Identification of DDX60 as a Regulator of MHC-I Class Molecules in Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Information and Data Collection
2.2. Tumor-Infiltrated Immune Cell Fraction Analysis
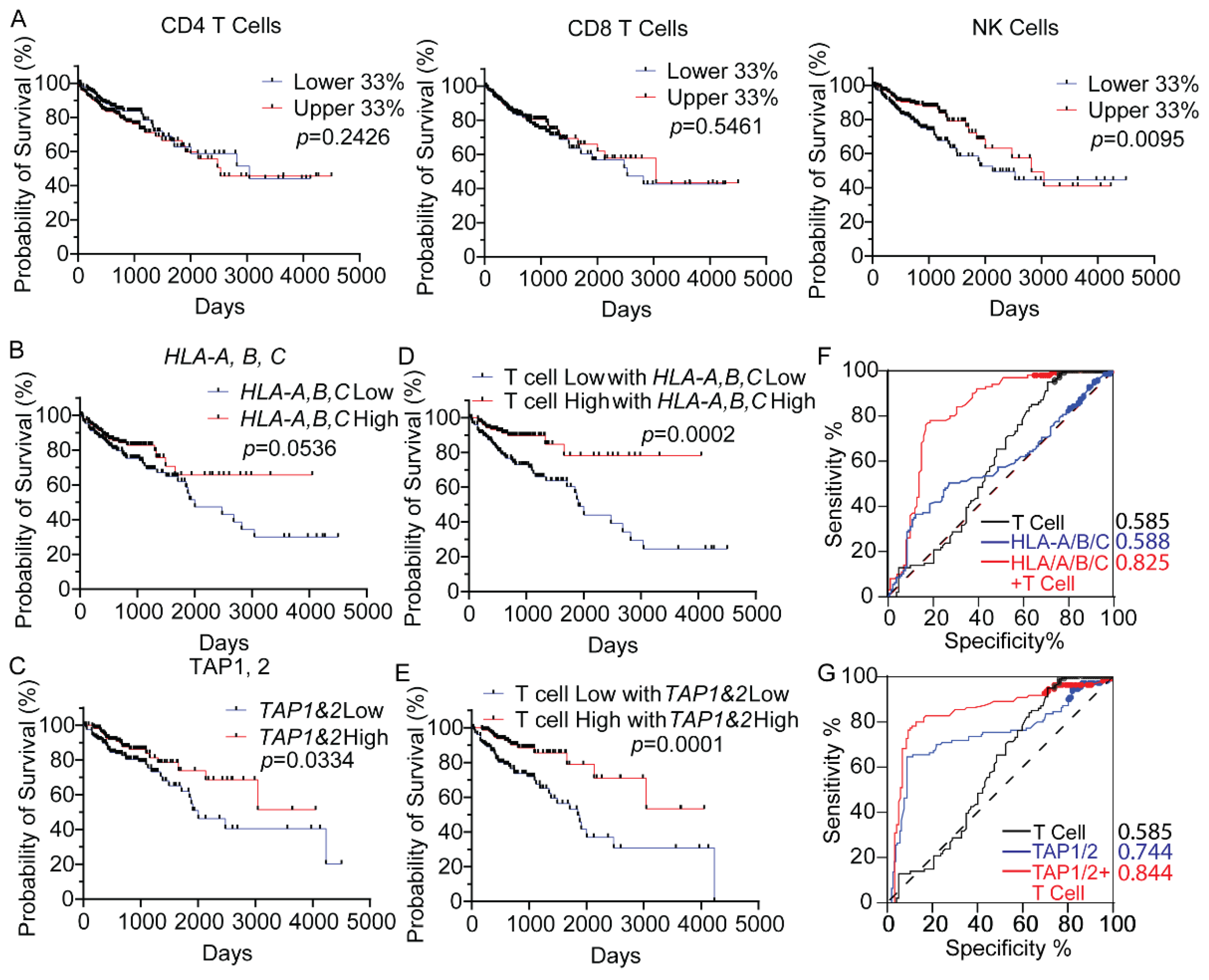
2.3. Survival Analysis Based on Cell Fractions and MHC-I Molecules
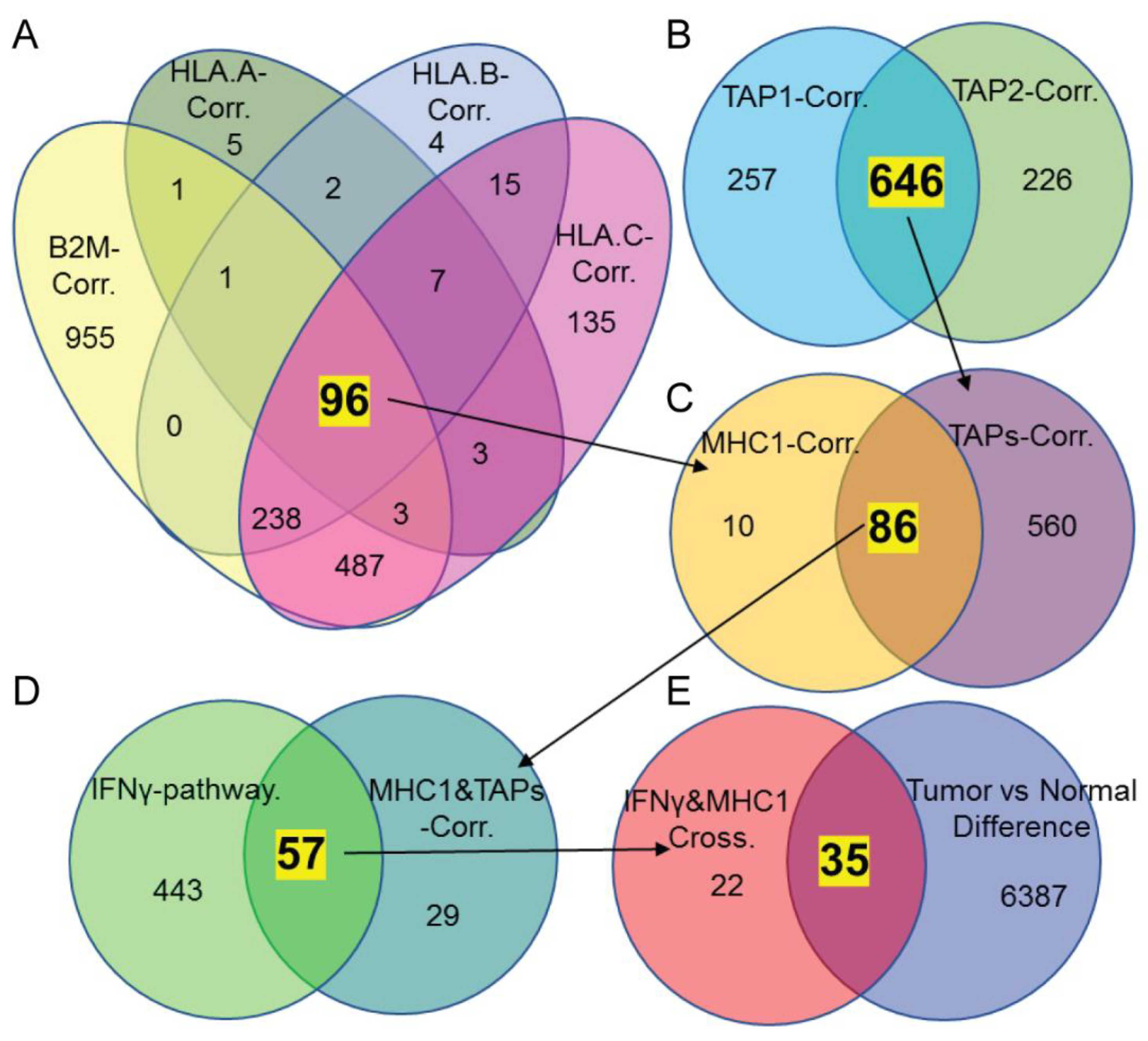
2.4. Identifying Molecules That Might Regulate MHC-I Expression Based on Silico Analysis
2.5. Gene Knockdown and Overexpression
2.6. Immunoblotting
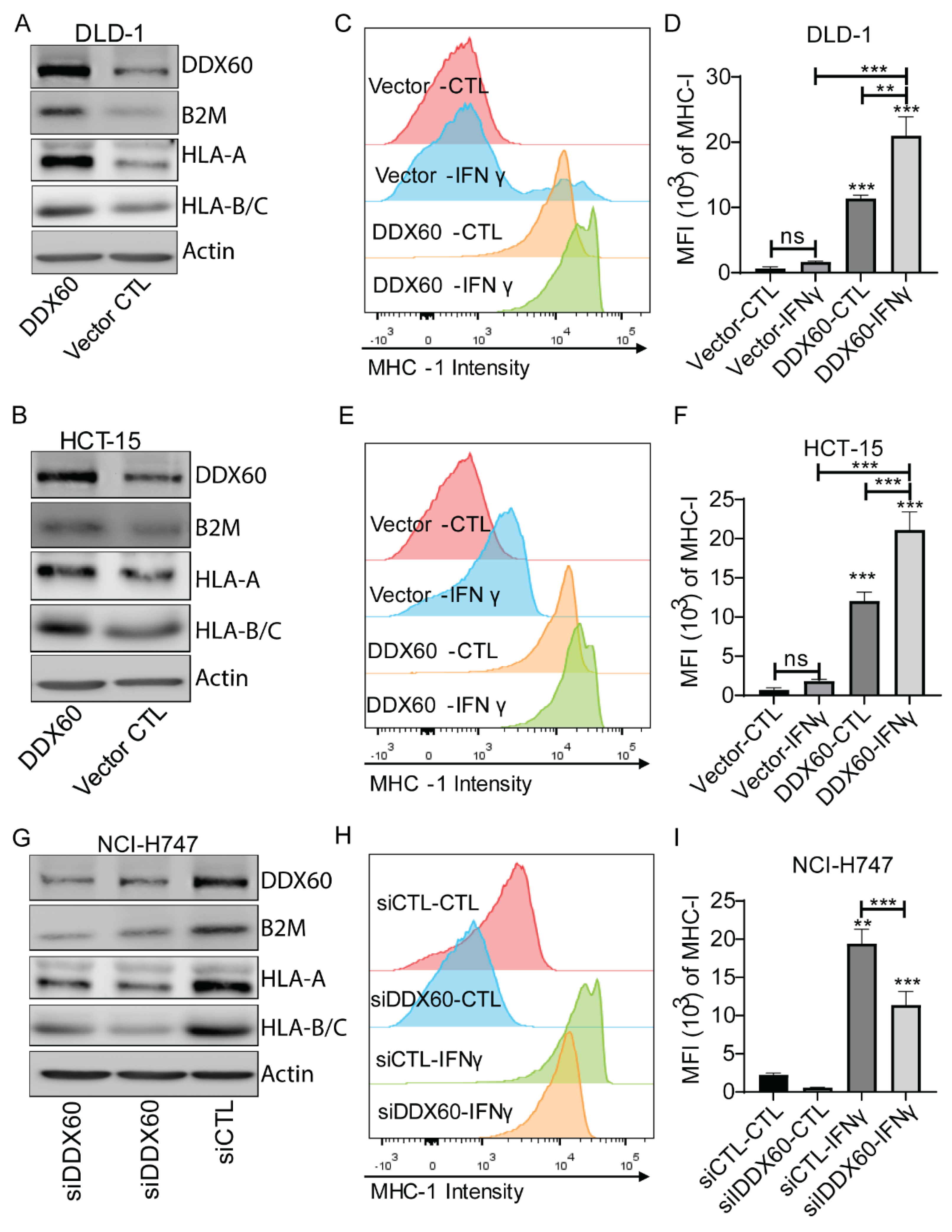
2.7. Fluorescence-Activated Cell Sorting (FACS) Analysis for MHC-I Expression Following Candidate Genes’ Modification
2.8. Statistical Analyses
3. Results
3.1. MHC-I Expression Is Correlated to Responders in Anti-PD1 Immunotherapy and CD8+ T Cell Infiltration in CRC
3.2. DDX60 and ISG15 Are Potential Candidates Correlated with MHC-I Expression
3.3. DDX60 regulates MHC-I expression in CRC cells
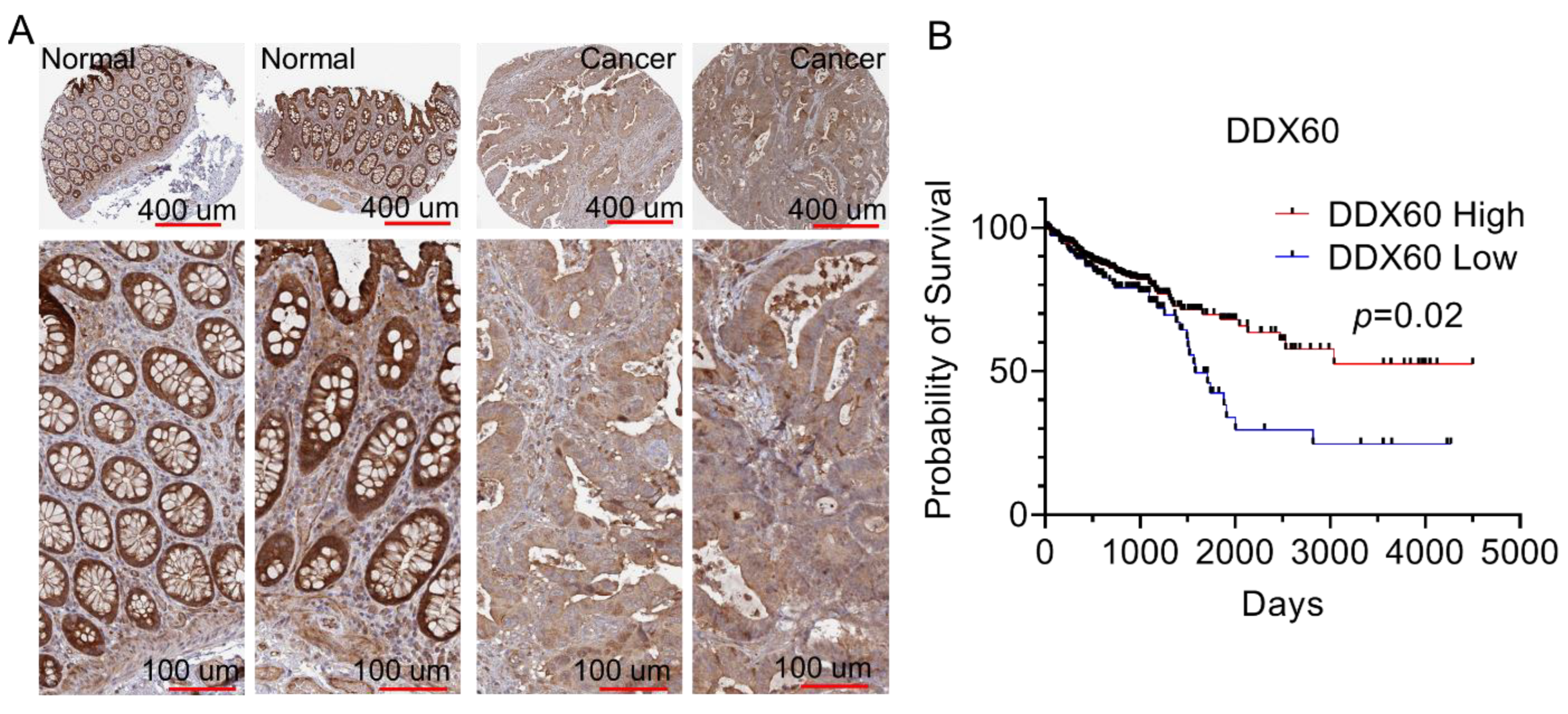
3.4. DDX60 Is a Favorable Prognostic Maker and Is Downregulated in CRC
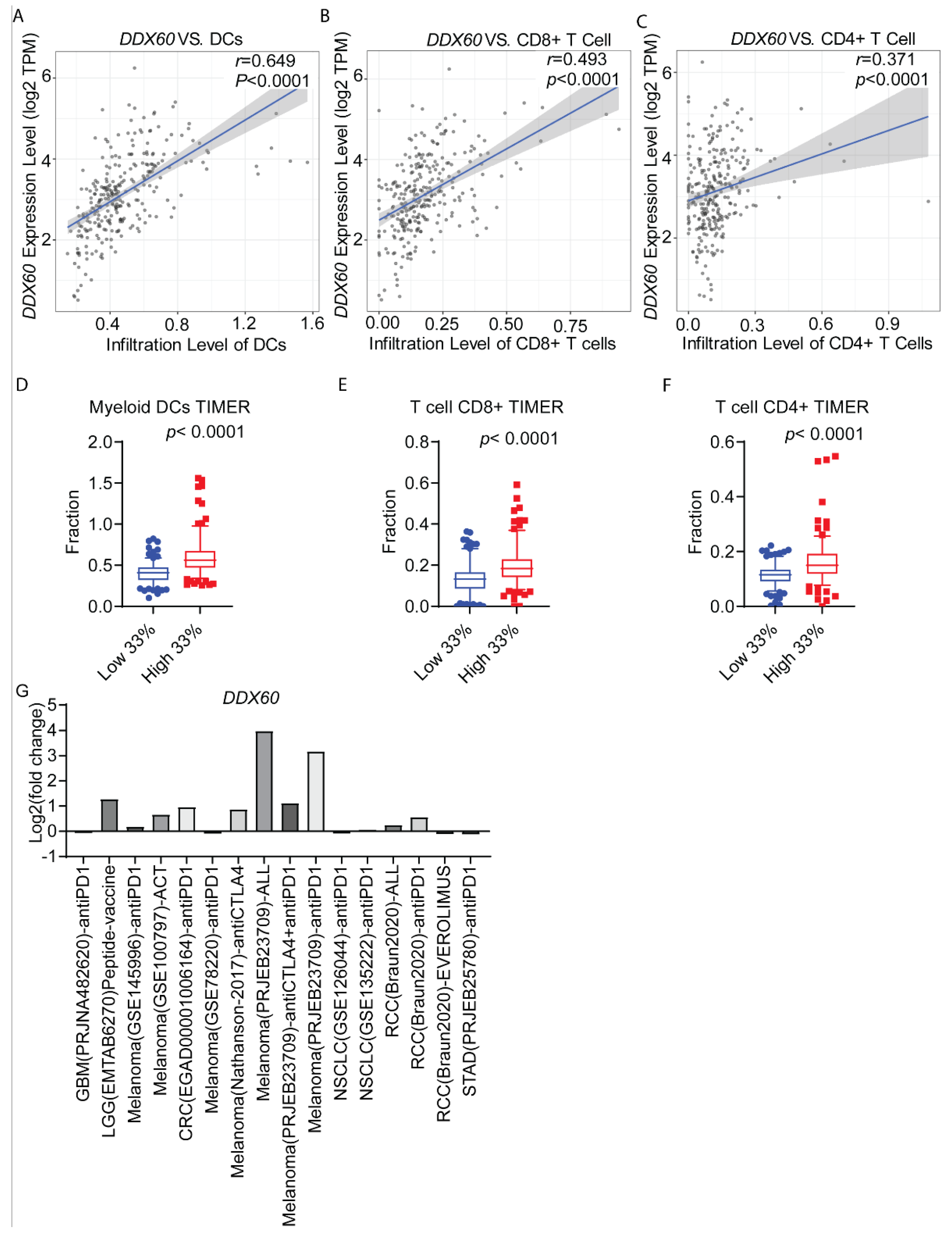
3.5. DDX60 Expression Is Correlated with Immune Cell Infiltration in CRC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elez, E.; Baraibar, I. Immunotherapy in colorectal cancer: An unmet need deserving of change. Lancet Oncol. 2022, 23, 830–831. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.R. Immunotherapy for microsatellite-instability-high advanced colorectal cancer. Zhonghua Wei Chang Wai Ke Za Zhi 2022, 25, 199–204. [Google Scholar] [PubMed]
