Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culturing and sEV Isolation
2.2. Differential Ultracentrifugation (UC)
2.3. Quantification of sEVs by Nanoparticle Tracking Analysis (NTA)
2.4. Sample Preparation for Phosphoproteomics
2.5. Phosphopeptide Enrichment by IMAC and TiO2
2.6. Nano-LC-MS/MS
2.7. MS Spectra Processing
2.8. Data Filtering and Phosphorylation Site Localization
2.9. Disease and Functional Annotation Analysis
2.10. Data Availability
2.11. ATP-Citrate Synthase Activity Assay
2.12. Phosphofructokinase Activity Assay
2.13. SIRT1 and SIRT6 Activity Assay
2.14. Western Blot
3. Results
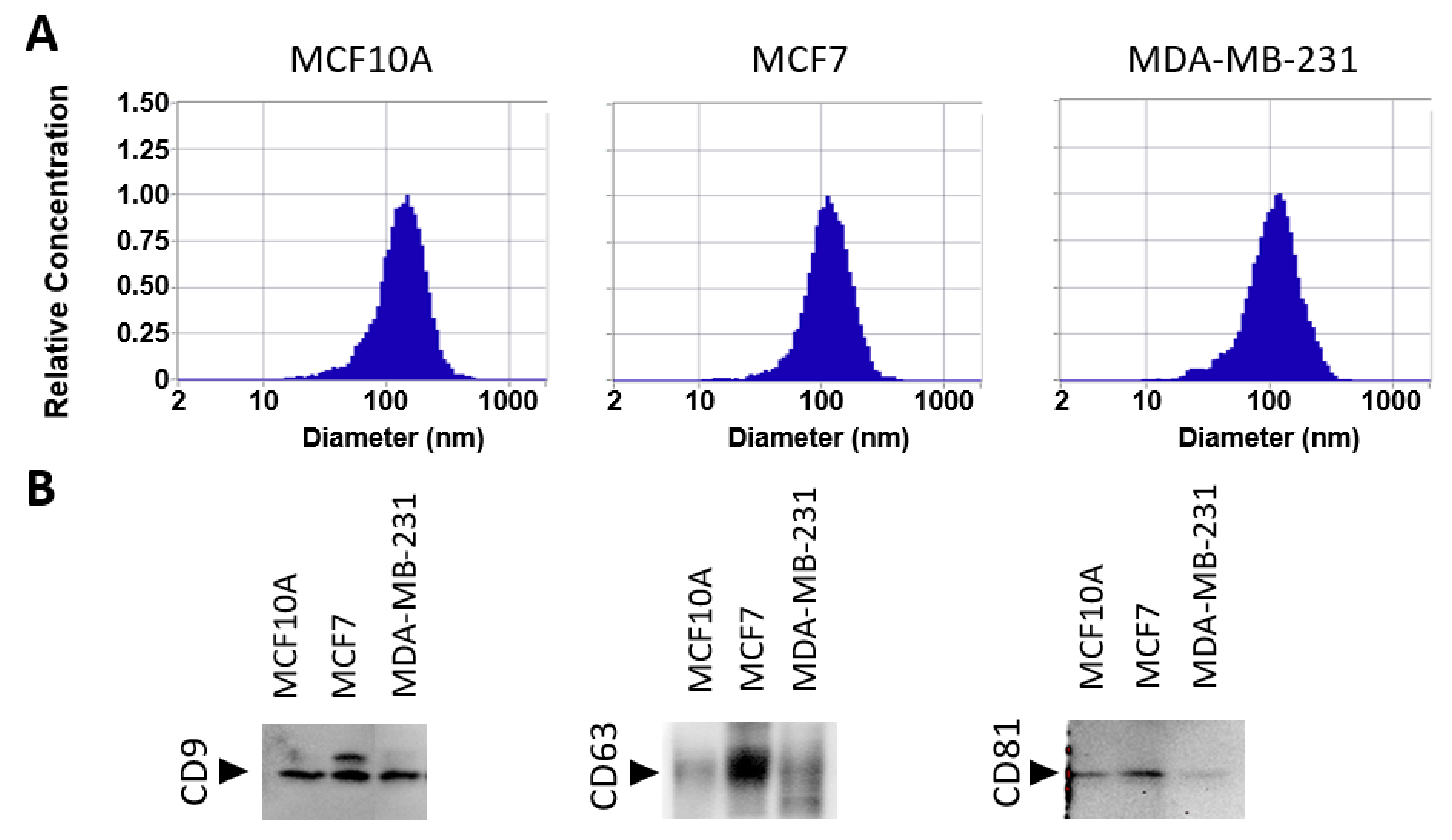
3.1. Isolation of sEV
3.2. Overall Phosphoproteome Profiling
3.3. Phosphorylation Site Distributions, Phosphorylation Motifs and Predicted Potential Kinases for Identified Phosphorylation Sites
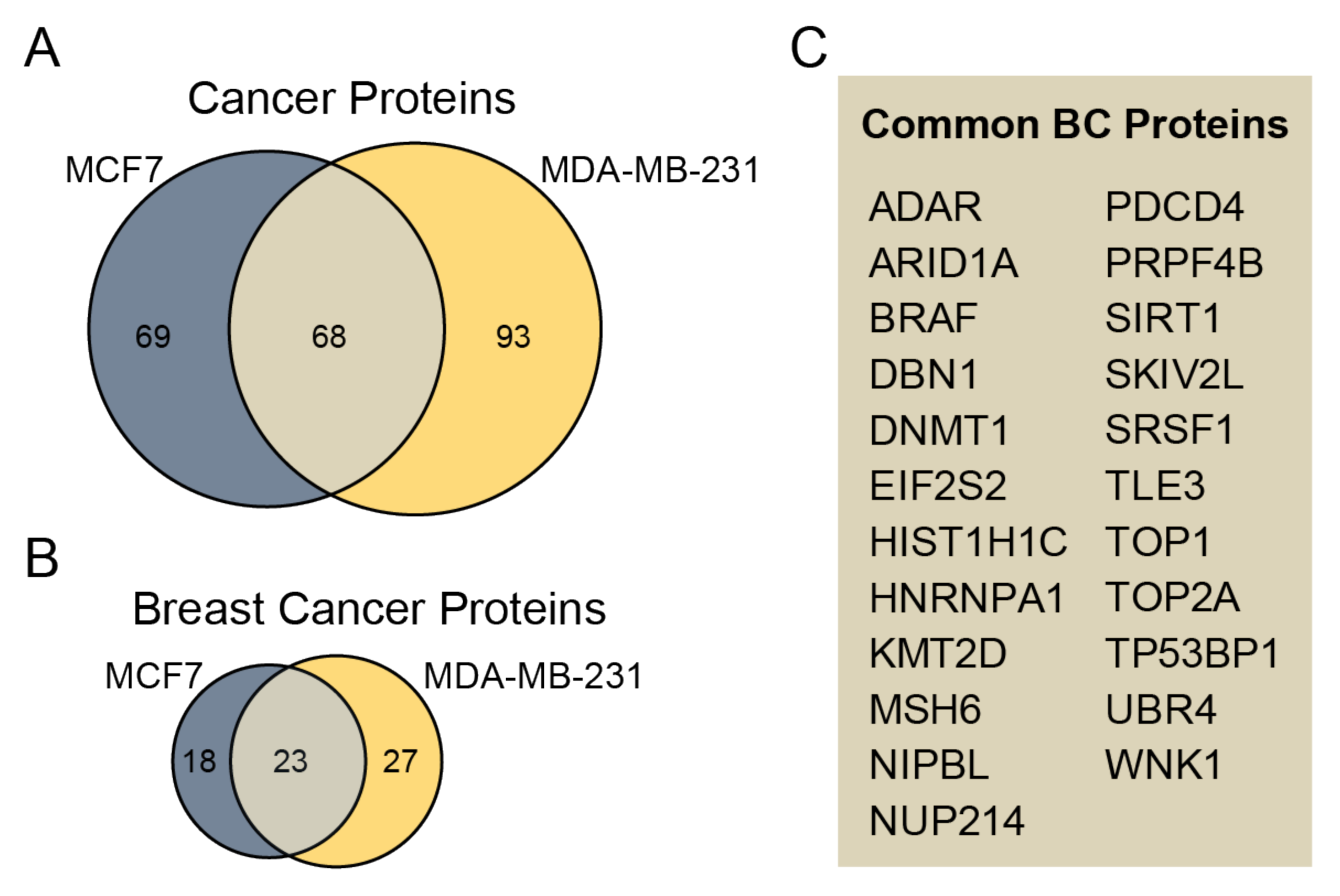
3.4. Identified Phosphoproteins in the Context of Cancer/Breast Cancer
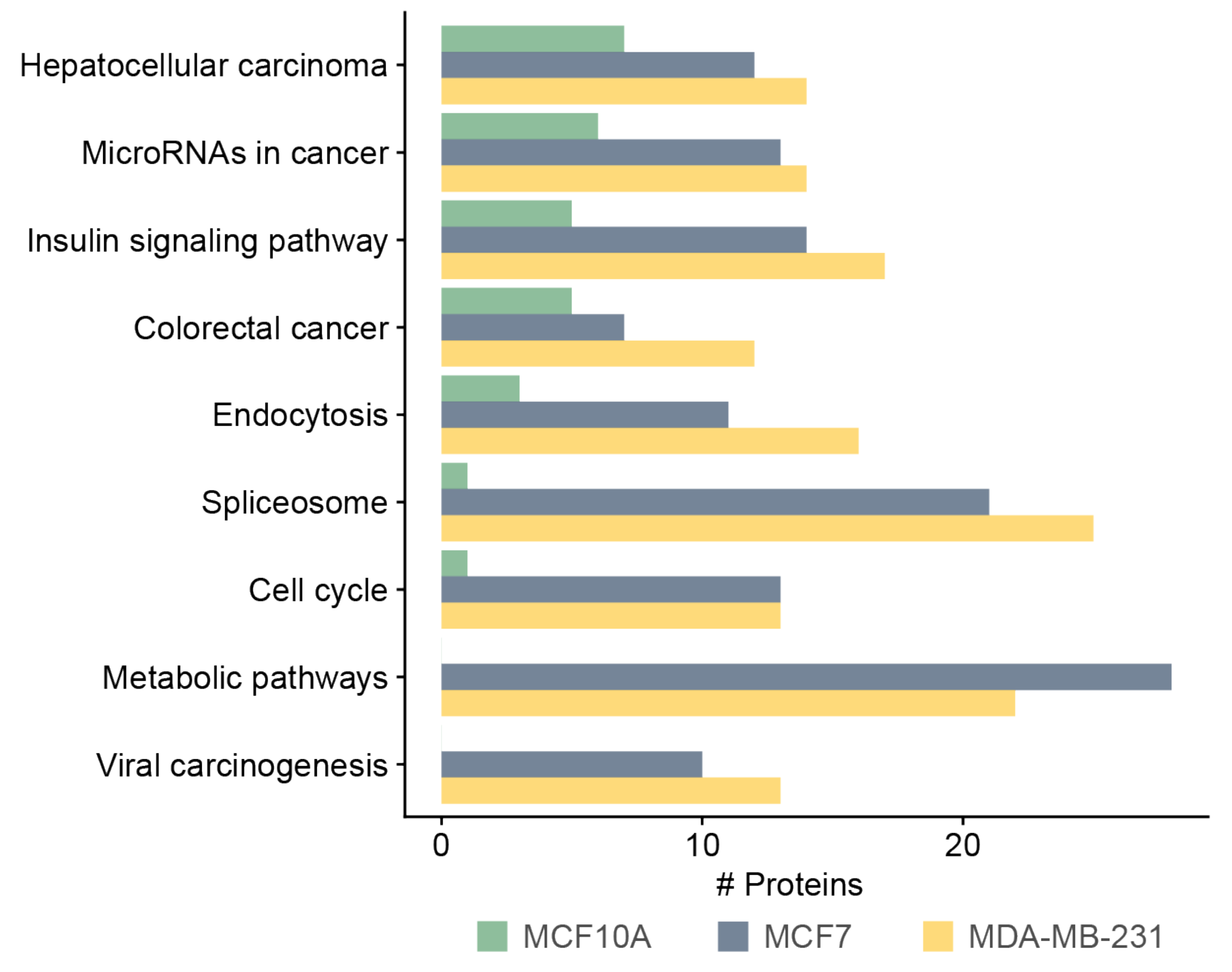
3.5. Functional and Pathway Analysis of Identified Phosphoproteins
3.6. Analysis of ACLY, PKFM, SIRT1, and SIRT6 in Cells and Their sEVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruger, S.; Abd Elmageed, Z.Y.; Hawke, D.H.; Wörner, P.M.; Jansen, D.A.; Abdel-Mageed, A.B.; Alt, E.U.; Izadpanah, R. Molecular Characterization of Exosome-Like Vesicles from Breast Cancer Cells. BMC Cancer 2014, 14, 44. [Google Scholar] [CrossRef] [Green Version]
- Palazzolo, G.; Albanese, N.N.; DI Cara, G.; Gygax, D.; Vittorelli, M.L.; Pucci-Minafra, I. Proteomic Analysis of Exosome-Like Vesicles Derived from Breast Cancer Cells. Anticancer Res. 2012, 32, 847–860. [Google Scholar] [PubMed]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic Profiling of NCI-60 Extracellular Vesicles Uncovers Common Protein Cargo and Cancer Type-Specific Biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef] [PubMed]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic Analysis of Exosomes from Mutant KRAS Colon Cancer Cells Identifies Intercellular Transfer of Mutant