Protein-Functionalized Microgel for Multiple Myeloma Cells’ 3D Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Polymerization and Microspheres Production
2.2. Sterilization and Functionalization
2.3. Characterization
2.4. Cell Culture
2.5. Proliferation Assay
2.6. Cell Cycle Assay and CAM-DR Effect on RPMI 8226 Cell Line
2.7. Statistical Analysis
3. Results
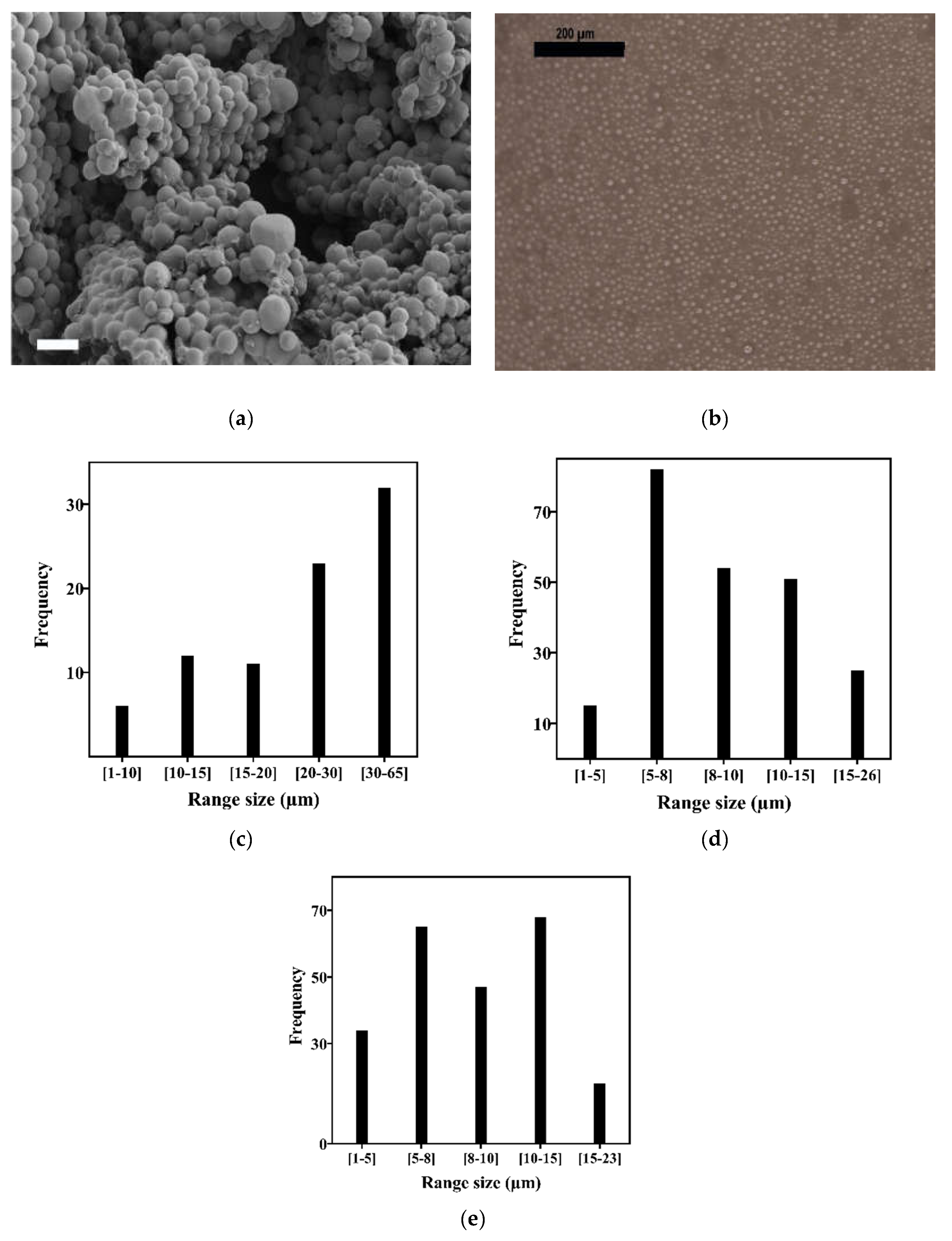
3.1. Acrylate Microsphere Production and Characterization
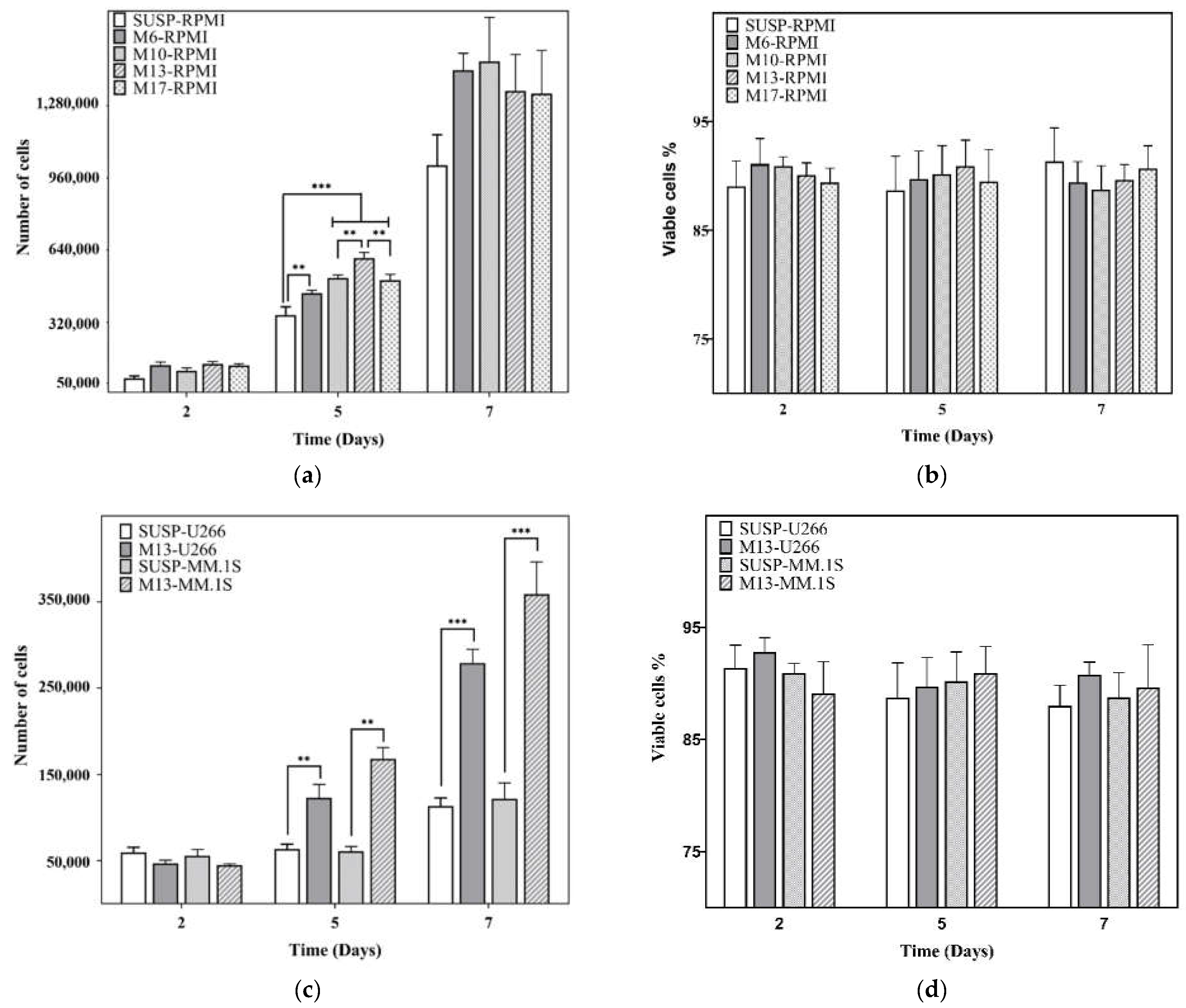
3.2. Proliferation Assay
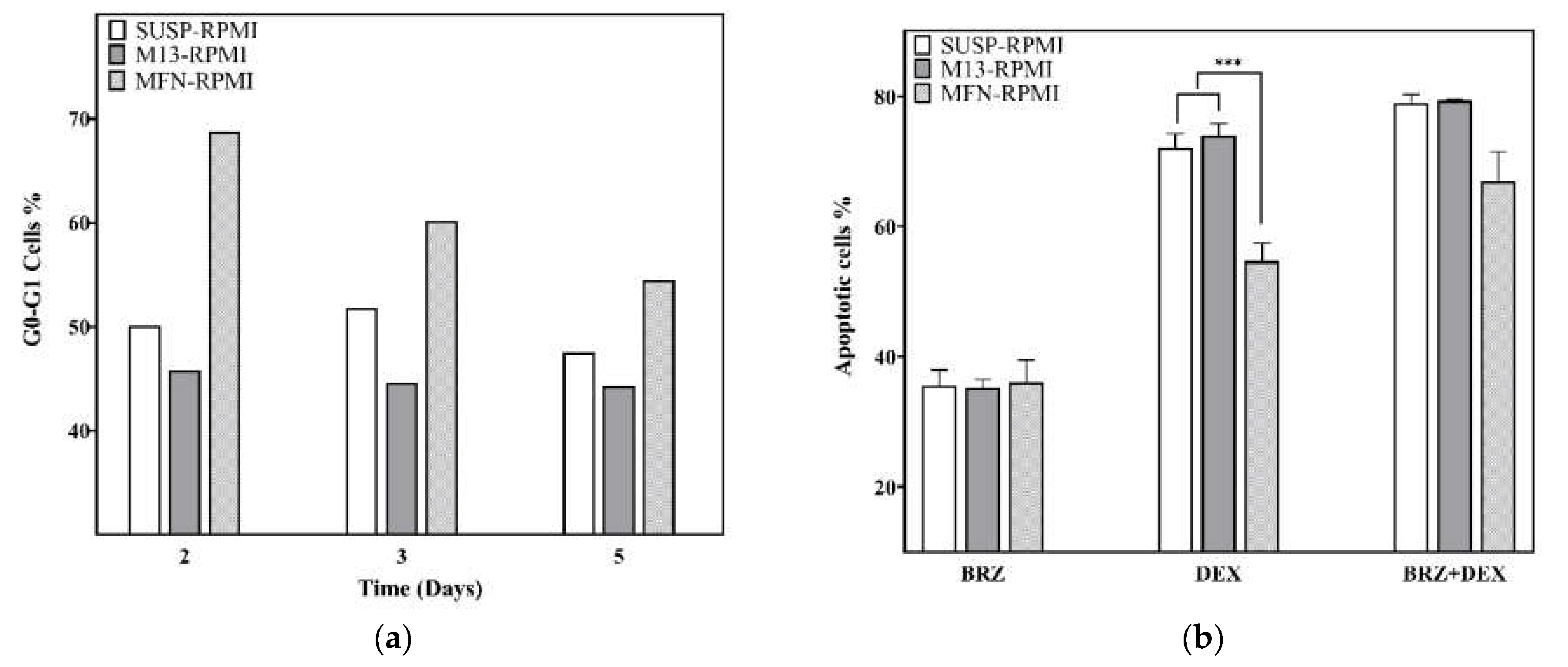
3.3. Cell Cycle Analysis on the RPMI 8226 Cell Line
3.4. Drug Resistance on RPMI8226 Cell Line
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piechotta, V.; Jakob, T.; Langer, P.; Monsef, I.; Scheid, C.; Estcourt, L.J.; Ocheni, S.; Theurich, S.; Kuhr, K.; Scheckel, B.; et al. Multiple Drug Combinations of Bortezomib, Lenalidomide, and Thalidomide for First-Line Treatment in Adults with Transplant-Ineligible Multiple Myeloma: A Network Meta-Analysis. Cochrane Database Syst. Rev. 2019, 11, CD013487. [Google Scholar] [CrossRef]
- Firth, J. Haematology: Multiple Myeloma. Clin. Med. J. R. Coll. Physicians Lond. 2019, 19, 58–60. [Google Scholar]
- Márk, Á.; Varga, G.; Timár, B.; Kriston, C.; Szabó, O.; Deák, L.; Matolcsy, A.; Barna, G. The Effect of Microenvironmental Factors on the Development of Myeloma Cells. Hematol. Oncol. 2017, 35, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; De Veirman, K.; De Beule, N.; Maes, K.; De Bruyne, E.; Van Valckenborgh, E.; Vanderkerken, K.; Menu, E. The Bone Marrow Microenvironment Enhances Multiple Myeloma Progression by Exosome-Mediated Activation of Myeloid-Derived Suppressor Cells. Oncotarget 2015, 6, 43992–44004. [Google Scholar] [CrossRef] [PubMed]