- Chakravarthy, A.; Khan, L.; Bensler, N.P.; Bose, P.; De Carvalho, D.D. TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 2018, 9, 4692. [Google Scholar] [CrossRef] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Saleh, R.; Elkord, E. Treg-mediated acquired resistance to immune checkpoint inhibitors. Cancer Lett. 2019, 457, 168–179. [Google Scholar] [CrossRef]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef] [Green Version]
- De Palma, M.; Lewis, C.E. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell 2013, 23, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Kronbichler, A.; Eisenhut, M.; Hong, S.H.; van der Vliet, H.J.; Kang, J.; Shin, J.I.; Gamerith, G. Tumor Mutational Burden and Efficacy of Immune Checkpoint Inhibitors: A Systematic Review and Meta-Analysis. Cancers 2019, 11, 1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.B.; Estrada, M.V.; Salgado, R.; Sanchez, V.; Doxie, D.B.; Opalenik, S.R.; Vilgelm, A.E.; Feld, E.; Johnson, A.S.; Greenplate, A.R.; et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 2016, 7, 10582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef]
- Kriegsman, B.A.; Vangala, P.; Chen, B.J.; Meraner, P.; Brass, A.L.; Garber, M.; Rock, K.L. Frequent Loss of IRF2 in Cancers Leads to Immune Evasion through Decreased MHC Class I Antigen Presentation and Increased PD-L1 Expression. J. Immunol. 2019, 203, 1999–2010. [Google Scholar] [CrossRef]
- Cornel, A.M.; Mimpen, I.L.; Nierkens, S. MHC Class I Downregulation in Cancer: Underlying Mechanisms and Potential Targets for Cancer Immunotherapy. Cancers 2020, 12, 1760. [Google Scholar] [CrossRef]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-beta-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018, 48, 147–160.e7. [Google Scholar] [CrossRef] [Green Version]
- Kearney, C.J.; Vervoort, S.J.; Hogg, S.J.; Ramsbottom, K.M.; Freeman, A.J.; Lalaoui, N.; Pijpers, L.; Michie, J.; Brown, K.K.; Knight, D.A.; et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 2018, 3, eaar3451. [Google Scholar] [CrossRef] [Green Version]
