Abstract
Diabetes is known to delay wound healing, and this delay is attributed to prolonged inflammation. We found that microRNAs (miRNAs) might be involved in the dysfunction of diabetic-derived neutrophils, and dynamics of neutrophil and chronic inflammation might be initiated by miRNA-regulated genes. Moreover, studies of miRNA function in nephropathy have suggested that circular RNAs (circRNAs), which function as sponges of miRNA to regulate their expression, are potential biomarkers and new therapeutic targets for the diagnosis of diabetic nephropathy. Accordingly, to investigate the molecular mechanism of the regulation of inflammation in diabetic-derived neutrophils, we identified circRNAs in diabetic-derived neutrophils obtained from BKS.Cg-Dock7m +/+ Leprdb/J (Leprdb/db and Leprdb/+) mice using microarrays. Neutrophils from pooled bone marrow of three diabetic and three non-diabetic mice were isolated and total RNA was extracted. Microarray analysis was performed using the Arraystar Mouse Circular RNA Array. The results showed that three circRNAs were significantly increased and six circRNAs were significantly decreased in diabetic-derived neutrophils compared with non-diabetic-derived neutrophils. The expressions of some circRNAs in diabetic-derived neutrophils were more than double those in non-diabetic-derived neutrophils. The circRNAs contain binding sites of miRNAs, which were differentially expressed in diabetic-derived neutrophils. Our results suggest that circRNAs may be involved in the regulation of inflammation in diabetic-derived neutrophils.
1. Introduction
As population growth continues to increase, the number of diabetic patients is also expected to increase, with estimates reaching up to approximately 300 million by 2030 [1]. Diabetes causes various complications such as neuropathy and diabetic kidney disease. Diabetic nephropathy is a major chronic microvascular complication that occurs in about 25–40% of type 2 diabetes mellitus cases [2]. Moreover, diabetes can delay the healing of wounds, cause infection, and make it more severe, resulting in an intractable ulcer. Approximately 15% of diabetic patients require amputation of a toe, foot or part of a leg in more than 80% of those cases [3]. The development of intractable ulcers involves the functional decline of immune cells from persistent hyperglycemia, vascular insufficiency, and neuropathy [4,5,6]. Research aimed at developing new therapeutic methods and identifying new therapeutic targets for diabetes is being conducted, which can be facilitated by clarifying the molecular mechanisms involved in delayed diabetic wound healing. However, these mechanisms remain largely unknown.
We previously found that diabetic wounds had a prolonged inflammatory phase and impaired transition to the proliferative phase [7], and that the cause of prolonged inflammation was from the dysfunction of diabetic-derived neutrophils rather than the diabetic environment [8]. Furthermore, our results revealed that microRNAs (miRNAs) exhibit differential expression in diabetic-derived neutrophils compared with non-diabetic-derived neutrophils, and that miR-129-2-3p overexpression at the wound site of type 2 diabetic mice accelerated wound healing [9]. These results suggest that upregulating miR-129-2-3p may be a strategy to improve the dysfunction of diabetic-derived neutrophils. miRNA may thus be a new therapeutic target for diabetic wounds and may be developed into a new treatment method.
Circular RNAs (circRNAs) are key molecules that are involved in the regulation of miRNA expression. circRNAs are synthesized by back splicing and have been shown to function as miRNA sponges and upregulate the expression of mRNAs targeted by miRNAs [10,11,12]. Multiple studies have shown that circRNAs play important roles in the biological processes of various diseases [13]. circSDHC functions as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma [14]. Circ100084 functions as a sponge of miR-23a-5p to regulate IGF2 expression in hepatocellular carcinoma [15]. circDiaph3 regulates vascular smooth muscle cell differentiation, proliferation, and migration in rat [16]. Screening for circRNAs associated with inflammation in peripheral leukocytes of patients with type 2 diabetes mellitus was performed [17], and circRNAs have shown potential applications as disease biomarkers and novel therapeutic targets [18].
We hypothesized that circRNAs might be involved in regulating miRNAs involved in chronic inflammation in diabetic-derived neutrophils. To explore the potential molecular mechanism of inflammatory control in diabetic-derived neutrophils, we screened for circRNA expressions in diabetic-derived neutrophils using microarrays.
2. Materials and Methods
2.1. Animals
The Animal Care Committee of Nagasaki University and University of Occupational and Environmental Health approved the protocol for this study (approval numbers: 1906191539 and AE22-013). BKS.Cg-Dock7m +/+ Leprdb/J (Leprdb/db and Leprdb/+) mice (5 weeks old) were purchased from Charles River Laboratories (Yokohama, Japan). Animals were housed under a 12/12 h light/dark cycle (light on: 07:00, light off: 19:00) at constant temperature and humidity and allowed free access to food and water. After euthanasia by overanesthesia, male mice at 8 weeks of age were used and were age-matched to controls. To eliminate the effect of hormonal action related to sexual maturation on skin wound healing, we used only male mice.
2.2. Microarray Analysis
Bone marrow (BM) was flushed from femurs and tibiae. Neutrophils from the pooled BM of three diabetic (db) and three non-diabetic (non-db) mice were isolated using an anti-Ly-6G microbead kit (Miltenyi Biotec Inc., Bergisch Gladbach, Germany). Microarray analysis was performed on six pools (three pools of three db BM samples and three pools of three non-db BM samples) using Arraystar Mouse Circular RNA Microarray (Arraystar Inc., Rockville, MD, USA), in accordance with the manufacturer’s instructions. Bioinformatic analyses were performed using Miranda and TargetScan (Miranda, Lewis) (Arraystar Inc.). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE213297.
