An Integrative Approach to Characterize the Early Phases of Dimethylhydrazine-Induced Colorectal Carcinogenesis in the Rat
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Chemicals
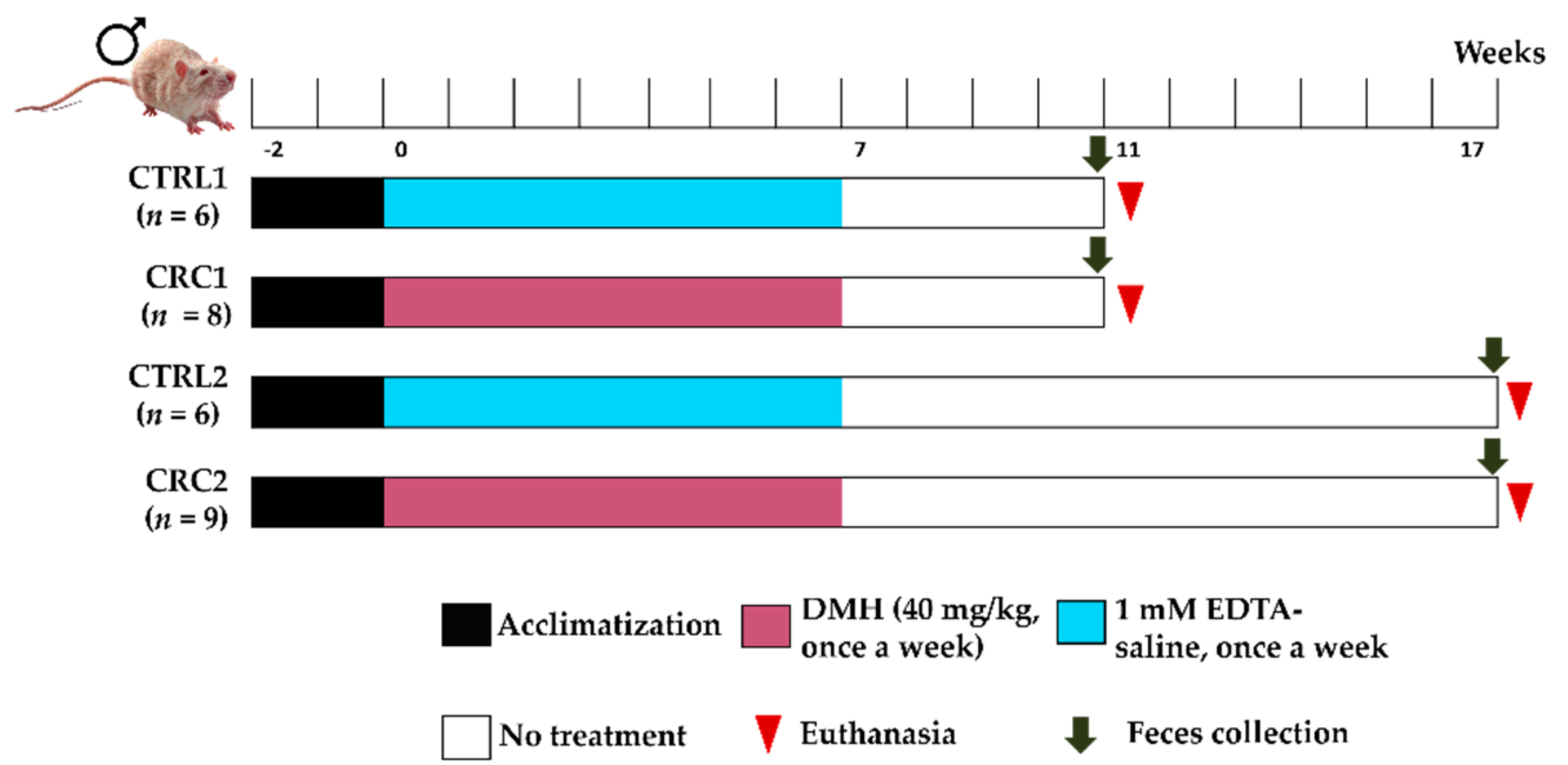
2.2. Experimental Groups and Treatments
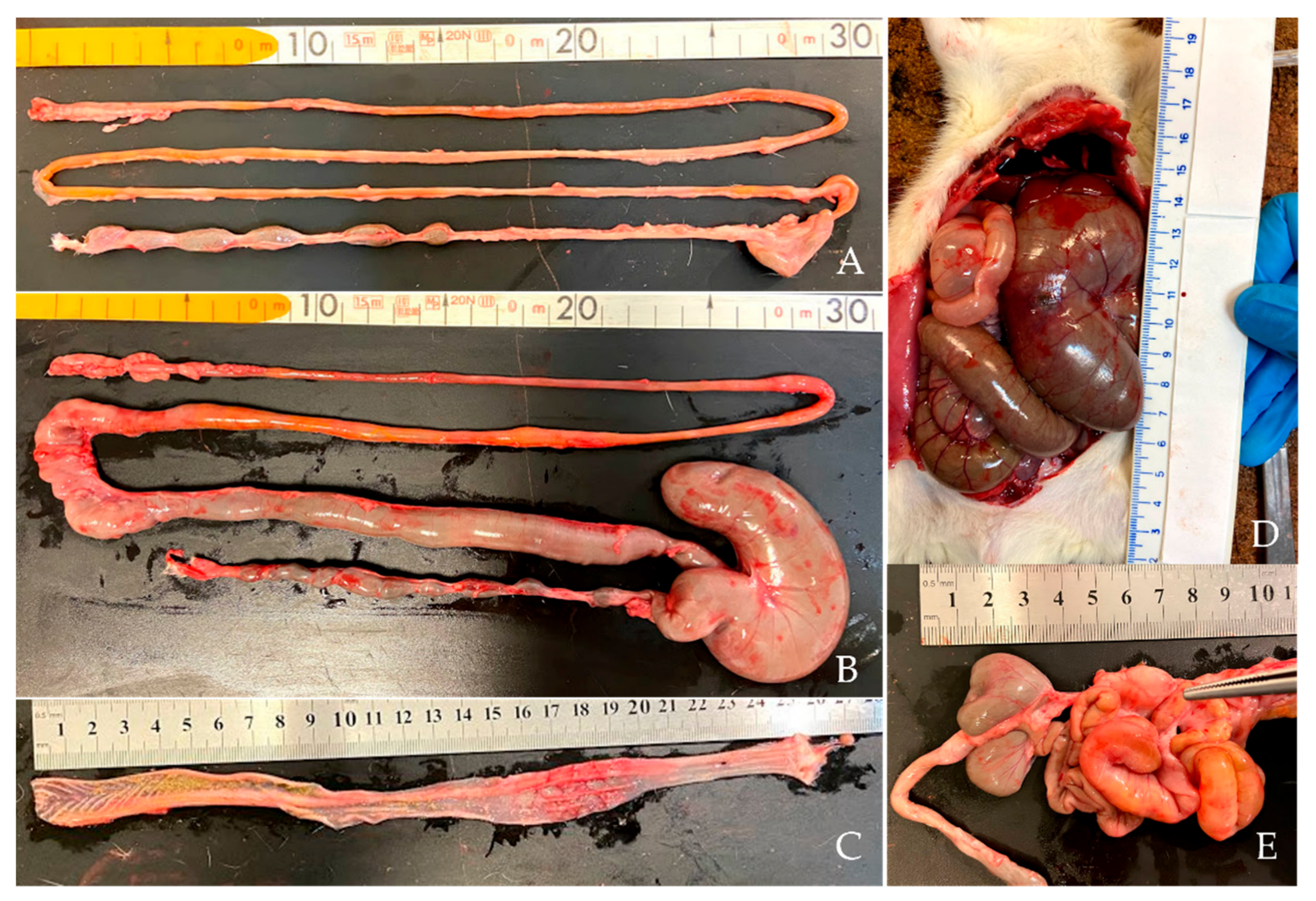
2.3. Samples Collection and Necropsy
2.4. Microhematocrit Analysis
2.5. Serum Biochemical Analysis and Immunoblotting
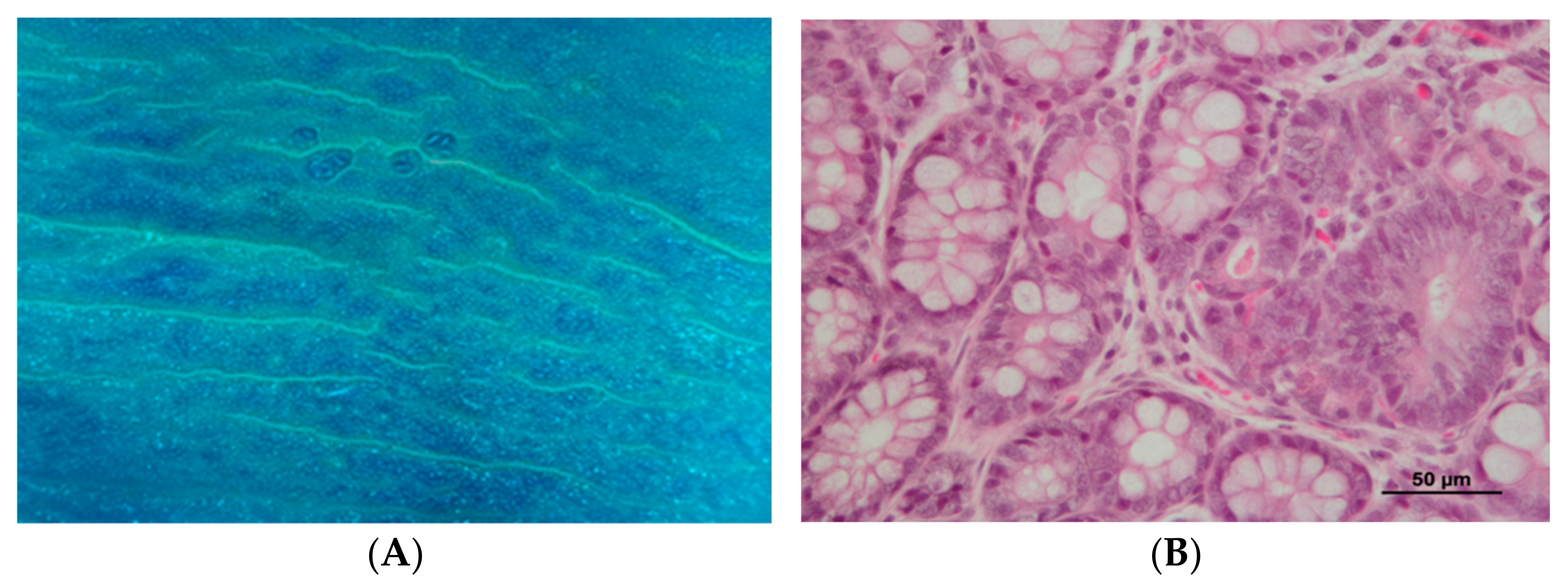
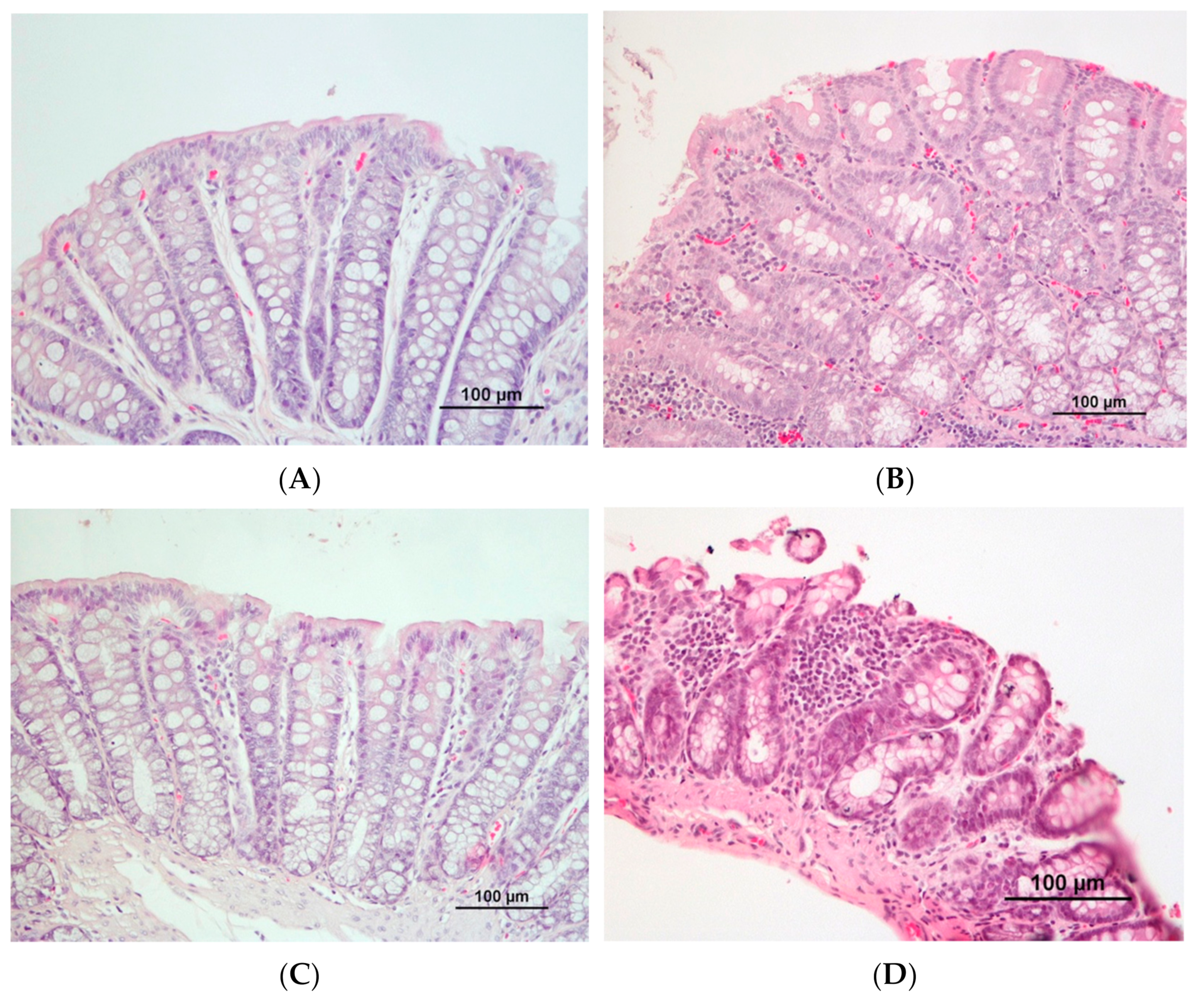
2.6. Histopathological Analysis
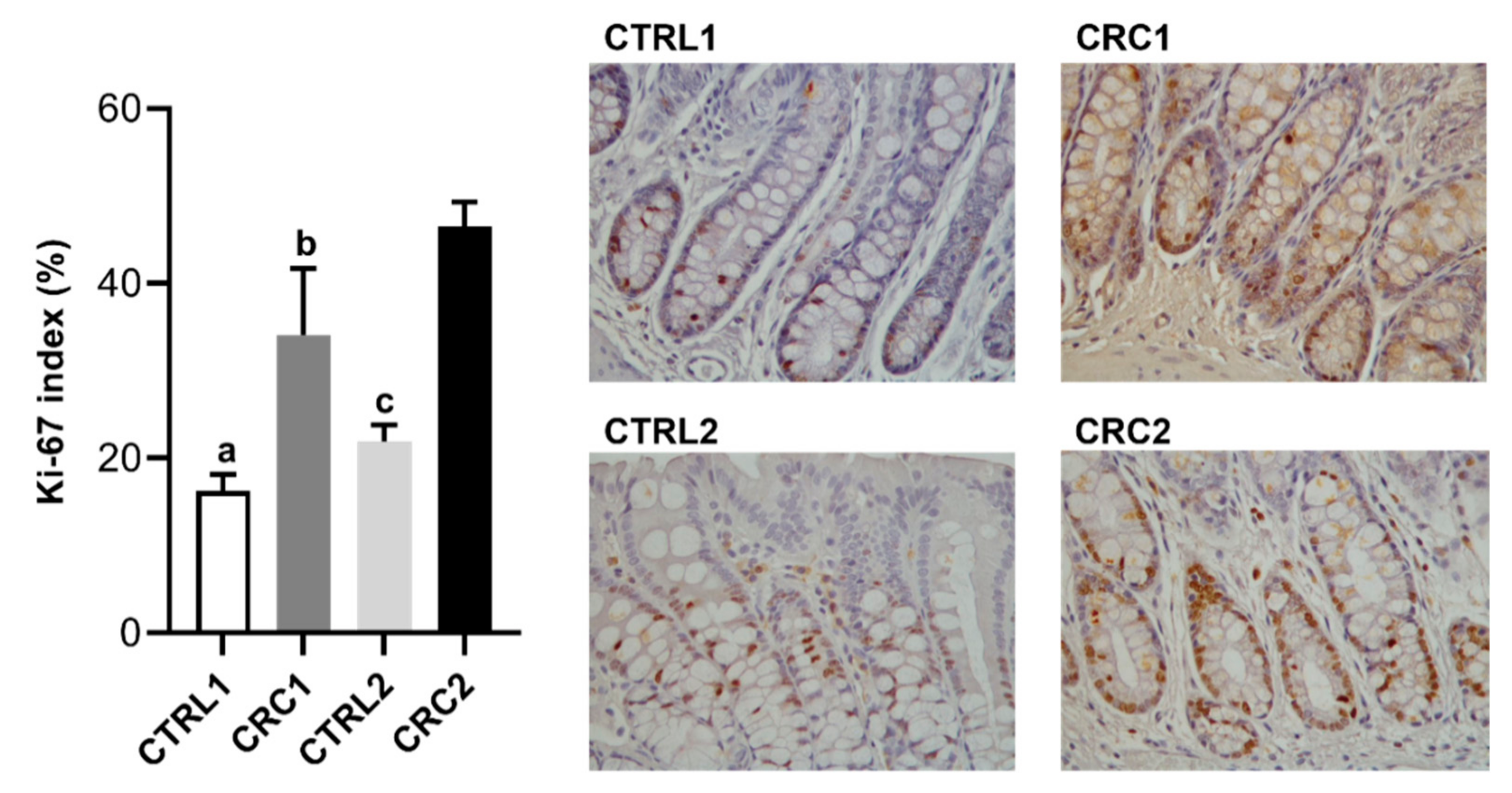
2.7. Ki-67 Immunostaining
2.8. Antioxidant Enzymes Activity
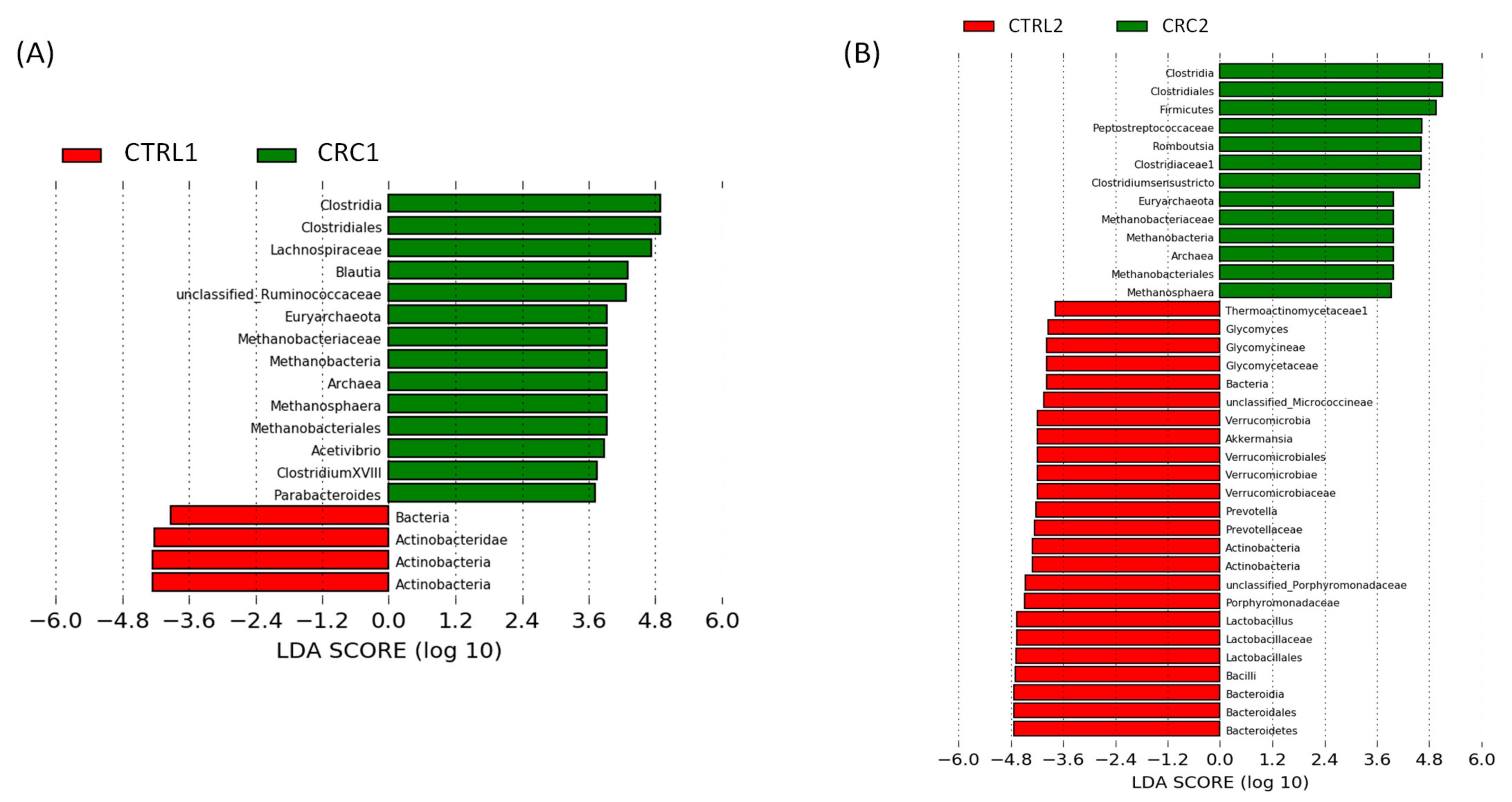
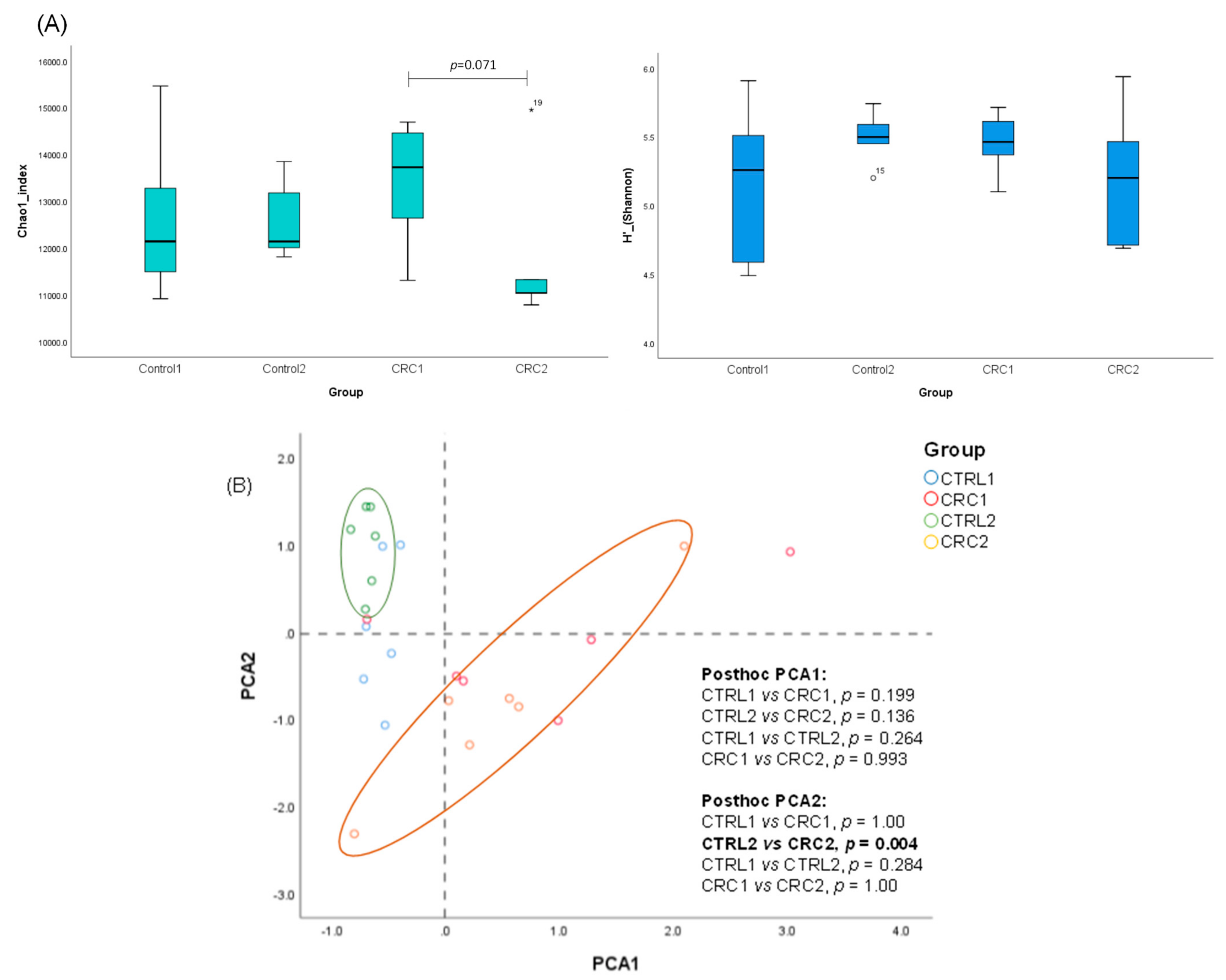
2.9. Gut Microbiota Analysis
2.10. Statistical Analysis
3. Results
3.1. Characterization of Animals’ Systemic Adaptations to CRC Induction
3.2. DMH Induced Preneoplastic Lesions Associated with CRC
3.3. Effect of DMH on the Activity of Colonic Antioxidant Enzymes
3.4. Impact of DMH-Induced CRC on the Rat Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.L.; Fleet, J.C. Animal Models of Colorectal Cancer. Cancer Metastasis Rev. 2013, 32, 39–61. [Google Scholar] [CrossRef] [PubMed]
- Nascimento-Gonçalves, E.; Mendes, B.A.L.; Silva-Reis, R.; Faustino-Rocha, A.I.; Gama, A.; Oliveira, P.A. Animal Models of Colorectal Cancer: From Spontaneous to Genetically Engineered Models and Their Applications. Vet. Sci. 2021, 8, 59. [Google Scholar] [CrossRef]
- Ochiai, M.; Watanabe, M.; Nakanishi, M.; Taguchi, A.; Sugimura, T.; Nakagama, H. Differential Staining of Dysplastic Aberrant Crypt Foci in the Colon Facilitates Prediction of Carcinogenic Potentials of Chemicals in Rats. Cancer Lett. 2005, 220, 67–74. [Google Scholar] [CrossRef]
- Caderni, G.; Femia, A.P.; Giannini, A.; Favuzza, A.; Luceri, C.; Salvadori, M.; Dolara, P. Identification of Mucin-Depleted Foci in the Unsectioned Colon of Azoxymethane-Treated Rats: Correlation with Carcinogenesis. Cancer Res. 2003, 63, 2388–2392. [Google Scholar]
- Bird, R.P. Observation and Quantification of Aberrant Crypts in the Murine Colon Treated with a Colon Carcinogen: Preliminary Findings. Cancer Lett. 1987, 37, 147–151. [Google Scholar] [CrossRef]