- Holtzhausen, A.; Zhao, F.; Evans, K.S.; Tsutsui, M.; Orabona, C.; Tyler, D.S.; Hanks, B.A. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotolerance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer Immunol. Res. 2015, 3, 1082–1095. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Li, Y.; Ran, L.; Dong, Y.; Zhou, X.; Tang, J.; Han, L.; Wang, M.; Pang, L.; Qi, Y.; et al. Screening and identification of HLA-A2-restricted neoepitopes for immunotherapy of non-microsatellite instability-high colorectal cancer. Sci. China Life Sci. 2022, 65, 572–587. [Google Scholar] [CrossRef]
- Talaat, I.M.; Elemam, N.M.; Saber-Ayad, M. Complement System: An Immunotherapy Target in Colorectal Cancer. Front. Immunol. 2022, 13, 810993. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fu, Y.; Wang, P.-P.; Ding, Z.-Y. Neoantigen: A Promising Target for the Immunotherapy of Colorectal Cancer. Dis. Markers 2022, 2022, 8270305. [Google Scholar] [CrossRef] [PubMed]
- Gorzo, A.; Galos, D.; Volovat, S.R.; Lungulescu, C.V.; Burz, C.; Sur, D. Landscape of Immunotherapy Options for Colorectal Cancer: Current Knowledge and Future Perspectives beyond Immune Checkpoint Blockade. Life 2022, 12, 229. [Google Scholar] [CrossRef] [PubMed]
- Choo, S.Y. The HLA system: Genetics, immunology, clinical testing, and clinical implications. Yonsei Med. J. 2007, 48, 11–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, M.; Fremont, D.H.; Peterson, P.A.; Wilson, L.A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 1992, 257, 927–934. [Google Scholar] [CrossRef]