2.3. Isolation of Neutrophils, Macrophages, T Cells, and B Cells
Neutrophils from BM of six non-db mice were isolated using a neutrophil isolation kit (Miltenyi Biotec Inc.). Macrophages, T cells, and B cells from BM of six non-db mice were isolated with a Microbead Kit (Miltenyi Biotec Inc.) in accordance with the manufacturer’s instructions. Cells were incubated with anti-CD11b Ab, anti-CD5 Ab, and anti-CD19 Ab to isolate macrophages, T cells, and B cells, respectively [9].
2.4. RNA Isolation for Real-Time Quantitative PCR
Neutrophils were isolated from BM of six db and six non-db mice using a neutrophil isolation kit and anti-Ly-6G Microbead kit (Miltenyi Biotec). Total RNA, including miRNA, was extracted using an miRNeasy Mini kit and miRNeasy Micro kit (QIAGEN, Germantown, MD, USA) in accordance with the manufacturer’s instructions. Total RNA was quantified using a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA samples were stored at −80 °C until use.
2.5. cDNA Synthesis for circRNA and microRNA, and Quantitative Real-Time PCR (qRT-PCR)
Complementary DNA (cDNA) was synthesized from RNA (850 ng) using the circRNA cDNA synthesis kit (Applied Biological Materials Inc., Richmond, BC, Canada) in accordance with the manufacturer’s instructions. qRT-PCR was performed in a 20 µL reaction using EmeraldAmp PCR Master Mix (Takara Bio, Kusatsu, Shiga, Japan) and a Thermal Cycler Dice Real Time System (Takara Bio). The amplification mix and thermal cycling conditions were established as described by Panda et al. [19]. Primers for circRNA were designed for the 100 bp sequence before and after the junction sequence by Primer 3 (Table 1). DNA synthesis of circRNAs was performed by Hokkaido System Science Co., Ltd. (Sapporo, Japan). Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was purchased from Takara Bio Inc.

Table 1.
Primers of circRNAs.
Total RNA (350 ng) was used as a template for complementary DNA (cDNA) synthesis for miRNA expression analysis using TaqMan MicroRNA Assay (Thermo Fisher Scientific). qRT-PCR was performed using TaqMan MicroRNA Assay for miRNA expression analysis (Thermo Fisher Scientific). Primers (mmu-miR-92a-2-3p (Catalog #: 4427975, Assay ID: 002496), mmu-miR-450b (Catalog #: 4427975, Assay ID: 001962) and U6 snRNA (Catalog #: 4427975, Assay ID: 001973)) were purchased from Thermo Fisher Scientific. The relative quantification of mRNA transcripts and miRNA was performed using the ∆∆Ct method [20].