- Vodicka, P.; Urbanova, M.; Makovicky, P.; Tomasova, K.; Kroupa, M.; Stetina, R.; Opattova, A.; Kostovcikova, K.; Siskova, A.; Schneiderova, M.; et al. Oxidative Damage in Sporadic Colorectal Cancer: Molecular Mapping of Base Excision Repair Glycosylases in Colorectal Cancer Patients. Int. J. Mol. Sci. 2020, 21, 2473. [Google Scholar] [CrossRef]
- Hamiza, O.O.; Rehman, M.U.; Tahir, M.; Khan, R.; Khan, A.Q.; Lateef, A.; Ali, F.; Sultana, S. Amelioration of 1,2 Dimethylhydrazine (DMH) Induced Colon Oxidative Stress, Inflammation and Tumor Promotion Response by Tannic Acid in Wistar Rats. Asian Pac. J. Cancer Prev. 2012, 13, 4393–4402. [Google Scholar] [CrossRef]
- Zhu, Q.; Jin, Z.; Wu, W.; Gao, R.; Guo, B.; Gao, Z.; Yang, Y.; Qin, H. Analysis of the Intestinal Lumen Microbiota in an Animal Model of Colorectal Cancer. PLoS ONE 2014, 9, e90849. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Vinayagam, R.; Anand, M.A.V.; Isa, N.M.; Ponnaiyan, R. Biochemical and Molecular Aspects of 1,2-Dimethylhydrazine (DMH)-Induced Colon Carcinogenesis: A Review. Toxicol. Res. 2020, 9, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, D.W.; Giardina, C.; Tanaka, T. Mouse Models for the Study of Colon Carcinogenesis. Carcinogenesis 2009, 30, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Juan, M.E.; Lozano-Mena, G.; Sánchez-González, M.; Planas, J.M. Reduction of Preneoplastic Lesions Induced by 1,2-Dimethylhydrazine in Rat Colon by Maslinic Acid, a Pentacyclic Triterpene from Olea Europaea L. Molecules 2019, 24, 1266. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.A.M.; Silva, L.A.G.; Salvadori, D.M.F.; de Camargo, J.L.V.; Montenegro, M.R. Aberrant Crypt Foci and Colon Cancer: Comparison between a Short- and Medium-Term Bioassay for Colon Carcinogenesis Using Dimethylhydrazine in Wistar Rats. Braz. J. Med. Biol. Res. 2002, 35, 351–355. [Google Scholar] [CrossRef]
- Ramos Caetano, B.F.; Baptista Tablas, M.; Ribeiro Romualdo, G.; Marchesan Rodrigues, M.A.; Barbisan, L.F. Early Molecular Events Associated with Liver and Colon Sub-Acute Responses to 1,2-Dimethylhydrazine: Potential Implications on Preneoplastic and Neoplastic Lesion Development. Toxicol. Lett. 2020, 329, 67–79. [Google Scholar] [CrossRef]