KRAS. Mol. Cell Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and Proteomic Analysis of Ovarian Cancer-Derived Exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The Proteomic Analysis of Breast Cell Line Exosomes Reveals Disease Patterns and Potential Biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- Risha, Y.; Susevski, V.; Hüttmann, N.; Poolsup, S.; Minic, Z.; Berezovski, M.V. Breast Cancer-Derived Microvesicles Are the Source of Functional Metabolic Enzymes as Potential Targets for Cancer Therapy. Biomedicines. 2021, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.S.; Henrique Baeta, H.; Moraes, M.C.S.; Beck, H.C.; Rodriguez, M.S.; Saraswat, M.; Pandey, A.; Matthiesen, R. Extra-Cellular Vesicles Carry Proteome of Cancer Hallmarks. Front. Biosci. 2020, 25, 398–436. [Google Scholar]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; Mccague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic Profiling of Extracellular Vesicles Derived from Prostate and Prostate Cancer Cell Lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular Vesicle Isolation and Characterization: Toward Clinical Application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minciacchi, V.R.; Freeman, M.R.; Vizio, D.D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizio, D.D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large Oncosomes in Human Prostate Cancer Tissues and in the Circulation of Mice with Metastatic Disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Dahms, T.E.S.; Babu, M. Chromatographic Separation Strategies for Precision Mass Spectrometry to Study Protein-Protein Interactions and Protein Phosphorylation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1102-1103, 96–108. [Google Scholar] [CrossRef]
- Yu, S.; Cao, H.; Shen, B.; Feng, J. Tumor-Derived Exosomes in Cancer Progression and Treatment Failure. Oncotarget 2015, 6, 37151–37168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Cui, Y.; Yan, Z.; Luo, Y.; Zhang, W.; Deng, S.; Tang, S.; Zhang, G.; He, Q.Y.; Wang, T. Phosphoproteome Characterization of Human Colorectal Cancer SW620 Cell-Derived Exosomes and New Phosphosite Discovery for C-HPP. J. Proteome Res. 2016, 15, 4060–4072. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ding, Y.; Liu, G.; Yang, X.; Zhao, R.; Zhang, Y.; Zhao, X.; Anderson, G.J.; Nie, G. Cancer Cell-derived Exosomes Induce Mitogen-activated Protein Kinase-dependent Monocyte Survival by Transport of Functional Receptor Tyrosine Kinases. J. Biol. Chem. 2016, 291, 8453–8464. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in Extracellular Vesicles as Candidate Markers for Breast Cancer. Proc Natl Acad Sci U S A. 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [Green Version]
- Iliuk, A.; Wu, X.; Li, L.; Sun, J.; Hadisurya, M.; Boris, R.S.; Tao, W.A. Plasma-Derived Extracellular Vesicle Phosphoproteomics through Chemical Affinity Purification. J. Proteome Res. 2020, 19, 2563–2574. [Google Scholar] [CrossRef]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2016, 14, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- R. Core Team. R: A Language and Environment for Statistical Computing. R. Core Team: Vienna, Austria, 2021. [Google Scholar]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and Recalibrations. Nucleic Acids Res. 2015, 43, D512-520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Xu, H.; Lin, S.; Deng, W.; Zhou, J.; Zhang, Y.; Shi, Y.; Peng, D.; Xue, Y. GPS 5.0: An Update on the Prediction of Kinase-Specific Phosphorylation Sites in Proteins. Genomics Proteomics Bioinformatics 2020, 18, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48 (D1), D845–D855. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gene Ontology Consortium. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49 (D1), D325–D334.
- Carlson, M. Org.Hs.Eg.Db: Genome Wide Annotation for Human, R Package Version 3.8.2. 2019.
- Adams, J.A. Kinetic and Catalytic Mechanisms of Protein Kinases. Chem. Rev. 2001, 101, 2271–2290. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, N.; Imamura, H.; Ishihama, Y. Large-scale Discovery of Substrates of the Human Kinome. Sci Rep. 2019, 9, 10503. [Google Scholar] [CrossRef] [PubMed]