- de la Puente, P.; Muz, B.; Gilson, R.C.; Azab, F.; Luderer, M.; King, J.; Achilefu, S.; Vij, R.; Azab, A.K. 3D Tissue-Engineered Bone Marrow as a Novel Model to Study Pathophysiology and Drug Resistance in Multiple Myeloma. Biomaterials 2015, 73, 70–84. [Google Scholar] [CrossRef]
- Waldschmidt, J.M.; Fruttiger, S.J.; Wider, D.; Jung, J.; Thomsen, A.R.; Hartmann, T.N.; Duyster, J.; Hug, M.J.; Azab, K.A.; Jung, M.; et al. Ex Vivo Propagation in a Novel 3D High-Throughput Co-Culture System for Multiple Myeloma. J. Cancer Res. Clin. Oncol. 2022, 148, 1045–1055. [Google Scholar] [CrossRef]
- Fei, M.; Hang, Q.; Hou, S.; He, S.; Ruan, C. Adhesion to Fibronectin Induces P27Kip1 Nuclear Accumulation through Down-Regulation of Jab1 and Contributes to Cell Adhesion-Mediated Drug Resistance (CAM-DR) in RPMI 8,226 Cells. Mol. Cell. Biochem. 2014, 386, 177–187. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Leng, Y.; Dai, Y.; Kmieciak, M.; Kramer, L.; Sharma, K.; Wang, Y.; Craun, W.; Grant, S. The IAP Antagonist Birinapant Potentiates Bortezomib Anti-Myeloma Activity in Vitro and in Vivo. J. Hematol. Oncol. 2019, 12, 25. [Google Scholar] [CrossRef]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The Bone Marrow Microenvironment as a Tumor Sanctuary and Contributor to Drug Resistance. Clin. Cancer Res. 2008, 14, 2519–2526. [Google Scholar] [CrossRef]
- Salvatore, V.; Teti, G.; Focaroli, S.; Mazzotti, M.C.; Mazzotti, A.; Falconi, M. Oncotarget 9608 Www.Impactjournals.Com/Oncotarget The Tumor Microenvironment Promotes Cancer Progression and Cell Migration. Oncotarget 2017, 8, 9608–9616. [Google Scholar] [CrossRef]
- Wu, M.; Melody, A.S. Modeling Tumor Microenvironments in Vitro. J. Biomech. Eng. 2014, 136, 021011. [Google Scholar] [CrossRef]
