biomater. Sci. 2017, 5, 1567–1578. [Google Scholar] [CrossRef]
- Russo, V.; Mauro, A.; Martelli, A.; Di Giacinto, O.; Di Marcantonio, L.; Nardinocchi, D.; Berardinelli, P.; Barboni, B. Cellular and Molecular Maturation in Fetal and Adult Ovine Calcaneal Tendons. J. Anat. 2015, 226, 126–142. [Google Scholar] [CrossRef] [Green Version]
- Murchison, N.D.; Price, B.A.; Conner, D.A.; Keene, D.R.; Olson, E.N.; Tabin, C.J.; Schweitzer, R. Regulation of Tendon Differentiation by Scleraxis Distinguishes Force-Transmitting Tendons from Muscle-Anchoring Tendons. Development 2007, 134, 2697–2708. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, R.; Chyung, J.H.; Murtaugh, L.C.; Brent, A.E.; Rosen, V.; Olson, E.N.; Lassar, A.; Tabin, C.J. Analysis of the Tendon Cell Fate Using Scleraxis, a Specific Marker for Tendons and Ligaments. Development 2001, 128, 3855–3866. [Google Scholar] [CrossRef]
- Liu, C.-F.; Aschbacher-Smith, L.; Barthelery, N.J.; Dyment, N.; Butler, D.; Wylie, C. What We Should Know before Using Tissue Engineering Techniques to Repair Injured Tendons: A Developmental Biology Perspective. Tissue Eng. Part B Rev. 2011, 17, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.H.; Yang, R.S.; al-Shaikh, R.; Lane, J.M. Collagen in Tendon, Ligament, and Bone Healing. A Current Review. Clin. Orthop. Relat. Res. 1995, 265–278. [Google Scholar]
- Manuelpillai, U.; Moodley, Y.; Borlongan, C.V.; Parolini, O. Amniotic Membrane and Amniotic Cells: Potential Therapeutic Tools to Combat Tissue Inflammation and Fibrosis? Placenta 2011, 32 (Suppl. 4), S320–S325. [Google Scholar] [CrossRef] [PubMed]
- Saberianpour, S.; Heidarzadeh, M.; Geranmayeh, M.H.; Hosseinkhani, H.; Rahbarghazi, R.; Nouri, M. Tissue Engineering Strategies for the Induction of Angiogenesis Using Biomaterials. J. Biol. Eng. 2018, 12, 36. [Google Scholar] [CrossRef] [Green Version]
- Thomopoulos, S.; Das, R.; Sakiyama-Elbert, S.; Silva, M.J.; Charlton, N.; Gelberman, R.H. BFGF and PDGF-BB for Tendon Repair: Controlled Release and Biologic Activity by Tendon Fibroblasts In Vitro. Ann. Biomed. Eng. 2010, 38, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomopoulos, S.; Zaegel, M.; Das, R.; Harwood, F.L.; Silva, M.J.; Amiel, D.; Sakiyama-Elbert, S.; Gelberman, R.H. PDGF-BB Released in Tendon Repair Using a Novel Delivery System Promotes Cell Proliferation and Collagen Remodeling. J. Orthop. Res. 2007, 25, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Evrova, O.; Kellenberger, D.; Calcagni, M.; Vogel, V.; Buschmann, J. Supporting Cell-Based Tendon Therapy: Effect of PDGF-BB and Ascorbic Acid on Rabbit Achilles Tenocytes In Vitro. Int. J. Mol. Sci. 2020, 21, 458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stålman, A.; Bring, D.; Ackermann, P.W. Chemokine Expression of CCL2, CCL3, CCL5 and CXCL10 during Early Inflammatory Tendon Healing Precedes Nerve Regeneration: An Immunohistochemical Study in the Rat. Knee Surg. Sport. Traumatol. Arthrosc. 2015, 23, 2682–2689. [Google Scholar] [CrossRef]
- Ross, D.; Maerz, T.; Kurdziel, M.; Hein, J.; Doshi, S.; Bedi, A.; Anderson, K.; Baker, K. The Effect of Granulocyte-Colony Stimulating Factor on Rotator Cuff Healing after Injury and Repair. Clin. Orthop. Relat. Res. 2015, 473, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, P.W.; Domeij-Arverud, E.; Leclerc, P.; Amoudrouz, P.; Nader, G.A. Anti-Inflammatory Cytokine Profile in Early Human Tendon Repair. Knee Surg. Sport. Traumatol. Arthrosc. 2013, 21, 1801–1806. [Google Scholar] [CrossRef]
- Lin, T.W.; Cardenas, L.; Glaser, D.L.; Soslowsky, L.J. Tendon Healing in Interleukin-4 and Interleukin-6 Knockout Mice. J. Biomech. 2006, 39, 61–69. [Google Scholar] [CrossRef]
- Chisari, E.; Rehak, L.; Khan, W.S.; Maffulli, N. Tendon Healing Is Adversely Affected by Low-Grade Inflammation. J. Orthop. Surg. Res. 2021, 16, 700. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Product Size (bp) |
|---|---|---|---|
| GAPDH | CCTGCACCACCAACTGCTTG | TTGAGCTCAGGGATGACCTTG | 224 |
| IL-10 | CCAGGATGGTGACTCGACTAG | TGGCTCTGCTCTCCCAGAAC | 76 |
| IL-12b | ACAAAGGAGGCGAGGTTCTG | CTGTGGTCCATGCTGACCTT | 283 |
| TNMD | TGGTGAAGACCTTCACTTTCC | TTAAACCCTCCCCAGCATGC | 352 |
| SCX | AACAGCGTGAACACGGCTTTC | TTTCTCTGGTTGCTGAGGCAG | 299 |
| COL1 | CGTGATCTGCGACGAACTTAA | GTCCAGGAAGTCCAGGTTGT | 212 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, V.; El Khatib, M.; Prencipe, G.; Mauro, A.; Di Giacinto, O.; Haidar-Montes, A.A.; Pulcini, F.; Dufrusine, B.; Cerveró-Varona, A.; Faydaver, M.; et al. Tendon 3D Scaffolds Establish a Tailored Microenvironment Instructing Paracrine Mediated Regenerative Amniotic Epithelial Stem Cells Potential. Biomedicines 2022, 10, 2578. https://doi.org/10.3390/biomedicines10102578
Russo V, El Khatib M, Prencipe G, Mauro A, Di Giacinto O, Haidar-Montes AA, Pulcini F, Dufrusine B, Cerveró-Varona A, Faydaver M, et al. Tendon 3D Scaffolds Establish a Tailored Microenvironment Instructing Paracrine Mediated Regenerative Amniotic Epithelial Stem Cells Potential. Biomedicines. 2022; 10(10):2578. https://doi.org/10.3390/biomedicines10102578
Chicago/Turabian StyleRusso, Valentina, Mohammad El Khatib, Giuseppe Prencipe, Annunziata Mauro, Oriana Di Giacinto, Arlette A. Haidar-Montes, Fanny Pulcini, Beatrice Dufrusine, Adrián Cerveró-Varona, Melisa Faydaver, and et al. 2022. "Tendon 3D Scaffolds Establish a Tailored Microenvironment Instructing Paracrine Mediated Regenerative Amniotic Epithelial Stem Cells Potential" Biomedicines 10, no. 10: 2578. https://doi.org/10.3390/biomedicines10102578