- Nolte, T.; Brander-Weber, P.; Dangler, C.; Deschl, U.; Elwell, M.R.; Greaves, P.; Hailey, R.; Leach, M.W.; Pandiri, A.R.; Rogers, A.; et al. Nonproliferative and Proliferative Lesions Ofthe Gastrointestinal Tract, Pancreas AndSalivary Glands of the Rat and Mouse. J. Toxicol. Pathol. 2016, 29, 1S–125S. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, K.; Wang, Z.; Li, N.; Ji, G. Chemopreventive Effect of Dietary Glutamine on Colitis-Associated Colon Tumorigenesis in Mice. Carcinogenesis 2013, 34, 1593–1600. [Google Scholar] [CrossRef]
- Payá, M.; Halliwell, B.; Hoult, J.R.S. Interactions of a Series of Coumarins with Reactive Oxygen Species: Scavenging of Superoxide, Hypochlorous Acid and Hydroxyl Radicals. Biochem. Pharmacol. 1992, 44, 205–214. [Google Scholar] [CrossRef]
- del Río, L.A.; Gómez, M.; López-Gorgé, J. Catalase and Peroxidase Activities, Chlorophyll and Proteins during Storage of Pea Plants of Chilling Temperatures. Rev. Esp. Fisiol. 1977, 33, 143–148. [Google Scholar]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; Cortés-Martín, A.; Loria-Kohen, V.; Ramírez-de-Molina, A.; García-Mantrana, I.; Collado, M.C.; Espín, J.C.; Selma, M.V. Deciphering the Human Gut Microbiome of Urolithin Metabotypes: Association with Enterotypes and Potential Cardiometabolic Health Implications. Mol. Nutr. Food Res. 2019, 63, 1800958. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Team, G. Manipulation of FASTQ Data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Silva-Reis, R.; Faustino-Rocha, A.I.; Gonçalves, M.; Ribeiro, C.C.; Ferreira, T.; Ribeiro-Silva, C.; Gonçalves, L.; Antunes, L.; Venâncio, C.; Ferreira, R.; et al. Refinement of Animal Model of Colorectal Carcinogenesis through the Definition of Novel Humane Endpoints. Animals 2021, 11, 985. [Google Scholar] [CrossRef]
- Wallace, J. Humane Endpoints and Cancer Research. ILAR J. 2000, 41, 87–93. [Google Scholar] [CrossRef]
- Murphy, G.; Cross, A.J.; Dawsey, S.M.; Stanczyk, F.Z.; Kamangar, F.; Weinstein, S.J.; Taylor, P.R.; Männistö, S.; Albanes, D.; Abnet, C.C.; et al. Serum Ghrelin Is Associated with Risk of Colorectal Adenocarcinomas in the ATBC Study. Gut 2018, 67, 1646–1651. [Google Scholar] [CrossRef]