- Croft, N.P.; Smith, S.A.; Pickering, J.; Sidney, J.; Peters, B.; Faridi, P.; Witney, M.J.; Sebastian, P.; Flesch, I.E.A.; Heading, S.L.; et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA 2019, 116, 3112–3117. [Google Scholar] [CrossRef] [Green Version]
- Lobigs, M.; Müllbacher, A. Recognition of vaccinia virus-encoded major histocompatibility complex class I antigens by virus immune cytotoxic T cells is independent of the polymorphism of the peptide transporters. Proc. Natl. Acad. Sci. USA 1993, 90, 2676–2680. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.; Testi, M.G.; Pasetto, M.; Picchio, M.C.; Innamorati, G.; Mazzocco, M.; Ugel, S.; Cingarlini, S.; Bronte, V.; Zanovello, P.; et al. IFN-gamma-mediated upmodulation of MHC class I expression activates tumor-specific immune response in a mouse model of prostate cancer. Vaccine 2010, 28, 3548–3557. [Google Scholar] [CrossRef]
- Zhang, S.; Kohli, K.; Black, R.G.; Yao, L.; Spadinger, S.M.; He, Q.; Pillarisetty, V.G.; Cranmer, L.D.; Van Tine, B.A.; Yee, C.; et al. Systemic Interferon-gamma Increases MHC Class I Expression and T-cell Infiltration in Cold Tumors: Results of a Phase 0 Clinical Trial. Cancer Immunol. Res. 2019, 7, 1237–1243. [Google Scholar] [CrossRef] [Green Version]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.-P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1136. [Google Scholar] [CrossRef]
- Liu, D.; Jenkins, R.W.; Sullivan, R.J. Mechanisms of Resistance to Immune Checkpoint Blockade. Am. J. Clin. Dermatol. 2019, 20, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, D.; Xiao, B.; Zhou, C.; Jiang, W.; Tang, J.; Li, Y.; Zhang, R.; Han, K.; Hou, Z.; et al. B2M and JAK1/2-mutated MSI-H Colorectal Carcinomas Can Benefit From Anti-PD-1 Therapy. J. Immunother. 