Polymer Thin Film Promotes Tumor Spheroid Formation via JAK2-STAT3 Signaling Primed by Fibronectin-Integrin α5 and Sustained by LMO2-LDB1 Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Condition
2.2. Spheroid Formation on pV4D4 and Polymer X and Treatment of Inhibitors
2.3. Plasmids
2.4. Lentiviral Transduction and Gene Transfections
2.5. Establishment of Reporter Gene-Introduced Cell Lines
2.6. Immunocytochemistry/Immunofluorescence (ICC/IF)
2.7. RNA Extraction and Quantitative RT-PCR
2.8. Western Blot Analysis
2.9. Reporter Gene Assay
2.10. In Silico Analysis
2.11. Quantification and Statistical Analysis
3. Results
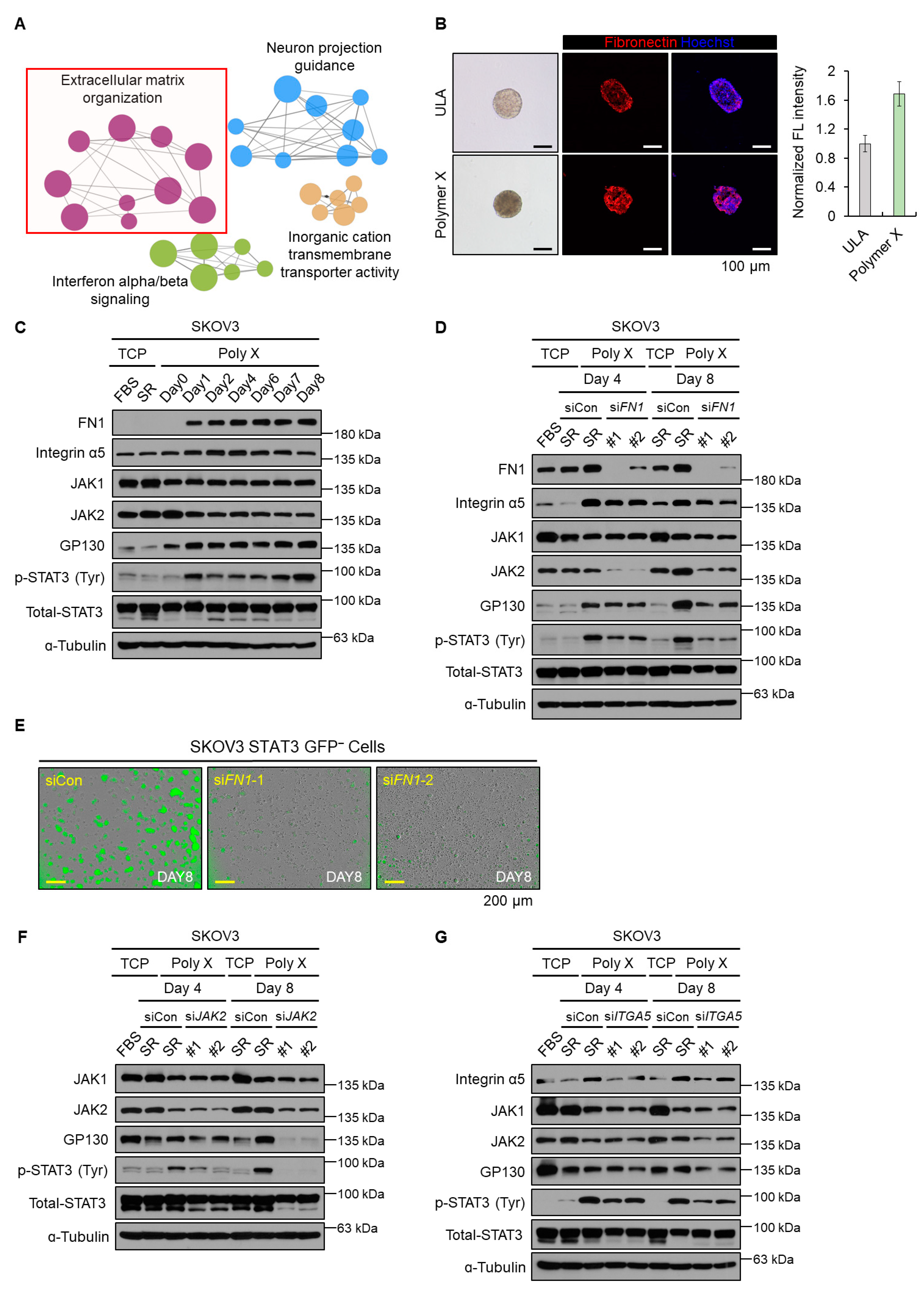
3.1. Polymer X Allowed Formation of Uniform Spheroids and Activated Cellular STAT3 Signaling
3.2. Initial Activation of STAT3 Signaling Was Induced by Fibronectin-JAK2 Axis
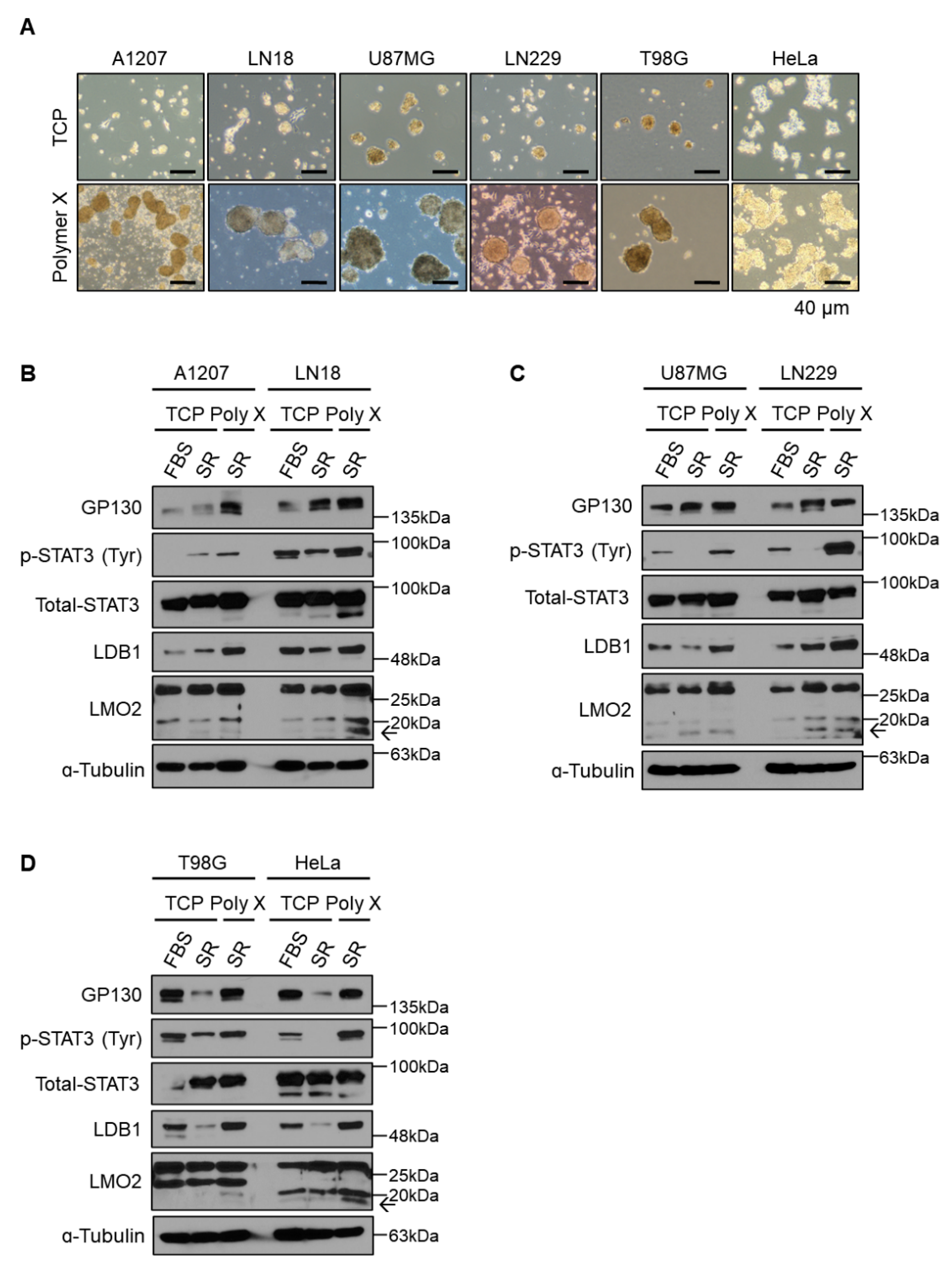
3.3. Long-Term Activation of STAT3 Signaling via LMO2-LDB1 Complex
3.4. Polymer X-Induced Tumor Spheroids Acquired Cancer Stem-Like Properties via STAT3 Signaling
3.5. General Application of Polymer X Using Various Cancer Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in Cancer Treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, J.; Xu, B.; Zhang, X. Tumor metastasis: Mechanistic insights and therapeutic interventions. MedComm 2021, 2, 587–617. [Google Scholar] [CrossRef] [PubMed]
- Labrie, M.; Brugge, J.S.; Mills, G.B.; Zervantonakis, I.K. Therapy resistance: Opportunities created by adaptive responses to targeted therapies in cancer. Nat. Rev. Cancer 2022, 22, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Atashzar, M.R.; Baharlou, R.; Karami, J.; Abdollahi, H.; Rezaei, R.; Pourramezan, F.; Zoljalali Moghaddam, S.H. Cancer stem cells: A review from origin to therapeutic implications. J. Cell. Physiol. 2020, 235, 790–803. [Google Scholar] [CrossRef]
- Matsui, W.H. Cancer stem cell signaling pathways. Medicine 2016, 95, S8–S19. [Google Scholar] [CrossRef]
- Duan, J.J.; Qiu, W.; Xu, S.L.; Wang, B.; Ye, X.Z.; Ping, Y.F.; Zhang, X.; Bian, X.W.; Yu, S.C. Strategies for isolating and enriching cancer stem cells: Well begun is half done. Stem Cells Dev. 2013, 22, 2221–2239. [Google Scholar] [CrossRef]
- Masciale, V.; Grisendi, G.; Banchelli, F.; D’Amico, R.; Maiorana, A.; Sighinolfi, P.; Stefani, A.; Morandi, U.; Dominici, M.; Aramini, B. Isolation and Identification of Cancer Stem-Like Cells in Adenocarcinoma and Squamous Cell Carcinoma of the Lung: A Pilot Study. Front. Oncol. 2019, 9, 1394. [Google Scholar] [CrossRef]