2.6. Statistical Analysis
Data are shown as means ± SD. The statistical significance of differences between means was assessed by Mann–Whitney U test, unpaired t-test, and one-way ANOVA, followed by Tukey’s multiple comparison test and two-way ANOVA, followed by Bonferroni post-tests to compare replicate means (GraphPad Software, San Diego, CA, USA). A p-value < 0.05 was considered significant.
3. Results
3.1. circRNA Expression Is Altered in Neutrophils from Diabetic Mice
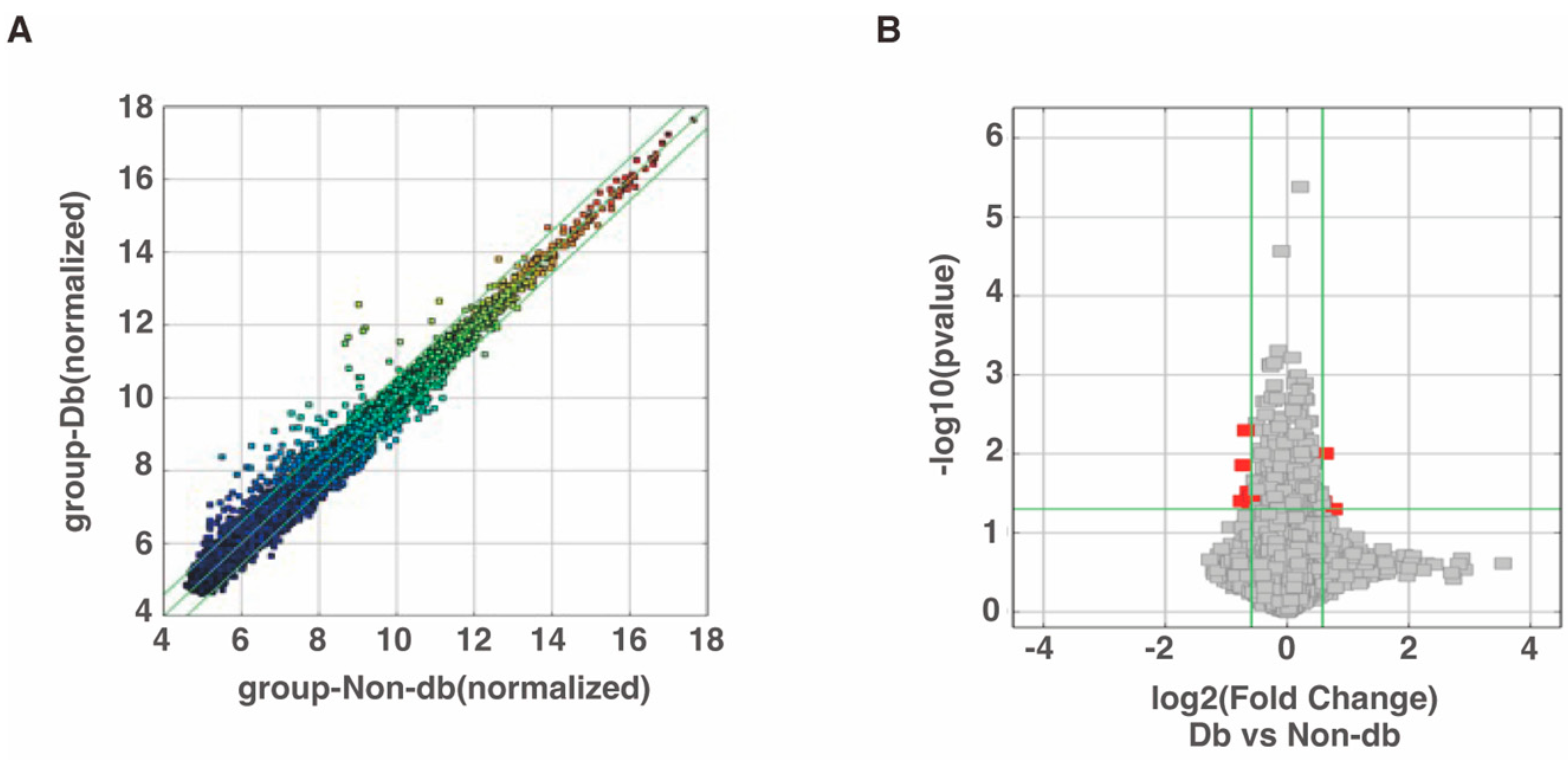
Microarray analysis was performed to evaluate circRNA expression and the results identified 206 circRNAs that were expressed more than 1.5-fold higher in diabetic-derived neutrophils compared with non-diabetic-derived neutrophils (Figure 1A, Supplementary Table S1). Among the 206 total circRNAs, 3 circRNAs were significantly increased in diabetic-derived neutrophils (Figure 1B, Table 2). We also identified 247 circRNAs that were decreased to two-thirds or less in fold change analysis (Figure 1A, Supplementary Table S2). Of the 247 circRNAs, 6 circRNAs were significantly decreased in diabetic-derived neutrophils (Figure 1B, Table 2).
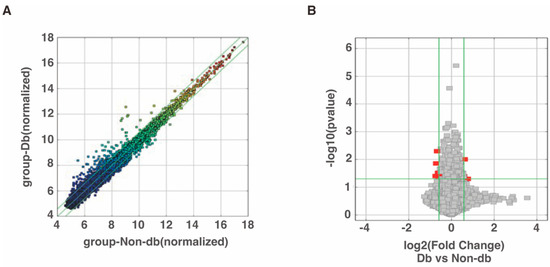
Figure 1.
Changes in circRNA expressions in diabetic-derived neutrophils. (A) Scatter plot was used for assessing the circRNA expression variation between two samples or two groups of samples. The X- and Y-axes are the normalized signal values of the samples (log2 scaled). The green lines indicate fold change. CircRNAs above the top green line and below the bottom green line are circRNAs with more than 1.5-fold change between the two compared samples. (B) Volcano plots were used for visualizing differential expression between two different conditions. The vertical lines correspond to 1.5-fold increase and decrease, respectively, and the horizontal line represents a p-value of 0.05. The red points represent the differentially expressed circRNAs with statistical significance.

Table 2.
Differentially expressed circRNAs.