2022, 45, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E.; et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016, 167, 397–404.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Burr, M.L.; van den Boomen, D.J.H.; Bye, H.; Antrobus, R.; Wiertz, E.J.; Lehner, P.J. MHC class I molecules are preferentially ubiquitinated on endoplasmic reticulum luminal residues during HRD1 ubiquitin E3 ligase-mediated dislocation. Proc. Natl. Acad. Sci. USA 2013, 110, 14290–14295. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Brägelmann, J.; Kryukov, I.; Saraiva-Agostinho, N.; Perner, S. FirebrowseR: An R client to the Broad Institute’s Firehose Pipeline. Database 2017, 2017, baw160. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Vasaikar, S.; Huang, C.; Wang, X.; Petyuk, V.A.; Savage, S.R.; Wen, B.; Dou, Y.; Zhang, Y.; Shi, Z.; Arshad, O.A.; et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 2019, 177, 1035–1049.e19. [Google Scholar] [CrossRef] [Green Version]
- Plattner, C.; Finotello, F.; Rieder, D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzym. 2020, 636, 261–285. [Google Scholar]
- Lin, G.; Chai, J.; Yuan, S.; Mai, C.; Cai, L.; Murphy, R.W.; Zhou, W.; Luo, J. VennPainter: A Tool for the Comparison and Identification of Candidate Genes Based on Venn Diagrams. PLoS ONE 2016, 11, e0154315. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Lv, W.; Zhang, Q.; Wang, Y.; Tao, L.; Liu, D. Gene set enrichment analysis of pathways and transcription factors associated with diabetic retinopathy using a microarray dataset. Int. J. Mol. Med. 2015, 36, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Ortenberg, R.; Varanasi, S.K.; Mangalhara, K.C.; Mardamshina, M.; Markovits, E.; Baruch, E.N.; Tripple, V.; Arama-Chayoth, M.; Greenberg, E.; et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell 2019, 179, 236–250.e18. [Google Scholar] [CrossRef] [PubMed]