- Boschat, A.C.; Minet, N.; Martin, E.; Barouki, R.; Latour, S.; Sanquer, S. CTP Synthetase Activity Assay by Liquid Chromatography Tandem Mass Spectrometry in the Multiple Reaction Monitoring Mode. J. Mass Spectrum. 2019, 54, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Im, E.J.; Lee, C.H.; Moon, P.G.; Rangaswamy, G.G.; Lee, B.; Lee, J.M.; Lee, J.C.; Jee, J.G.; Bae, J.S.; Kwon, T.K.; et al. Sulfisoxazole Inhibits the Secretion of Small Extracellular Vesicles by Targeting the Endothelin Receptor A. Nat. Commun. 2019, 10, 1387. [Google Scholar] [PubMed]
- Cordover, E.; Wei, J.; Patel, C.; Shan, N.L.; Gionco, J.; Sargsyan, D.; Wu, R.; Cai, L.; Kong, A.N.; Jacinto, E.; et al. KPT-9274, an Inhibitor of PAK4 and NAMPT, Leads to Downregulation of mTORC2 in Triple Negative Breast Cancer Cells. Chem. Res. Toxicol. 2020, 33, 482–491. [Google Scholar] [CrossRef]
- Roovers, K.; Wagner, S.; Storbeck, C.J.; O’Reilly, P.; Lo, V.; Northey, J.J.; Chmielecki, J.; Muller, W.J.; Siegel, P.M.; Sabourin, L.A. The Ste20-like Kinase SLK is Required for ErbB2-Driven Breast Cancer Cell Motility. Oncogene 2009, 28, 2839–4288. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.H.; Yoo, K.C.; An, Y.; Lee, H.J.; Lee, M.; Uddin, N.; Kim, M.J.; Kim, I.G.; Suh, Y.; Lee, S.J. FYN Promotes Mesenchymal Phenotypes of Basal Type Breast Cancer Cells Through STAT5/NOTCH2 Signaling Node. Oncogene 2018, 37, 1857–1868. [Google Scholar] [CrossRef]
- Kim, L.C.; Song, L.; Haura, E.B. Src Kinases as Therapeutic Targets for Cancer. Nat. Rev. Clin. Oncol. 2009, 6, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Singh, T.; Singh, A.K. Enzymes as Diagnostic Tools. Adv. Enzym. Technol 2019, 225–271. [Google Scholar]
- Catalano, S.; Barone, I.; Marsico, S.; Bruno, R.; Andò, S. Phosphorylation Processes Controlling Aromatase Activity in Br east Cancer: An Update. Mini Rev. Med. Chem. 2016, 16, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Seshacharyulu, P.; Pandey, P.; Datta, K.; Batra, S.K. Phosphatase: PP2A Structural Importance, Regulation and Its Aberrant Expression in Cancer. Cancer Lett. 2013, 335, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Longati, P.; Comoglio, P.M.; Bardelli, A. Receptor Tyrosine Kinases as Therapeutic Targets: The Model of the MET Oncogene. Curr. Drug Targets 2001, 2, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Richards, E.G.; Mukherjee, A.; Srere, P.A. Structure of ATP Citrate Lyase from Rat Liver. Physicochemical Studies and Proteolytic Modification. J. Biol. Chem. 1976, 251, 5242–5250. [Google Scholar] [CrossRef]
- Granchi, C. ATP Citrate Lyase (ACLY) Inhibitors: An Anti-Cancer Strategy at the Crossroads of Glucose and Lipid Metabolism. Eur. J. Med. Chem. 2018, 157, 1276–1291. [Google Scholar] [CrossRef]
- Chypre, M.; Zaidi, N.; Smans, K. ATP-Citrate Lyase: A Mini-Review. Biochem. Biophys. Res. Commun. 2012, 422, 1–4. [Google Scholar] [CrossRef]
- Migita, T.; Narita, T.; Nomura, K.; Miyagi, E.; Inazuka, F.; Matsuura, M.; Ushijima, M.; Mashima, T.; Seimiya, H.; Satoh, Y.; et al. ATP Citrate Lyase: Activation and Therapeutic Implications in Non-Small Cell Lung Cancer. Cancer Res. 2008, 68, 8547–8554. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; Tao, R.; Gao, X.; Li, T.; Zhou, X.; Guan, K.L.; Xiong, Y.; Lei, Q.Y. Acetylation Stabilizes ATP-Citrate Lyase to Promote Lipid Biosynthesis and Tumor Growth. Mol. Cell. 2013, 51, 506–518. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Huang, X.; Ye, T. MiR-22 Down-Regulates the Proto-Oncogene ATP Citrate Lyase to Inhibit the Growth and Metastasis of Breast Cancer. Am. J. Transl. Res. 2018, 10, 659–669. [Google Scholar] [PubMed]
- Zheng, Y.; Zhou, Q.; Zhao, C.; Li, J.; Yu, Z.; Zhu, Q. ATP Citrate Lyase Inhibitor Triggers Endoplasmic Reticulum Stress to Induce Hepatocellular Carcinoma Cell Apoptosis via p-eIF2α/ATF4/CHOP Axis. J. Cell Mol. Med. 2021, 25, 1468–1479. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, K.; Gong, D.; Zhang, J.; Li, Q.; Zhao, G.; Lin, P. ACLY: A Biomarker of Recurrence in Breast Cancer. Pathol. Res. Pract. 2020, 216, 153076. [Google Scholar] [CrossRef]