3.2. miR-129-2-3p May Be Regulated by circRNAs in Diabetic-Derived Neutrophils
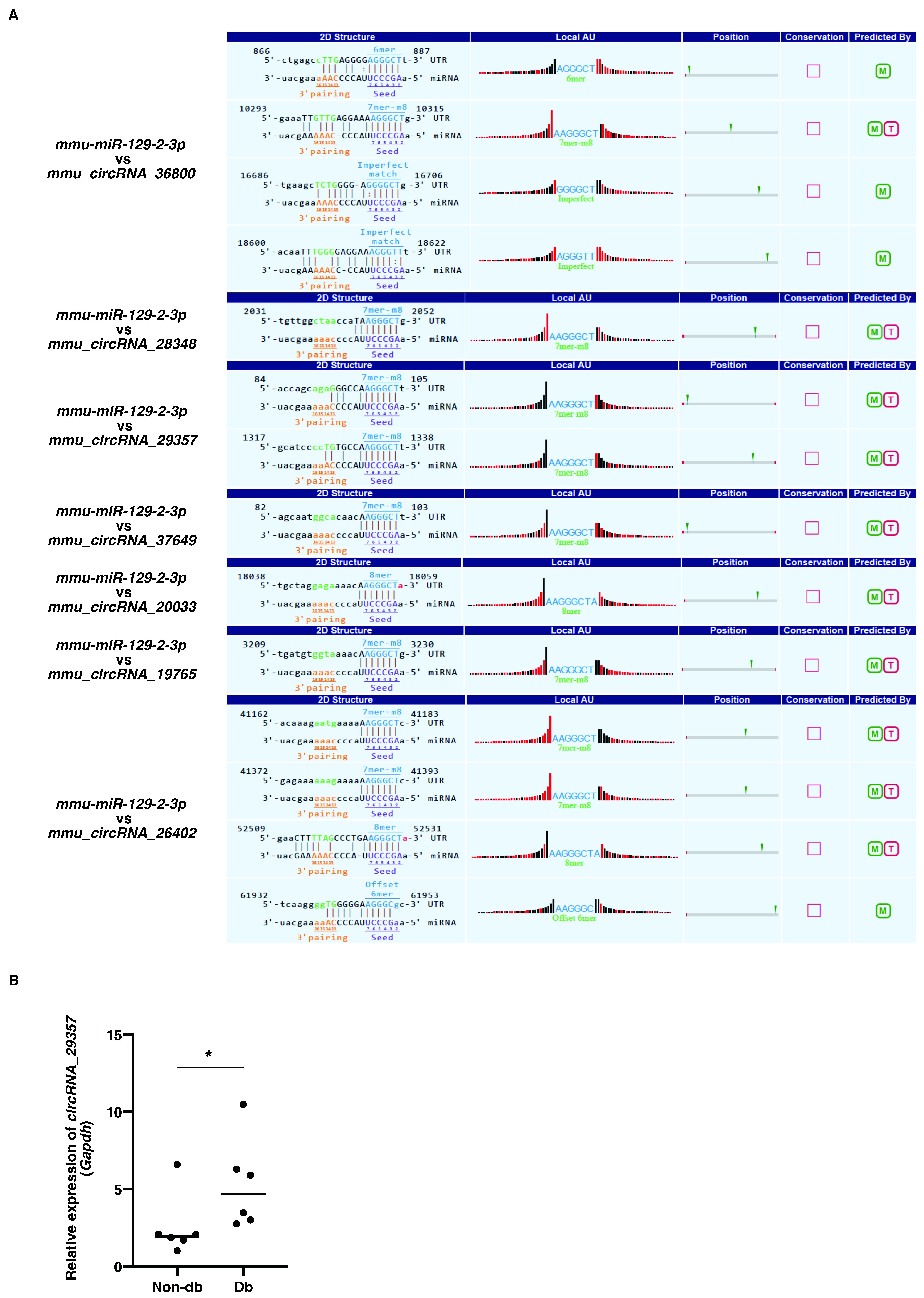
Previous studies showed that the expression of miR-129-2-3p was decreased in diabetic-derived neutrophils [9]. We next examined the upregulated circRNAs for miR-129-2-3p binding sites. However, the three circRNAs that were significantly increased in diabetic-derived neutrophils did not possess miR-129-2-3p binding sites. Therefore, we extracted the top seven circRNAs among the total circRNAs with miR-129-2-3p binding sites using Miranda and TargetScan (Figure 2A). The seven identified circRNAs each had one to four miRNA binding sites.
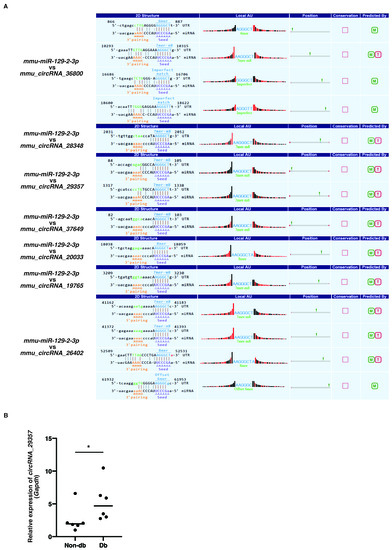
Figure 2.
Identification of circRNAs with putative miR-129-2-3p binding sites. (A). The top seven circRNAs with putative miR-129-2-3p binding sites using Miranda and TargetScan from 206 circRNAs were extracted. The binding sequences between miR-129-2-3p and circRNAs are shown. (B). Relative expression of circRNA_29357 in Db. Graphs show mean ± SD (n = 6). The statistical significance of differences between means was assessed by Mann–Whitney U test. * p < 0.05.
qRT-PCR showed that the expression of mmu_circRNA_29357 was significantly increased in diabetic-derived neutrophils compared with non-diabetic-derived neutrophils (Figure 2B). The expression of the other circRNAs could not be detected by qRT-PCR. These results, showing that the expression of miR-129-2-3p was significantly decreased whereas the expression of mmu_circRNA_29357 was significantly increased in diabetic-derived neutrophils, suggest that miR-129-2-3p may be a target of mmu_circRNA_29357.