- McCune, J.S. Rapid Advances in Immunotherapy to Treat Cancer. Clin. Pharmacol. Ther. 2018, 103, 540–544. [Google Scholar] [CrossRef]
- Amin, M.; Lockhart, A.C. The potential role of immunotherapy to treat colorectal cancer. Expert Opin. Investig. Drugs 2015, 24, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef] [PubMed]
- Jhunjhunwala, S.; Hammer, C.; Delamarre, L. Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 2021, 21, 298–312. [Google Scholar] [CrossRef]
- Balasubramanian, A.; John, T.; Asselin-Labat, M.-L. Regulation of the antigen presentation machinery in cancer and its implication for immune surveillance. Biochem. Soc. Trans. 2022, 50, 825–837. [Google Scholar] [CrossRef]
- Oshiumi, H.; Miyashita, M.; Okamoto, M.; Morioka, Y.; Okabe, M.; Matsumoto, M.; Seya, T. DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation. Cell Rep. 2015, 11, 1193–1207. [Google Scholar] [CrossRef]
- Miyashita, M.; Oshiumi, H.; Matsumoto, M.; Seya, T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011, 31, 3802–3819. [Google Scholar] [CrossRef] [Green Version]
- Goubau, D.; van der Veen, A.G.; Chakravarty, P.; Lin, R.; Rogers, N.; Rehwinkel, J.; Deddouche, S.; Rosewell, I.; Hiscott, J.; e Sousa, C.R. Mouse superkiller-2-like helicase DDX60 is dispensable for type I IFN induction and immunity to multiple viruses. Eur. J. Immunol. 2015, 45, 3386–3403. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.-Y.; Wu, C.-N.; Sie, H.-C.; Cheng, J.-T.; Lin, Y.-S.; Liou, H.-H.; Tseng, Y.-K.; Shu, C.-W.; Tsai, K.-W.; Yen, L.