- Loumaye, A.; de Barsy, M.; Nachit, M.; Lause, P.; Frateur, L.; van Maanen, A.; Trefois, P.; Gruson, D.; Thissen, J.P. Role of Activin A and Myostatin in Human Cancer Cachexia. J. Clin. Endocrinol. Metab. 2015, 100, 2030–2038. [Google Scholar] [CrossRef]
- Jrah Harzallah, H.; Grayaa, R.; Kharoubi, W.; Maaloul, A.; Hammami, M.; Mahjoub, T. Thymoquinone, the Nigella Sativa Bioactive Compound, Prevents Circulatory Oxidative Stress Caused by 1,2-Dimethylhydrazine in Erythrocyte during Colon Postinitiation Carcinogenesis. Oxidative Med. Cell. Longev. 2012, 2012, 854065. [Google Scholar] [CrossRef]
- Väyrynen, J.P.; Tuomisto, A.; Väyrynen, S.A.; Klintrup, K.; Karhu, T.; Mäkelä, J.; Herzig, K.H.; Karttunen, T.J.; Mäkinen, M.J. Preoperative Anemia in Colorectal Cancer: Relationships with Tumor Characteristics, Systemic Inflammation, and Survival. Sci. Rep. 2018, 8, 1126. [Google Scholar] [CrossRef]
- Liao, C.; Yu, Y.; Lin, Y.; Hsu, Y.; Chern, Y.; Chiang, J.; You, J.-F. Prognostic Value of the C-Reactive Protein to Albumin Ratio in Colorectal Cancer: An Updated Systematic Review and Meta-Analysis. World J. Surg. Oncol. 2021, 19, 139. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.; Bhadauria, A.S.; Singh, A.K.; Kumar, U.; Rai, A.; Keshari, A.K.; Kumar, P.; Kumar, D.; Maity, B.; Nath, S.; et al. Novel 1,3,4-Thiadiazoles Inhibit Colorectal Cancer via Blockade of IL-6/COX-2 Mediated JAK2/STAT3 Signals as Evidenced through Data-Based Mathematical Modeling. Cytokine 2019, 118, 144–159. [Google Scholar] [CrossRef] [PubMed]
- Jisha, N.; Vysakh, A.; Vijeesh, V.; Anand, P.S.; Latha, M.S. Methanolic Extract of Muntingia Calabura L. Mitigates 1,2-Dimethyl Hydrazine Induced Colon Carcinogenesis in Wistar Rats. Nutr. Cancer 2020, 73, 2363–2375. [Google Scholar] [CrossRef] [PubMed]
- Perše, M.; Cerar, A. Morphological and Molecular Alterations in 1,2 Dimethylhydrazine and Azoxymethane Induced Colon Carcinogenesis in Rats. J. Biomed. Biotechnol. 2011, 2011, 473964. [Google Scholar] [CrossRef] [PubMed]
- Chari, K.Y.; Polu, P.R.; Shenoy, R.R. An Appraisal of Pumpkin Seed Extract in 1, 2-Dimethylhydrazine Induced Colon Cancer in Wistar Rats. J. Toxicol. 2018, 2018, 6086490. [Google Scholar] [CrossRef]
- Jia, X.D.; Han, C. Chemoprevention of Tea on Colorectal Cancer Induced by Dimethylhydrazine in Wistar Rats. World J. Gastroenterol. 2000, 6, 699–703. [Google Scholar] [CrossRef]