3.3. The Function of Diabetic-Derived Neutrophils May Be Regulated by miRNA-circRNA Axis other than miR-129-2-3p-circRNA Axis
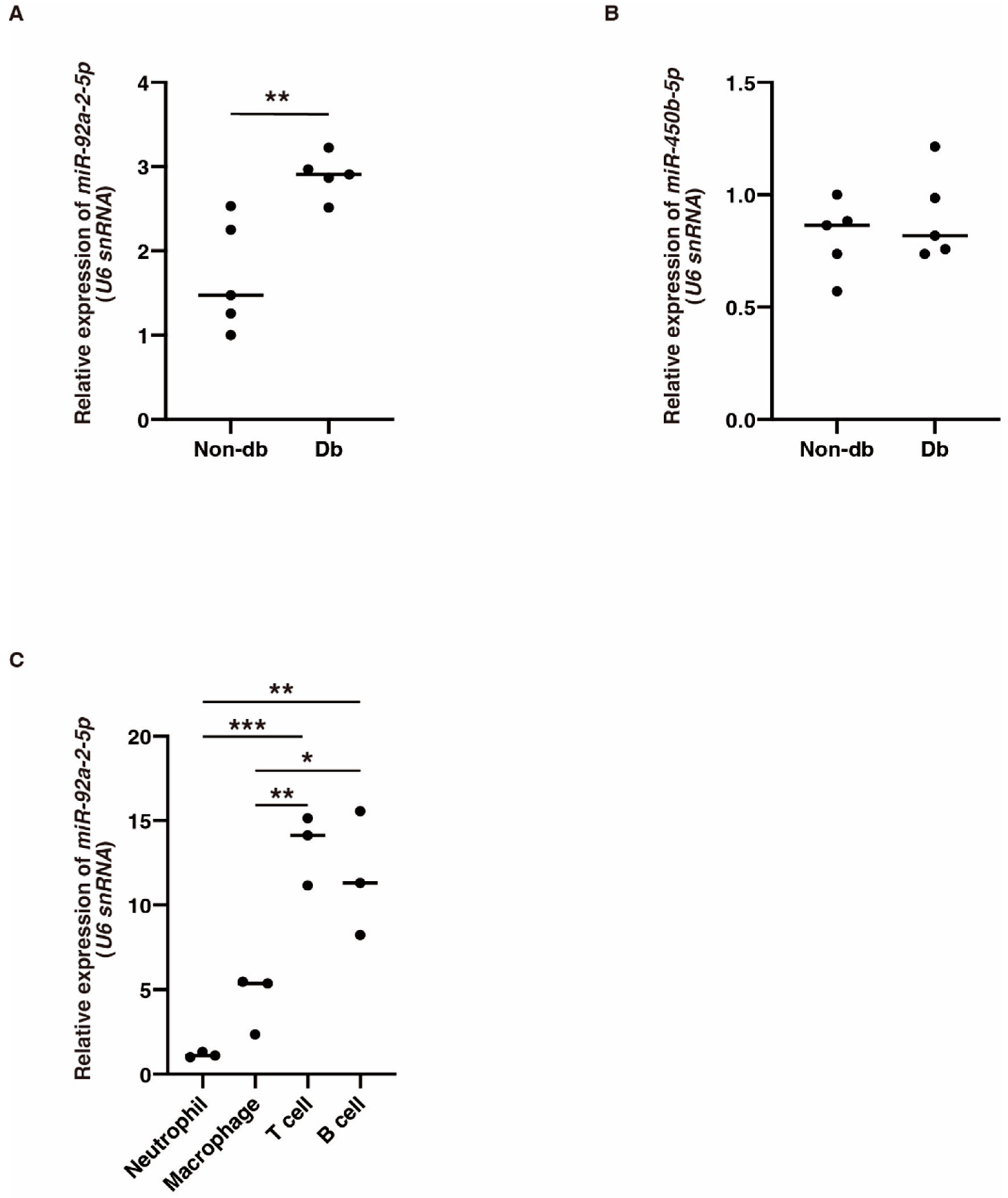
We next focused on the six circRNAs that were significantly downregulated in diabetic-derived neutrophils and investigated their potential miRNA targets (Table 3). The expression of mmu-miR-92a-2-5p, which was identified as a target of mmu_circRNA_22278, was significantly increased in diabetic-derived neutrophils (Figure 3A). The expression of mmu-miR-450b-5p, which was identified as a target of mmu_circRNA_001389, was not changed between diabetic-derived neutrophils and non-diabetic-derived neutrophils (Figure 3B). These results showed that the circRNA–miRNAs may or may not be involved or related to the function of cells from diabetic mice. Further examination of the interactions between these molecules is needed.

Table 3.
miRNA binding sites of circRNAs.
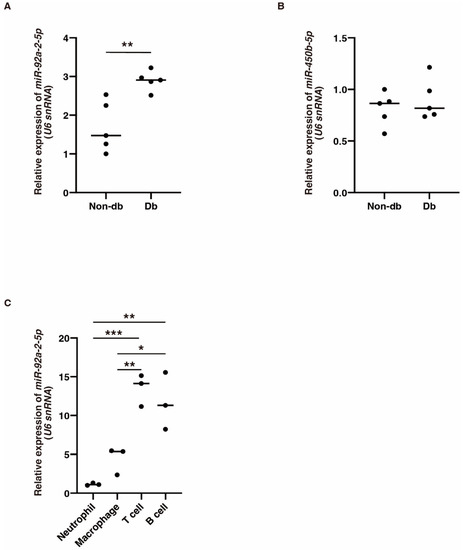
Figure 3.