- Musumeci, O.; Bruno, C.; Mongini, T.; Rodolico, C.; Aguennouz, M.; Barca, E.; Amati, A.; Cassandrini, D.; Serlenga, L.; Vita, G.; et al. Clinical Features and New Molecular Findings in Muscle Phosphofructokinase Deficiency (GSD Type VII). Neuromuscul. Disord. 2012, 22, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Mor, I.; Cheung, E.C.; Vousden, K.H. Control of Glycolysis Through Regulation of PFK1: Old Friends and Recent Additions. Cold Spring Harb Symp. Quant. Biol. 2011, 76, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Forouhar, F.; Szu, F.E.; Seetharaman, J.; Tong, L.; Barber, D.L. Structures of Human Phosphofructokinase-1 and Atomic Basis of Cancer-Associated Mutations. Nature 2015, 523, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Ismail, R.; Ul Hussain, M. The Up Regulation of Phosphofructokinase1 (PFK1) Protein During Chemically Induced Hypoxia is Mediated by the Hypoxia-Responsive Internal Ribosome Entry Site (IRES) Element, Present in Its 5’Untranslated Region. Biochimie 2017, 139, 38–45. [Google Scholar] [CrossRef]
- Lee, J.H.; Liu, R.; Li, J.; Zhang, C.; Wang, Y.; Cai, Q.; Qian, X.; Xia, Y.; Zheng, Y.; Piao, Y.; et al. Stabilization of Phosphofructokinase 1 Platelet Isoform by AKT Promotes Tumorigenesis. Nat. Commun. 2017, 8, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clem, B.; Telang, S.; Clem, A.; Yalcin, A.; Meier, J.; Simmons, A.; Rasku, M.A.; Arumugam, S.; Dean, W.L.; Eaton, J.; et al. Small-Molecule Inhibition of 6-Phosphofructo-2-Kinase Activity Suppresses Glycolytic Flux and Tumor Growth. Mol. Cancer Ther. 2008, 7, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Ahsan, H.; Halpern, J.; Kibriya, M.G.; Pierce, B.L.; Tong, L.; Gamazon, E.; McGuire, V.; Felberg, A.; Shi, J.; Jasmine, F.; et al. A Genome-Wide Association Study of Early-Onset Breast Cancer Identifies PFKM as a Novel Breast Cancer Gene and Supports a Common Genetic Spectrum for Breast Cancer at Any Age. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 658–669. [Google Scholar] [CrossRef] [Green Version]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [Green Version]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-Dependent Transcription and Cell Survival by the SIRT1 Deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, Y.W.; Kang, H.J.; Kim, H.J.; Kong, Y.; Brown, M.L.; Bae, I. Targeting Mutant p53 by a SIRT1 Activator YK-3-237 Inhibits the Proliferation of Triple-Negative Breast Cancer Cells. Oncotarget 2013, 4, 984–994. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.S.; Hyun, C.L.; Park, I.A.; Kim, J.Y.; Chung, Y.R.; Im, S.A.; Lee, K.H.; Moon, H.G.; Ryu, H.S. SIRT1 Induces Tumor Invasion by Targeting Epithelial Mesenchymal Transition-Related Pathway and is a Prognostic Marker in Triple Negative Breast Cancer. Tumour Biol. 2016, 37, 4743–4753. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, W.; Dong, F.; Guo, Y.; Tan, J.; Ruan, S.; Huang, T. The Prognostic Role of Sirt1 Expression in Solid Malignancies: A Meta-Analysis. Oncotarget 2017, 8, 66343–66351. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.-W.; Li, W.-Q.; Wan, G.-X.; Li, Y.-X.; Du, X.-M.; Li, Y.-C.; Feng, L. Correlation and Prognostic Value of SIRT1 and Notch1 Signaling in Breast Cancer. J. Exp. Clin. Cancer Res CR 2014, 33, 97. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.R.; Kim, H.; Park, S.Y.; Park, I.A.; Jang, J.J.; Choe, J.-Y.; Jung, Y.Y.; Im, S.A.; Moon, H.G.; Lee, K.H.; et al. Distinctive role of SIRT1 expression on tumor invasion and metastasis in breast cancer by molecular subtype. Hum. Pathol. 2015, 46, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.I.; Nirzhor, S.S.R.; Akter, R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules 2018, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebastián, C.; Zwaans, B.M.M.; Silberman, D.M.; Gymrek, M.; Goren, A.; Zhong, L.; Ram, O.; Truelove, J.; Guimaraes, A.R.; Toiber, D.; et al. The Histone Deacetylase SIRT6 Is a Tumor Suppressor that Controls Cancer Metabolism. Cell 2012, 151, 1185–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenhunen, J.; Kučera, T.; Huovinen, M.; Küblbeck, J.; Bisenieks, E.; Vigante, B.; Ogle, Z.; Duburs, G.; Doležal, M.; Moaddel, R.; et al. Screening of SIRT6 Inhibitors and Activators: A Novel Activator has an Impact on Breast Cancer Cells. Biomed. Pharmacother. 