- Salo, T.; Vered, M.; Bello, I.O.; Nyberg, P.; Bitu, C.C.; Zlotogorski Hurvitz, A.; Dayan, D. Insights into the Role of Components of the Tumor Microenvironment in Oral Carcinoma Call for New Therapeutic Approaches. Exp. Cell Res. 2014, 325, 58–64. [Google Scholar] [CrossRef]
- Clara-Trujillo, S.; Tolosa, L.; Cordón, L.; Sempere, A.; Gallego Ferrer, G.; Gómez Ribelles, J.L. Novel Microgel Culture System as Semi-Solid Three-Dimensional in Vitro Model for the Study of Multiple Myeloma Proliferation and Drug Resistance. Biomater. Adv. 2022, 135, 212749. [Google Scholar] [CrossRef]
- Marín-Payá, J.C.; Díaz-Benito, B.; Martins, L.A.; Trujillo, S.C.; Cordón, L.; Lanceros-Méndez, S.; Ferrer, G.G.; Sempere, A.; Ribelles, J.L.G. Biomimetic 3d Environment Based on Microgels as a Model for the Generation of Drug Resistance in Multiple Myeloma. Materials 2021, 14, 7121. [Google Scholar] [CrossRef]
- Liaw, C.Y.; Ji, S.; Guvendiren, M. Engineering 3D Hydrogels for Personalized In Vitro Human Tissue Models. Adv. Healthc. Mater. 2018, 7, 1701165. [Google Scholar] [CrossRef]
- Nii, T.; Tabata, Y. Immunosuppressive Mesenchymal Stem Cells Aggregates Incorporating Hydrogel Microspheres Promote an in Vitro Invasion of Cancer Cells. Regen. Ther. 2021, 18, 516–522. [Google Scholar] [CrossRef]
- Seeto, W.J.; Tian, Y.; Pradhan, S.; Minond, D.; Lipke, E.A. Droplet Microfluidics-Based Fabrication of Monodisperse Poly(Ethylene Glycol)-Fibrinogen Breast Cancer Microspheres for Automated Drug Screening Applications. ACS Biomater. Sci. Eng. 2022, 8, 3831–3841. [Google Scholar] [CrossRef]
- Pradhan, S.; Clary, J.M.; Seliktar, D.; Lipke, E.A. A Three-Dimensional Spheroidal Cancer Model Based on PEG-Fibrinogen Hydrogel Microspheres. Biomaterials 2017, 115, 141–154. [Google Scholar] [CrossRef]