- Rehman, M.U.; Rashid, S.; Arafah, A.; Qamar, W.; Alsaffar, R.M.; Ahmad, A.; Almatroudi, N.M.; Alqahtani, S.M.A.; Rashid, S.M.; Ahmad, S.B. Piperine Regulates Nrf-2/Keap-1 Signalling and Exhibits Anticancer Effect in Experimental Colon Carcinogenesis in Wistar Rats. Biology 2020, 9, 302. [Google Scholar] [CrossRef]
- Tanaka, T. Colorectal Carcinogenesis: Review of Human and Experimental Animal Studies. J. Carcinog. 2009, 8, 5. [Google Scholar] [CrossRef]
- LI, L.T.; JIANG, G.; CHEN, Q.; ZHENG, J.N. Ki67 Is a Promising Molecular Target in the Diagnosis of Cancer (Review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef]
- Ganaie, M.A.; Al Saeedan, A.; Madhkali, H.; Jan, B.L.; Khatlani, T.; Sheikh, I.A.; Rehman, M.U.; Wani, K. Chemopreventive Efficacy Zingerone (4-[4-Hydroxy-3-Methylphenyl] Butan-2-One) in Experimental Colon Carcinogenesis in Wistar Rats. Environ. Toxicol. 2019, 34, 610–625. [Google Scholar] [CrossRef]
- Alazzouni, A.S.; Dkhil, M.A.; Gadelmawla, M.H.A.; Gabri, M.S.; Farag, A.H.; Hassan, B.N. Ferulic Acid as Anticarcinogenic Agent against 1,2-Dimethylhydrazine Induced Colon Cancer in Rats. J. King Saud Univ.—Sci. 2021, 33, 101354. [Google Scholar] [CrossRef]
- Martin, M.S.; Martin, F.; Justrabo, E.; Knopf, J.F.; Bastien, H.; Knobel, S. Induction of colonic cancers in rats through a single injection of 1,2 dimethylhydrazine. Biol. Gastro-Enterol. 1974, 7, 37–42. [Google Scholar]
- Teague, C.A.; Gavin, J.B.; Herdson, P.B. The Response of Three Inbred Strains of Rat to the Carcinogen 1,2-Dimethylhydrazine. Pathology 1981, 13, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Sunter, J.P.; Senior, P. v Induction of Renal Tumours in Rats by the Administration of 1,2 Dimethylhydrazine. J. Pathol. 1983, 140, 69–76. [Google Scholar] [CrossRef]
- Jiminez, J.A.; Uwiera, T.C.; Douglas Inglis, G.; Uwiera, R.R.E. Animal Models to Study Acute and Chronic Intestinal Inflammation in Mammals. Gut Pathog. 2015, 7, 29. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Kraus, S.; Arber, N. Inflammation and Colorectal Cancer. Curr. Opin. Pharmacol. 2009, 9, 405–410. [Google Scholar] [CrossRef]
- Kurutas, E.B. The Importance of Antioxidants Which Play the Role in Cellular Response against Oxidative/Nitrosative Stress: Current State. Nutr. J. 2016, 15, 71. [Google Scholar] [CrossRef]