- Zhang, Y.; An, J.; Liu, M.; Li, N.; Wang, W.; Yao, H.; Li, N.; Yang, X.; Sun, Y.; Xu, N.; et al. Efficient isolation, culture, purification, and stem cell expression profiles of primary tumor cells derived from uterine cervical squamous cell carcinoma. Am. J. Reprod. Immunol. 2020, 84, e13251. [Google Scholar] [CrossRef]
- Ooki, A.; VandenBussche, C.J.; Kates, M.; Hahn, N.M.; Matoso, A.; McConkey, D.J.; Bivalacqua, T.J.; Hoque, M.O. CD24 regulates cancer stem cell (CSC)-like traits and a panel of CSC-related molecules serves as a non-invasive urinary biomarker for the detection of bladder cancer. Br. J. Cancer 2018, 119, 961–970. [Google Scholar] [CrossRef]
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Chen, D.; Yang, J.; Yang, C.; Zhang, Y.; Zhang, H.; Dou, J. Evaluation of characteristics of CD44+CD117+ ovarian cancer stem cells in three dimensional basement membrane extract scaffold versus two dimensional monocultures. BMC Cell Biol. 2013, 14, 7. [Google Scholar] [CrossRef]
- Fang, D.D.; Kim, Y.J.; Lee, C.N.; Aggarwal, S.; McKinnon, K.; Mesmer, D.; Norton, J.; Birse, C.E.; He, T.; Ruben, S.M.; et al. Expansion of CD133+ colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br. J. Cancer 2010, 102, 1265–1275. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, S.; Ren, J.; Yao, C.; Zhao, Z.; Qi, X.; Zhang, X.; Wang, S.; Li, L. Salinomycin may inhibit the cancer stem-like populations with increased chemoradioresistance that nasopharyngeal cancer tumorspheres contain. Oncol. Lett. 2018, 16, 2495–2500. [Google Scholar] [CrossRef]
- Serra, A.T.; Serra, M.; Silva, A.C.; Brckalo, T.; Seshire, A.; Brito, C.; Wolf, M.; Alves, P.M. Scalable Culture Strategies for the Expansion of Patient-Derived Cancer Stem Cell Lines. Stem Cells Int. 2019, 2019, 8347595. [Google Scholar] [CrossRef] [PubMed]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Hope, K.J.; Jin, L.; Dick, J.E. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 2004, 5, 738–743. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Yu, S.J.; Choi, Y.; Lee, H.R.; Lee, E.; Lee, E.; Lee, Y.; Song, J.; Son, J.G.; Lee, T.G.; et al. Polymer Thin Film-Induced Tumor Spheroids Acquire Cancer Stem Cell-like Properties. Cancer Res. 2018, 78, 6890–6902. [Google Scholar] [CrossRef] [PubMed]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef]