-M.; et al. Subsite-specific association of DEAD box RNA helicase DDX60 with the development and prognosis of oral squamous cell carcinoma. Oncotarget 2016, 7, 85097–85108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elion, D.L.; Jacobson, M.E.; Hicks, D.J.; Rahman, B.; Sanchez, V.; Gonzales-Ericsson, P.I.; Fedorova, O.; Pyle, A.M.; Wilson, J.T.; Cook, R.S. Therapeutically Active RIG-I Agonist Induces Immunogenic Tumor Cell Killing in Breast Cancers. Cancer Res. 2018, 78, 6183–6195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iurescia, S.; Fioretti, D.; Rinaldi, M. The Innate Immune Signalling Pathways: Turning RIG-I Sensor Activation Against Cancer. Cancers 2020, 12, 3158. [Google Scholar] [CrossRef] [PubMed]
- Mjelle, R.; Sjursen, W.; Thommesen, L.; Saetrom, P.; Hofsli, E. Small RNA expression from viruses, bacteria and human miRNAs in colon cancer tissue and its association with microsatellite instability and tumor location. BMC Cancer 2019, 19, 161. [Google Scholar] [CrossRef]
- Zapatka, M.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sultmann, H.; Moch, H.; Pathogens, P.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Fu, M.; Zhang, M.; Zhang, J.; Du, Z.; Zhang, H.; Hua, W.; Mao, Y. DDX60 Is Associated With Glioma Malignancy and Serves as a Potential Immunotherapy Biomarker. Front. Oncol. 2021, 11, 665360. [Google Scholar] [CrossRef]
- Xin, D.; Liu, J.; Gu, J.; Ji, Y.; Jin, J.; Sun, L.; Tai, Q.; Cao, J.; Tian, Y.; Qin, H.; et al. Low Expression of DDX60 Gene Might Associate with the Radiosensitivity for Patients with Breast Cancer. J. Oncol. 2020, 2020, 8309492. [Google Scholar] [CrossRef]

| Gene Name | High Expression in Normal | Cases of High Expression in Cancer | ||
|---|---|---|---|---|
| Cases | Fraction (%) | Cases | Fraction (%) | |
| GZMA | 77 | 79.38 | 20 | 20.62 |
| HLA-DPA1 | 76 | 78.35 | 21 | 21.65 |