- Hughes, V. Neighbourhood Watch. Nature 2011, 480, S48–S49. [Google Scholar] [CrossRef]
- Campillo-Fernández, A.J.; Unger, R.E.; Peters, K.; Halstenberg, S.; Santos, M.; Salmerón Sánchez, M.; Meseguer Dueñas, J.M.; Monleón Pradas, M.; Gómez Ribelles, J.L.; Kirkpatrick, C.J. Analysis of the Biological Response of Endothelial and Fibroblast Cells Cultured on Synthetic Scaffolds with Various Hydrophilic/Hydrophobic Ratios: Influence of Fibronectin Adsorption and Conformation. Tissue Eng. Part A 2009, 15, 1331–1341. [Google Scholar] [CrossRef]
- Ribeiro, C.; Panadero, J.A.; Sencadas, V.; Lanceros-M?ndez, S.; Tama?o, M.N.; Moratal, D.; Salmer?n-S?nchez, M.; G?mez Ribelles, J.L. Fibronectin Adsorption and Cell Response on Electroactive Poly(Vinylidene Fluoride) Films. Biomed. Mater. 2012, 7, 035004. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, L.; Faetti, M.; Salmerón Sanchez, M.; Gómez Ribelles, J.L. Phenomenological Theory of Structural Relaxation Based on a Thermorheologically Complex Relaxation Time Distribution. Eur. Phys. J. E 2008, 27, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Salmerón-Sánchez, M.; Rico, P.; Moratal, D.; Lee, T.T.; Schwarzbauer, J.E.; García, A.J. Role of Material-Driven Fibronectin Fibrillogenesis in Cell Differentiation. Biomaterials 2011, 32, 2099–2105. [Google Scholar] [CrossRef] [PubMed]
- Hazlehurst, L.A.; Damiano, J.S.; Buyuksal, I.; Pledger, W.J.; Dalton, W.S. Adhesion to Fibronectin via Β1 Integrins Regulates P27(Kip1) Levels and Contributes to Cell Adhesion Mediated Drug Resistance (CAM-DR). Oncogene 2000, 19, 4319–4327. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.; Mechti, N. Extracellular Matrix in Bone Marrow Can Mediate Drug Resistance in Myeloma. Leuk. Lymphoma 2005, 46, 803–811. [Google Scholar] [CrossRef]