- Carvalho, J. Cell Reversal From a Differentiated to a Stem-Like State at Cancer Initiation. Front. Oncol. 2020, 10, 541. [Google Scholar] [CrossRef]
- Hu, Y.B.; Yan, C.; Mu, L.; Mi, Y.L.; Zhao, H.; Hu, H.; Li, X.L.; Tao, D.D.; Wu, Y.Q.; Gong, J.P.; et al. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene 2019, 38, 1951–1965. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Kikushige, Y.; Miyawaki, K.; Kunisaki, Y.; Mizuno, S.; Takenaka, K.; Tamura, S.; Okumura, Y.; Ito, M.; Ariyama, H.; et al. Dedifferentiation process driven by TGF-beta signaling enhances stem cell properties in human colorectal cancer. Oncogene 2019, 38, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choi, S.I.; Kim, R.K.; Cho, E.W.; Kim, I.G. Tescalcin/c-Src/IGF1Rβ-mediated STAT3 activation enhances cancer stemness and radioresistant properties through ALDH1. Sci. Rep. 2018, 8, 10711. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshimura, J.; Morishita, S.; Ui-Tei, K. siDirect 2.0: Updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform. 2009, 10, 392. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Joshi, R.K.; Megha, S.; Basu, U.; Rahman, M.H.; Kav, N.N. Genome Wide Identification and Functional Prediction of Long Non-Coding RNAs Responsive to Sclerotinia sclerotiorum Infection in Brassica napus. PLoS ONE 2016, 11, e0158784. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Miao, Y.; Xu, P.; Qiu, X. IL-8 regulates the stemness properties of cancer stem cells in the small-cell lung cancer cell line H446. Onco Targets Ther. 2018, 11, 5723–5731. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; Wu, Y.H.; Chiu, W.T.; Weng, T.Y.; Huang, Y.F.; Chou, C.Y. All-trans retinoic acid downregulates ALDH1-mediated stemness and inhibits tumour formation in ovarian cancer cells. Carcinogenesis 2015, 36, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Balanis, N.; Wendt, M.K.; Schiemann, B.J.; Wang, Z.; Schiemann, W.P.; Carlin, C.R. Epithelial to mesenchymal transition promotes breast cancer progression via a fibronectin-dependent STAT3 signaling pathway. J. Biol. Chem. 2013, 288, 17954–17967. [Google Scholar] [CrossRef]
- Wendt, M.K.; Balanis, N.; Carlin, C.R.; Schiemann, W.P. STAT3 and epithelial-mesenchymal transitions in carcinomas. Jakstat 2014, 3, e28975. [Google Scholar] [CrossRef]