- Kuzma, J.; Chmelař, D.; Hájek, M.; Lochmanová, A.; Čižnár, I.; Rozložník, M.; Klugar, M. The Role of Intestinal Microbiota in the Pathogenesis of Colorectal Carcinoma. Folia Microbiol. 2020, 65, 17–24. [Google Scholar] [CrossRef]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 3100. [Google Scholar] [CrossRef]
- Reddy, B.S.; Narisawa, T.; Wright, P.; Vukusich, D.; Weisburger, J.H.; Wynder, E.L. Colon Carcinogenesis with Azoxymethane and Dimethylhydrazine in Germ-Free Rats. Cancer Res. 1975, 35, 287–290. [Google Scholar] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a Bacterial World, a New Imperative for the Life Sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Loke, Y.L.; Chew, M.T.; Ngeow, Y.F.; Lim, W.W.D.; Peh, S.C. Colon Carcinogenesis: The Interplay Between Diet and Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 733. [Google Scholar] [CrossRef] [PubMed]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.Y.; Yu, J. Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology 2020, 159, 1459–1470.e5. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.; Abe, F.; Osawa, R. Age-Related Changes in Gut Microbiota Composition from Newborn to Centenarian: A Cross-Sectional Study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Blais Lecours, P.; Marsolais, D.; Cormier, Y.; Berberi, M.; Haché, C.; Bourdages, R.; Duchaine, C. Increased Prevalence of Methanosphaera Stadtmanae in Inflammatory Bowel Diseases. PLoS ONE 2014, 9, e87734. [Google Scholar] [CrossRef]
- Hirano, A.; Umeno, J.; Okamoto, Y.; Shibata, H.; Ogura, Y.; Moriyama, T.; Torisu, T.; Fujioka, S.; Fuyuno, Y.; Kawarabayasi, Y.; et al. Comparison of the Microbial Community Structure between Inflamed and Non-Inflamed Sites in Patients with Ulcerative Colitis. J. Gastroenterol. Hepatol. 2018, 33, 1590–1597. [Google Scholar] [CrossRef]
- El-Baz, A.M.; Khodir, A.E.; Adel El-Sokkary, M.M.; Shata, A. The Protective Effect of Lactobacillus versus 5-Aminosalicylic Acid in Ulcerative Colitis Model by Modulation of Gut Microbiota and Nrf2/Ho-1 Pathway. Life Sci. 2020, 256, 117927. [Google Scholar] [CrossRef]
| Group | Relative Weight (g g−1) | ||
|---|---|---|---|