The expression of miR-92a-2-5p and miR-450b-5p in diabetic-derived neutrophils and miR-92a-2-5p in T and B cells. (A,B). Relative expression of miR-92a-2-5p and miR-450b-5p in Db. Graphs show mean ± SD (n = 5). The statistical significance of differences between means was assessed by unpaired t-test. ** p < 0.01. (C). Relative expression of miR-92a-2-5p in neutrophils, macrophages, T cells, and B cells isolated from BM from non-db mice. miR-92a-2-5p is mainly expressed in T cells and B cells. Graphs show mean ± SD (n = 3). The statistical significance of differences between means was assessed by one-way ANOVA, followed by Tukey’s multiple comparison test. * p < 0.05; ** p < 0.01; *** p < 0.001.
To examine the cellular expression of mmu-miR-92a-2-5p, we isolated neutrophils, macrophages, B cells, and T cells from BM in non-db mice and examined the expression of mmu-miR-92a-2-5p using qRT-PCR. Its expression was significantly increased in T cells and B cells compared with that in other cells (Figure 3C). This suggests that mmu-miR-92a-2-5p might be involved in various biological processes, not just inflammation.
4. Discussion
circRNA are circular RNA molecules that act as sponges for miRNAs, competing with miRNA response elements, and thereby controlling post-transcriptional regulation of miRNAs [21,22]. Recent studies have demonstrated the utility of miRNAs and circRNAs as new diagnostic markers and therapeutic targets for various diseases such as liver disease and cancer [23,24,25,26,27,28,29,30]. To successfully develop diagnostic markers and therapeutic agents, there is a need to elucidate the molecular mechanisms underlying the specific disease. Many discoveries have been made about the role of miRNAs in diabetic nephropathy, and studies of miRNAs have deepened our understanding of this condition. Although previous studies on diabetic nephropathy had the limitation of being restricted to animal experiments, the findings suggest that the circRNA_0001946–miR671-5p–CDR1 axis is a potential therapeutic target for diabetic nephropathy. Findings on other circRNA–miRNA axes such as circRNA_15698–miR-185 also suggested that miRNA regulation may trigger diabetic nephropathy or be involved in the progression of diabetic nephropathy [31,32,33]. On the other hand, Hsa_circ_0054633 were also shown to be useful as diagnostic markers of pre-diabetes and type 2 diabetes mellitus [34]. In an animal experiment, mmu_circ_0000250 suppressed miR-128-3p expression and increased SIRT1 expression, thereby promoting diabetic wound healing [35]. Other studies reported that circRNAs may exert therapeutic effects via regulation of miRNAs/mRNAs such as STAT3 and the miR-31–FBN1 axis in wound healing [36,37]. However, to the best of our knowledge, no studies have focused on circRNA expression in neutrophils as a factor associated with delayed wound healing in diabetes mellitus. Accordingly, in this study, we performed a comprehensive analysis of circRNAs in diabetic-derived neutrophils.
Wound healing in diabetic patients is known to be delayed, which can result in intractable skin ulcers [9]. Repair and regeneration of wound skin tissue consists of three phases: inflammation, proliferation/migration, and maturation/resolution. A balance between the appropriate recruitment and elimination of cells involved in inflammation, such as neutrophils and macrophages, from the BM to the wound site is important for effective tissue repair. We previously showed that, although myeloid cells, including granulocytes, gradually converge on wound sites and then disappear 7 days after wounding in non-diabetic mice, in diabetic mice the number of myeloid cells in wound sites continued to increase on days 2 to 10 after wounding [7]. Thus, the accumulation of chronic myeloid cells at wound sites, involving prolongation of the inflammatory phase, impedes the transition to the proliferative phase such as angiogenesis. In diabetic-derived neutrophils, many miRNAs are differentially expressed, and our results identified miR-129-2-3p as a downregulated miRNA. miR-129-2-3p directly regulates the translation of certain genes such as Casp6, Ccr2, and Dedd2 and is involved in various biological processes such as inflammatory responses and apoptosis [9]. We thus speculated that low-level expression of miR-129-2-3p may contribute to the dysfunction of diabetic-derived neutrophils. As a factor for the low expression of miR-129-2-3p in diabetic-derived neutrophils, other recent studies have shown that miR-129-2 is epigenetically regulated by DNA methylation [38]. In our previous study, analysis of ChIP data of the regulatory region in putative intron 1 of the gene, which is upstream from the sequence encoding the mature miRNA, showed that this region is bound by many transcription factors such as Pu.1 and Cebp transcription factors. Both of these genes are underexpressed in diabetic-derived Gr-1+CD11b+ myeloid cells [39], which include neutrophils. In this study, we speculated that miR-129-2-3p is regulated by circRNAs and performed circRNAs screening in diabetic-derived neutrophils. A comprehensive analysis of circRNAs in diabetic-derived neutrophils has not been performed so far. We also did not find any studies linking the nine circRNAs whose expression was significantly changed in this study with wound healing. Of the seven circRNAs with miR-129-2-3p binding sites, only circRNA_29357 showed significant expression changes in diabetic-derived neutrophils by qRT-PCR. Accordingly, further examination of the function of circRNA_29357 in diabetic-derived neutrophils is required.