- Meng, X.N.; Jin, Y.; Yu, Y.; Bai, J.; Liu, G.Y.; Zhu, J.; Zhao, Y.Z.; Wang, Z.; Chen, F.; Lee, K.Y.; et al. Characterisation of fibronectin-mediated FAK signalling pathways in lung cancer cell migration and invasion. Br. J. Cancer 2009, 101, 327–334. [Google Scholar] [CrossRef]
- Kim, N.G.; Gumbiner, B.M. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 2015, 210, 503–515. [Google Scholar] [CrossRef]
- Su, C.Y.; Li, J.Q.; Zhang, L.L.; Wang, H.; Wang, F.H.; Tao, Y.W.; Wang, Y.Q.; Guo, Q.R.; Li, J.J.; Liu, Y.; et al. The Biological Functions and Clinical Applications of Integrins in Cancers. Front. Pharmacol. 2020, 11, 579068. [Google Scholar] [CrossRef]
- Behera, R.; Kumar, V.; Lohite, K.; Karnik, S.; Kundu, G.C. Activation of JAK2/STAT3 signaling by osteopontin promotes tumor growth in human breast cancer cells. Carcinogenesis 2010, 31, 192–200. [Google Scholar] [CrossRef]
- Gu, J.; Taniguchi, N. Regulation of integrin functions by N-glycans. GlycoconJ. J. 2004, 21, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, F.; Ray, A.M.; Dontenwill, M. Integrin α5β1, the Fibronectin Receptor, as a Pertinent Therapeutic Target in Solid Tumors. Cancers 2013, 5, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Park, C.G.; Choi, S.H.; Lee, S.Y.; Eun, K.; Park, M.G.; Jang, J.; Jeong, H.J.; Kim, S.J.; Jeong, S.; Lee, K.; et al. Cytoplasmic LMO2-LDB1 Complex Activates STAT3 Signaling through Interaction with gp130-JAK in Glioma Stem Cells. Cells 2022, 11, 2031. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Bayik, D.; Lathia, J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Paluch, E.K.; Nelson, C.M.; Biais, N.; Fabry, B.; Moeller, J.; Pruitt, B.L.; Wollnik, C.; Kudryasheva, G.; Rehfeldt, F.; Federle, W. Mechanotransduction: Use the force(s). BMC Biol. 2015, 13, 47. [Google Scholar] [CrossRef]
- Wagh, K.; Ishikawa, M.; Garcia, D.A.; Stavreva, D.A.; Upadhyaya, A.; Hager, G.L. Mechanical Regulation of Transcription: Recent Advances. Trends Cell Biol. 2021, 31, 457–472. [Google Scholar] [CrossRef]
- Wang, K.; Seo, B.R.; Fischbach, C.; Gourdon, D. Fibronectin Mechanobiology Regulates Tumorigenesis. Cell. Mol. Bioeng. 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Erickson, H.P.; Carrell, N.; McDonagh, J. Fibronectin molecule visualized in electron microscopy: A long, thin, flexible strand. J. Cell Biol. 1981, 91, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, M.J.; Smith, M.L. Multiscale relationships between fibronectin structure and functional properties. Acta Biomater. 