| APOL1 | 74 | 76.29 | 23 | 23.71 |
| DDX60 | 73 | 75.26 | 24 | 24.74 |
| ISG15 | 68 | 70.10 | 29 | 29.90 |
| BTN3A3 | 64 | 65.98 | 33 | 34.02 |
| CMPK2 | 64 | 65.98 | 33 | 34.02 |
| HLA-DMB | 61 | 62.89 | 35 | 37.11 |
| ISG20 | 61 | 62.89 | 36 | 37.11 |
| DDX58 | 57 | 58.76 | 40 | 41.24 |
| APOL3 | 55 | 56.70 | 42 | 43.30 |
| HLA-DRA | 55 | 56.70 | 42 | 43.30 |
| HLA-DRB1 | 54 | 55.67 | 43 | 44.33 |
| HLA-F | 51 | 52.58 | 46 | 47.42 |
| IFI35 | 49 | 50.52 | 48 | 49.48 |
| TRIM22 | 49 | 50.52 | 48 | 49.48 |
| PARP12 | 48 | 49.48 | 49 | 50.52 |
| IFIT3 | 46 | 47.42 | 51 | 52.58 |
| OASL | 42 | 43.30 | 55 | 56.70 |
| APOL2 | 37 | 38.14 | 60 | 61.86 |
| CD74 | 36 | 37.11 | 61 | 62.89 |
| GBP4 | 34 | 35.05 | 63 | 64.95 |
| HLA-E | 34 | 35.05 | 63 | 64.95 |
| GBP2 | 34 | 35.05 | 63 | 64.95 |
| GBP1 | 33 | 34.02 | 64 | 65.98 |
| SERPINB9 | 33 | 34.02 | 64 | 65.98 |
| PARP14 | 30 | 30.93 | 67 | 69.07 |
| PARP9 | 27 | 27.84 | 70 | 72.16 |
| UBE2L6 | 27 | 27.84 | 70 | 72.16 |
| PSMB9 | 25 | 25.77 | 72 | 74.23 |
| TYMP | 21 | 21.65 | 76 | 78.35 |
| STAT1 | 15 | 15.46 | 82 | 84.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, N.; Hu, T.; He, C. Identification of DDX60 as a Regulator of MHC-I Class Molecules in Colorectal Cancer. Biomedicines 2022, 10, 3092. https://doi.org/10.3390/biomedicines10123092
Geng N, Hu T, He C. Identification of DDX60 as a Regulator of MHC-I Class Molecules in Colorectal Cancer. Biomedicines. 2022; 10(12):3092. https://doi.org/10.3390/biomedicines10123092
Chicago/Turabian StyleGeng, Nina, Tuo Hu, and Chunbo He. 2022. "Identification of DDX60 as a Regulator of MHC-I Class Molecules in Colorectal Cancer" Biomedicines 10, no. 12: 3092. https://doi.org/10.3390/biomedicines10123092