- Burwick, N.; Sharma, S. Glucocorticoids in Multiple Myeloma: Past, Present, and Future. Ann. Hematol. 2019, 98, 19–28. [Google Scholar] [CrossRef]
- Chauhan, D.; Hideshima, T.; Rosen, S.; Reed, J.C.; Kharbanda, S.; Anderson, K.C. Apaf-1/Cytochrome c-Independent and Smac-Dependent Induction of Apoptosis in Multiple Myeloma (MM) Cells. J. Biol. Chem. 2001, 276, 24453–24456. [Google Scholar] [CrossRef]
- Chauhan, D.; Auclair, D.; Robinson, E.K.; Hideshima, T.; Li, G.; Podar, K.; Gupta, D.; Richardson, P.; Schlossma, R.L.; Krett, N.; et al. Identification of Genes Regulated by Dexamethasone in Multiple Myeloma Cells Using Oligonucleotide Arrays. Oncogene 2002, 21, 1346–1358. [Google Scholar] [CrossRef]
- Hatano, K.; Kikuchi, J.; Takatoku, M.; Shimizu, R.; Wada, T.; Ueda, M.; Nobuyoshi, M.; Oh, I.; Sato, K.; Suzuki, T.; et al. Bortezomib Overcomes Cell Adhesion-Mediated Drug Resistance through Downregulation of VLA-4 Expression in Multiple Myeloma. Oncogene 2009, 28, 231–242. [Google Scholar] [CrossRef]
- Anderson, K. Proteasome Inhibitors in Multiple Myeloma. Semin. Oncol. 2009, 36, S20–S26. [Google Scholar] [CrossRef]
- Podar, K.; Gouill, S.L.; Zhang, J.; Opferman, J.T.; Zorn, E.; Tai, Y.T.; Hideshima, T.; Amiot, M.; Chauhan, D.; Harousseau, J.L.; et al. A Pivotal Role for Mcl-1 in Bortezomib-Induced Apoptosis. Oncogene 2008, 27, 721–731. [Google Scholar] [CrossRef] [PubMed][Green Version]