2021, 138, 111452. [Google Scholar] [CrossRef] [PubMed]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Atelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alghanem, B.; Ali, R.; Nehdi, A.; Al Zahrani, H.; Altolayyan, A.; Shaibah, H.; Baz, O.; Alhallaj, A.; Moresco, J. Proteomics profiling of KAIMRC1 in comparison to MDA-MB231 and MCF-7. Int. J. Mol. Sci. 2020, 21, 4328. [Google Scholar]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Enzyme | Gene | Number of Phosphorpeptide Identifications | MCF7 sEV | MDA-MB-231 sEV | ||
|---|---|---|---|---|---|---|
| CID | HCD | CID | HCD | |||
| Found in sample replicates (Number/3) | ||||||
| ATP citrate lyase | ACLY | 8 | 3 | 3 | 1 | 1 |
| 6-Phosphofructokinase | PFKM | 8 | 1 | 0 | 3 | 3 |
| Sirtuin 1 | SIRT1 | 7 | 3 | 1 | 0 | 3 |
| CTP synthetase 1 | CTPS1 | 7 | 1 | 0 | 3 | 3 |
| Sirtuin 6 | SIRT6 | 6 | 0 | 3 | 0 | 3 |
| Acetyl-CoA synthetase 2 | ACSS2 | 5 | 2 | 3 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minic, Z.; Hüttmann, N.; Poolsup, S.; Li, Y.; Susevski, V.; Zaripov, E.; Berezovski, M.V. Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes. Biomedicines 2022, 10, 408. https://doi.org/10.3390/biomedicines10020408
Minic Z, Hüttmann N, Poolsup S, Li Y, Susevski V, Zaripov E, Berezovski MV. Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes. Biomedicines. 2022; 10(2):408. https://doi.org/10.3390/biomedicines10020408
Chicago/Turabian StyleMinic, Zoran, Nico Hüttmann, Suttinee Poolsup, Yingxi Li, Vanessa Susevski, Emil Zaripov, and Maxim V. Berezovski. 2022. "Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes" Biomedicines 10, no. 2: 408. https://doi.org/10.3390/biomedicines10020408
APA StyleMinic, Z., Hüttmann, N., Poolsup, S., Li, Y., Susevski, V., Zaripov, E., & Berezovski, M. V. (2022). Phosphoproteomic Analysis of Breast Cancer-Derived Small Extracellular Vesicles Reveals Disease-Specific Phosphorylated Enzymes. Biomedicines, 10(2), 408. https://doi.org/10.3390/biomedicines10020408