| Small Intestine | Cecum | Colon + Rectum | |
| CTRL1 (n = 6) | 2.10 ± 0.40 | 0.42 ± 0.05 | 0.86 ± 0.37 |
| CRC1 (n = 6) | 3.54 ± 1.41 a,b | 1.04 ± 0.70 | 0.85 ± 0.12 |
| CTRL2 (n = 6) | 2.27 ± 0.48 | 0.45 ± 0.04 | 0.77 ± 0.05 |
| CRC2 (n = 6) | 3.48 ± 1.48 a,b | 1.30 ± 1.00 c,d | 1.01 ± 1.48 |
| Group | Microhematocrit (%) |
|---|---|
| CTRL1 (n = 6) | 50.2 ± 2.4 |
| CRC1 (n = 6) | 50.4 ± 1.8 a |
| CTRL2 (n = 6) | 48.8 ± 1.5 |
| CRC2 (n = 6) | 45.5 ± 2.6 b |
| Serum Markers | CTRL1 (n = 6) | CRC1 (n = 6) | CTRL2 (n = 6) | CRC2 (n = 6) |
|---|---|---|---|---|
| Albumin (g/L) | 18.72 ± 4.07 | 29.25 ± 6.40 | 21.25 ± 8.64 | 27.62 ± 5.44 |
| Total protein (g/L) | 34.45 ± 5.26 | 43.28 ± 9.90 | 35.48 ± 10.04 | 39.35 ± 12.18 |
| Cholesterol (mg/dL) | 124.97 ± 11.15 | 131.92 ± 10.62 | 146.68 ± 55.07 | 135.65 ± 34.27 |
| Glucose (mg/dL) | 252.15 ± 44.52 | 270.52 ± 76.91 | 267.68 ± 68.75 | 261.42 ± 67.45 |
| ALT (U/L) | 40.75 ± 7.72 | 44.70 ± 9.36 | 39.57 ± 9.93 | 40.10 ± 10.01 |
| IL-6 (AU) | 6.45 ± 2.39 | 5.66 ± 1.85 b | 13.36 ± 2.92 a | 8.25 ± 4.64 |
| CRP (AU) | 9.70 ± 2.27 | 7.48 ± 1.76 | 16.46 ± 4.35 | 12.28 ± 7.33 |
| Myostatin (AU) | 8.84 ± 3.78 | 7.47 ± 2.68 | 18.35 ± 4.82 | 11.87 ± 6.85 |
| Ghrelin (AU) | 9.80 ± 4.20 | 8.12 ± 2.00 c | 21.41 ± 5.61 a | 14.16 ± 6.92 |
| Group | Animal | Small Intestine | Cecum |
|---|---|---|---|
| CTRL1 (n = 6) | 1 | 1 | 2 |
| 2 | 1 | 1 | |
| 3 | 2 | 2 | |
| 4 | 2 | 2 | |
| 5 | 1 | 1 | |
| 6 | 1 | 2 | |
| Mean ± SD | 1.33 ± 0.47 | 1.67 ± 0.47 | |
| CRC 1 (n = 6) | 1 | 2 | 1 |
| 2 | 2 | 2 | |
| 3 | 2 | 2 | |
| 4 | 2 | 2 | |
| 5 | 2 | 2 | |
| 6 | 2 | 2 | |
| Mean ± SD | 2.00 ± 0.00 a | 1.83 ± 0.37 | |
| CTRL2 (n = 6) | 1 | 2 | 2 |
| 2 | 1 | 2 | |
| 3 | 1 | 2 | |
| 4 | 2 | 2 | |
| 5 | 1 | 2 | |
| 6 | 2 | 1 | |
| Mean ± SD | 1.50 ± 0.50 | 1.83 ± 0.37 | |
| CRC2 (n = 6) | 1 | 2 | 2 |
| 2 | 2 | 2 | |
| 3 | 2 | 2 | |
| 4 | 2 | 2 | |
| 5 | 2 | 2 | |
| 6 | 2 | 2 | |
| Mean ± SD | 2.00 ± 0.00 a | 2.00 ± 0.00 |
| Group | Normal | Mild to Moderate Dysplasia | Severe Dysplasia | Adenoma |
|---|---|---|---|---|
| CTRL1 (n = 6) | 6/6 (100%) | --- | --- | --- |
| CRC1 (n = 6) | 3/6 (50%) | 3/6 (50%) | --- | --- |
| CTRL2 (n = 6) | 6/6 (100%) | --- | --- | --- |
| CRC2 (n = 6) | 2/6 (33.3%) | 2/6 (33.3%) | 1/6 (16.7%) | 1/6 (16.7%) |