- San Miguel, J.F.; Weisel, K.C.; Song, K.W.; Delforge, M.; Karlin, L.; Goldschmidt, H.; Moreau, P.; Banos, A.; Oriol, A.; Garderet, L.; et al. Impact of Prior Treatment and Depth of Response on Survival in MM-003, a Randomized Phase 3 Study Comparing Pomalidomide plus Low-Dose Dexamethasone versus High-Dose Dexamethasone in Relapsed/Refractory Multiple Myeloma. Haematologica 2015, 100, 1334–1339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tabchi, S.; Nair, R.; Kunacheeva, C.; Patel, K.; Lee, H.; Thomas, S.; Amini, B.; Ahmed, S.; Mehta, R.; Bashir, Q.; et al. Retrospective Review of the Use of High-Dose Cyclophosphamide, Bortezomib, Doxorubicin, and Dexamethasone for the Treatment of Multiple Myeloma and Plasma Cell Leukemia. Clin. Lymphoma Myeloma Leuk. 2019, 19, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Qiu, G.Q.; Wu, H.Q.; Wang, Z.L.; Lin, Y.; Wu, W.; Xie, X.B.; Gu, W.Y. Decitabine Enhances Bortezomib Treatment in RPMI 8226 Multiple Myeloma Cells. Mol. Med. Rep. 2016, 14, 3469–3475. [Google Scholar] [CrossRef][Green Version]
- Ramakrishnan, V.; Mager, D.E. Network-Based Analysis of Bortezomib Pharmacodynamic Heterogeneity in Multiple Myeloma Cells. J. Pharmacol. Exp. Ther. 2018, 365, 734–751. [Google Scholar] [CrossRef]
- Ribatti, D. The Discovery of Plasma Cells: An Historical Note. Immunol. Lett. 2017, 188, 64–67. [Google Scholar] [CrossRef]
- Urdeitx, P.; Clara-Trujillo, S.; Gómez Ribelles, J.L.; Doweidar, M.H. Computational Modeling of Multiple Myeloma Growth and Tumor Aggregate Formation. Comput. Methods Programs Biomed. Updat. 2022, in press. [Google Scholar] [CrossRef]
- Kirshner, J.; Thulien, K.J.; Martin, L.D.; Marun, C.D.; Reiman, T.; Belch, A.R.; Pilarski, L.M. A Unique Three-Dimensional Model for Evaluating the Impact of Therapy on Multiple Myeloma. Blood 2008, 112, 2935–2945. [Google Scholar] [CrossRef]