2014, 10, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Broders-Bondon, F.; Nguyen Ho-Bouldoires, T.H.; Fernandez-Sanchez, M.E.; Farge, E. Mechanotransduction in tumor progression: The dark side of the force. J. Cell Biol. 2018, 217, 1571–1587. [Google Scholar] [CrossRef] [PubMed]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Eun, K.; Ham, S.W.; Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC targeting. BMB Rep. 2017, 50, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Granados, K.; Poelchen, J.; Novak, D.; Utikal, J. Cellular Reprogramming-A Model for Melanoma Cellular Plasticity. Int. J. Mol. Sci. 2020, 21, 8274. [Google Scholar] [CrossRef]
- Cinquin, O.; Demongeot, J. Positive and negative feedback: Striking a balance between necessary antagonists. J. Theor. Biol. 2002, 216, 229–241. [Google Scholar] [CrossRef]
- Wang, L.S.; Li, N.X.; Chen, J.J.; Zhang, X.P.; Liu, F.; Wang, W. Modulation of dynamic modes by interplay between positive and negative feedback loops in gene regulatory networks. Phys Rev. E 2018, 97, 042412. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, S.; Hong, N.; Song, J.; Kim, D.; Choi, Y.; Lee, D.; Jon, S.; Kim, H. Polymer Thin Film Promotes Tumor Spheroid Formation via JAK2-STAT3 Signaling Primed by Fibronectin-Integrin α5 and Sustained by LMO2-LDB1 Complex. Biomedicines 2022, 10, 2684. https://doi.org/10.3390/biomedicines10112684
Seo S, Hong N, Song J, Kim D, Choi Y, Lee D, Jon S, Kim H. Polymer Thin Film Promotes Tumor Spheroid Formation via JAK2-STAT3 Signaling Primed by Fibronectin-Integrin α5 and Sustained by LMO2-LDB1 Complex. Biomedicines. 2022; 10(11):2684. https://doi.org/10.3390/biomedicines10112684
Chicago/Turabian StyleSeo, Sunyoung, Nayoung Hong, Junhyuk Song, Dohyeon Kim, Yoonjung Choi, Daeyoup Lee, Sangyong Jon, and Hyunggee Kim. 2022. "Polymer Thin Film Promotes Tumor Spheroid Formation via JAK2-STAT3 Signaling Primed by Fibronectin-Integrin α5 and Sustained by LMO2-LDB1 Complex" Biomedicines 10, no. 11: 2684. https://doi.org/10.3390/biomedicines10112684
APA StyleSeo, S., Hong, N., Song, J., Kim, D., Choi, Y., Lee, D., Jon, S., & Kim, H. (2022). Polymer Thin Film Promotes Tumor Spheroid Formation via JAK2-STAT3 Signaling Primed by Fibronectin-Integrin α5 and Sustained by LMO2-LDB1 Complex. Biomedicines, 10(11), 2684. https://doi.org/10.3390/biomedicines10112684