We performed prediction analysis and identified miR-450b-5p as a potential target miRNA for circRNA_001389 and miR-92a-2-5p as a potential target microRNA for circRNA_22278. miR-450b-5p was previously reported to be associated with inflammation and shown to reverse endothelial cell damage through regulation of circRNA_0005699 [40]. In this study, no significant changes in miR-450b-5p expression were observed in diabetic-derived neutrophils, suggesting that miR-450b-5p may not be involved in the dysfunction of diabetic-derived neutrophils. In contrast, miR-92a-2-5p showed significant expression changes in diabetic-derived neutrophils. miR-92a-2-5p was reported to induce apoptosis by post-transcriptional repression of miR-92a-2-5p through regulation of circAMOTL1L in renal cell carcinoma [41]. Diabetic wounds in type 2 diabetic mice have myeloid cell accumulation in the wound area, and these cells cause abnormal apoptosis. Consequently, these results showed that these causes were caused by an increase in mmu-miR-92a-2-5p, suggesting that mmu_circRNA_22278 may be involved in regulation of apoptosis.
5. Conclusions
Our results showed that multiple circRNAs exhibited altered expressions in diabetic-derived neutrophils. circRNAs function as sponges for many miRNAs and affect downstream mRNA expression, suggesting that the aberrant levels of these circRNAs may be involved in the dysfunction of diabetic-derived neutrophils. Further investigation is required to determine whether the identified circRNAs are involved in the function of diabetic-derived neutrophils.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10123129/s1, Table S1: Upregulated circRNAs, Table S2: Downregulated circRNAs.
Author Contributions
Conceptualization, T.U., R.M., K.T. and K.A.M.; validation, T.U. and R.M.; formal analysis, T.U.; investigation, T.U., N.S., K.I. and H.S.; resources, T.U. and R.M.; data curation, T.U.; writing—original draft preparation, T.U.; writing—review and editing, T.U.; visualization, T.U.; supervision, T.U.; project administration, T.U.; funding acquisition, T.U., K.T., N.S., K.I. and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Japan Society for the Promotion of Science, Grant-in-Aid for Scientific Research B, grant number 21H03100.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care Committee of Nagasaki University and University of Occupational and Environmental Health (protocol code 1906191539 and 19 June 2019, protocol code AE22-013 and 9 June 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: GSE213297.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wild, S.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global Prevalence of Diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care 2004, 27, 1047–1053. [Google Scholar] [CrossRef]
- Amatruda, M.; Gembillo, G.; Giuffrida, A.E.; Santoro, D.; Conti, G. The Aggressive Diabetic Kidney Disease in Youth-Onset Type 2 Diabetes: Pathogenetic Mechanisms and Potential Therapies. Medicina 2021, 57, 868. [Google Scholar] [CrossRef] [PubMed]
- Brem, H.; Tomic-Canic, M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Investig. 2007, 117, 1219–1222. [Google Scholar] [CrossRef] [PubMed]
- Faselis, C.; Katsimardou, A.; Imprialos, K.; Deligkaris, P.; Kallistratos, M.S.; Dimitriadis, K. Microvascular Complications of Type 2 Diabetes Mellitus. Curr. Vasc. Pharmacol. 2020, 18, 117–124. [Google Scholar] [CrossRef]
- Javed, S.; Hayat, T.; Menon, L.; Alam, U.; Malik, R.A. Diabetic peripheral neuropathy in people with type 2 diabetes: Too little too late. Diabet. Med. 2020, 37, 573–579. [Google Scholar] [CrossRef]
- Zhou, T.; Hu, Z.; Yang, S.; Sun, L.; Yu, Z.; Wang, G. Role of Adaptive and Innate Immunity in Type 2 Diabetes Mellitus. J. Diabetes Res. 2018, 2018, 7457269. [Google Scholar] [CrossRef] [PubMed]
- Mahdipour, E.; Charnock, J.C.; Mace, K.A. Hoxa3 promotes the differentiation of hematopoietic progenitor cells into proangiogenic Gr-1+CD11b+ myeloid cells. Blood 2011, 117, 815–826. [Google Scholar] [CrossRef]
- Torbica, T.; Wicks, K.; Umehara, T.; Gungordu, L.; Alrdahe, S.; Wemyss, K.; Grainger, J.R.; Mace, K.A. Chronic Inflammation in Response to Injury: Retention of Myeloid Cells in Injured Tissue Is Driven by Myeloid Cell Intrinsic Factors. J. Investig. Dermatol. 2019, 139, 1583–1592. [Google Scholar] [CrossRef]
- Umehara, T.; Mori, R.; Mace, K.A.; Murase, T.; Abe, Y.; Yamamoto, T.; Ikematsu, K. Identification of Specific miRNAs in Neutrophils of Type 2 Diabetic Mice: Overexpression of miRNA-129-2-3p Accelerates Diabetic Wound Healing. Diabetes 2019, 68, 617–630. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Kjems, J.; Hansen, T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Han, B.; Chao, J.; Yao, H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018, 187, 31–44. [Google Scholar] [CrossRef]