- Belloni, D.; Heltai, S.; Ponzoni, M.; Villa, A.; Vergani, B.; Pecciarini, L.; Marcatti, M.; Girlanda, S.; Tonon, G.; Ciceri, F.; et al. Modeling Multiple Myeloma-Bone Marrow Interactions and Response to Drugs in a 3d Surrogate Microenvironment. Haematologica 2018, 103, 707–716. [Google Scholar] [CrossRef]
- Zdzisińska, B.; Roliński, J.; Piersiak, T.; Kandefer-Szerszeń, M. A Comparison of Cytokine Production in 2-Dimensional and 3-Dimensional Cultures of Bone Marrow Stromal Cells of Muliple Myeloma Patients in Response to RPMI8226 Myeloma Cells. Folia Histochem. Cytobiol. 2009, 47, 69–74. [Google Scholar] [CrossRef]
- Spicer, C.D.; Pashuck, E.T.; Stevens, M.M. Achieving Controlled Biomolecule-Biomaterial Conjugation. Chem. Rev. 2018, 118, 7702–7743. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marín-Payá, J.C.; Clara-Trujillo, S.; Cordón, L.; Gallego Ferrer, G.; Sempere, A.; Gómez Ribelles, J.L. Protein-Functionalized Microgel for Multiple Myeloma Cells’ 3D Culture. Biomedicines 2022, 10, 2797. https://doi.org/10.3390/biomedicines10112797
Marín-Payá JC, Clara-Trujillo S, Cordón L, Gallego Ferrer G, Sempere A, Gómez Ribelles JL. Protein-Functionalized Microgel for Multiple Myeloma Cells’ 3D Culture. Biomedicines. 2022; 10(11):2797. https://doi.org/10.3390/biomedicines10112797
Chicago/Turabian StyleMarín-Payá, Juan Carlos, Sandra Clara-Trujillo, Lourdes Cordón, Gloria Gallego Ferrer, Amparo Sempere, and José Luis Gómez Ribelles. 2022. "Protein-Functionalized Microgel for Multiple Myeloma Cells’ 3D Culture" Biomedicines 10, no. 11: 2797. https://doi.org/10.3390/biomedicines10112797
APA StyleMarín-Payá, J. C., Clara-Trujillo, S., Cordón, L., Gallego Ferrer, G., Sempere, A., & Gómez Ribelles, J. L. (2022). Protein-Functionalized Microgel for Multiple Myeloma Cells’ 3D Culture. Biomedicines, 10(11), 2797. https://doi.org/10.3390/biomedicines10112797