- Cen, J.; Liang, Y.; Huang, Y.; Pan, Y.; Shu, G.; Zheng, Z.; Liao, X.; Zhou, M.; Chen, D.; Fang, Y.; et al. Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol. Cancer 2021, 20, 19. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Yu, Z.; Zhou, Y.; Tu, J.; Lou, J.; Wang, Y. Circular RNA Circ100084 functions as sponge of miR-23a-5p to regulate IGF2 expression in hepatocellular carcinoma. Mol. Med. Rep. 2020, 21, 2395–2404. [Google Scholar] [CrossRef]
- Xu, J.-Y.; Chang, N.-B.; Rong, Z.-H.; Li, T.; Xiao, L.; Yao, Q.-P.; Jiang, R.; Jiang, J. circDiaph3 regulates rat vascular smooth muscle cell differentiation, proliferation, and migration. FASEB J. 2019, 33, 2659–2668. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, X.; Li, W.; Han, J.; Jin, J.; Su, F.; Zhang, J.; Huang, W.; Xiao, F.; Pan, Q.; et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int. J. Mol. Med. 2018, 42, 1865–1874. [Google Scholar] [CrossRef]
- Altesha, M.; Ni, T.; Khan, A.; Liu, K.; Zheng, X. Circular RNA in cardiovascular disease. J. Cell. Physiol. 2019, 234, 5588–5600. [Google Scholar] [CrossRef]
- Panda, A.C.; Gorospe, M. Detection and Analysis of Circular RNAs by RT-PCR. Bio-protocol 2018, 8, e2775. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Chien, Y.; Tsai, P.-H.; Lai, Y.-H.; Lu, K.-H.; Liu, C.-Y.; Lin, H.-F.; Huang, C.-S.; Wu, W.-W.; Wang, C.-Y. CircularRNA as novel biomarkers in liver diseases. J. Chin. Med. Assoc. 2020, 83, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, W.; Wang, X.; Wang, T.; Chen, Y.; Chen, B.; Liu, R.; Bai, P.; Xing, J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol. Med. 2018, 24, 40. [Google Scholar] [CrossRef]
- Li, R.-C.; Ke, S.; Meng, F.-K.; Lu, J.; Zou, X.-J.; He, Z.-G.; Wang, W.-F.; Fang, M.-H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018, 9, 838. [Google Scholar] [CrossRef]
- Zhu, G.; Chang, X.; Kang, Y.; Zhao, X.; Tang, X.; Ma, C.; Fu, S. CircRNA: A novel potential strategy to treat thyroid cancer (Review). Int. J. Mol. Med. 2021, 48, 201. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in Cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef]
- Jin, X.; Feng, C.-Y.; Xiang, Z.; Chen, Y.-P.; Li, Y.-M. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016, 7, 66455–66467. [Google Scholar] [CrossRef]
- Zahari Sham, S.Y.; Abdullah, M.; Osman, M.; Seow, H.F. An insight of dysregulation of microRNAs in the pathogenesis of diabetic kidney disease. Malays. J. Pathol. 2022, 44, 187–201. [Google Scholar]
- Li, X.; Diao, H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. 2019, 234, 13807–13819. [Google Scholar] [CrossRef]
- Hu, W.; Han, Q.; Zhao, L.; Wang, L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J. Cell. Physiol. 2019, 234, 1469–1476. [Google Scholar] [CrossRef]
- Tu, C.; Wang, L.; Wei, L.; Jiang, Z. The role of circular RNA in Diabetic Nephropathy. Int. J. Med. Sci. 2022, 19, 916–923. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Jian, D.; Hao, P.; Rao, L.; Li, M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017, 54, 237–245. [Google Scholar] [CrossRef]
- Shi, R.; Jin, Y.; Hu, W.; Lian, W.; Cao, C.; Han, S.; Zhao, S.; Yuan, H.; Yang, X.; Shi, J.; et al. Exosomes derived from mmu_circ_0000250-modified adipose-derived mesenchymal stem cells promote wound healing in diabetic mice by inducing miR-128-3p/SIRT1-mediated autophagy. Am. J. Physiol. Cell Physiol. 2020, 318, C848–C856. [Google Scholar] [CrossRef]
- Yang, Z.-G.; Awan, F.M.; Du, W.W.; Zeng, Y.; Lyu, J.; Wu, D.; Gupta, S.; Yang, W.; Yang, B.B. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol. Ther. 2017, 25, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liu, W.; Li, G.; Liu, L. Circ_PRKDC knockdown promotes skin wound healing by enhancing keratinocyte migration via miR-31/FBN1 axis. J. Mol. Histol. 2021, 52, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, X.; Wang, H.; Wen, R.; He, J.; Tang, J. Epigenetic regulation of miR-129-2 and its effects on the proliferation and invasion in lung cancer cells. J. Cell. Mol. Med. 2015, 19, 2172–2180. [Google Scholar] [CrossRef] [PubMed]
- Wicks, K.; Torbica, T.; Umehara, T.; Amin, S.; Bobola, N.; Mace, K.A. Diabetes Inhibits Gr-1+ Myeloid Cell Maturation via Cebpa Deregulation. Diabetes 2015, 64, 4184–4197. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, L.; Ye, B.; Chen, W.; Zheng, G.; Xie, H.; Guo, Y. Knockdown of hsa_circ_0005699 attenuates inflammation and apoptosis induced by ox-LDL in human umbilical vein endothelial cells through regulation of the miR-450b-5p/NFKB1 axis. Mol. Med. Rep. 2022, 26, 290. [Google Scholar] [CrossRef]
- Gao, L.; Shao, X.; Yue, Q.; Wu, W.; Yang, X.; He, X.; Li, L.; Hou, F.; Zhang, R. circAMOTL1L Suppresses Renal Cell Carcinoma Growth by Modulating the miR-92a-2-5p/KLLN Pathway. Oxidative Med. Cell. Longev. 2021, 2021, 9970272. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).