| Group | Animal | Severity | Thickness | Epithelial Damage | Extension |
|---|---|---|---|---|---|
| CTRL1 (n = 6) | 1 | 1 | 1 | 0 | 1 |
| 2 | 1 | 1 | 0 | 1 | |
| 3 | 1 | 1 | 1 | 2 | |
| 4 | 1 | 1 | 0 | 1 | |
| 5 | 1 | 1 | 1 | 2 | |
| 6 | 1 | 1 | 0 | 1 | |
| Mean ± SD | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.33 ± 0.47 | 1.33 ± 0.47 | |
| CRC 1 (n = 6) | 1 | 1 | 1 | 0 | 2 |
| 2 | 2 | 1 | 1 | 2 | |
| 3 | 2 | 1 | 2 | 3 | |
| 4 | 2 | 2 | 1 | 2 | |
| 5 | 2 | 2 | 1 | 2 | |
| 6 | 2 | 2 | 2 | 2 | |
| Mean ± SD | 1.83 ± 0.37 a | 1.50 ± 0.50 | 1.17 ± 0.69 a | 2.17 ± 0.37 a | |
| CTRL2 (n = 6) | 1 | 1 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 | 1 | |
| 3 | 1 | 1 | 1 | 1 | |
| 4 | 1 | 1 | 1 | 2 | |
| 5 | 2 | 1 | 2 | 2 | |
| 6 | 2 | 1 | 2 | 2 | |
| Mean ± SD | 1.33 ± 0.47 | 1.00 ± 0.00 | 1.33 ± 0.47 b | 1.50 ± 0.50 | |
| CRC2 (n = 6) | 1 | 2 | 2 | 2 | 3 |
| 2 | 2 | 2 | 2 | 2 | |
| 3 | 1 | 1 | 1 | 2 | |
| 4 | 2 | 1 | 1 | 2 | |
| 5 | 2 | 2 | 3 | 2 | |
| 6 | 2 | 1 | 1 | 2 | |
| Mean ± SD | 1.83 ±0.37 a | 1.50 ± 0.50 | 1.67 ± 0.75 c | 2.17 ± 0.37 a |
| Group | SOD (U/min/mg of Protein) | CAT (H2O2/min/mg of Protein) |
|---|---|---|
| CTRL1 (n = 6) | 19.42 ± 2.39 | 51.10 ± 12.65 |
| CRC1 (n = 6) | 16.12 ± 1.43 | 37.77 ± 16.61 |
| CTRL2 (n = 6) | 15.27 ± 3.73 | 40.50 ± 1.43 |
| CRC2 (n = 6) | 10.91 ± 2.11 | 34.94 ± 3.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Reis, R.; Castro-Ribeiro, C.; Gonçalves, M.; Ferreira, T.; Pires, M.J.; Iglesias-Aguirre, C.E.; Cortés-Martín, A.; Selma, M.V.; Espín, J.C.; Nascimento-Gonçalves, E.; et al. An Integrative Approach to Characterize the Early Phases of Dimethylhydrazine-Induced Colorectal Carcinogenesis in the Rat. Biomedicines 2022, 10, 409. https://doi.org/10.3390/biomedicines10020409
Silva-Reis R, Castro-Ribeiro C, Gonçalves M, Ferreira T, Pires MJ, Iglesias-Aguirre CE, Cortés-Martín A, Selma MV, Espín JC, Nascimento-Gonçalves E, et al. An Integrative Approach to Characterize the Early Phases of Dimethylhydrazine-Induced Colorectal Carcinogenesis in the Rat. Biomedicines. 2022; 10(2):409. https://doi.org/10.3390/biomedicines10020409
Chicago/Turabian StyleSilva-Reis, Rita, Catarina Castro-Ribeiro, Mariana Gonçalves, Tiago Ferreira, Maria João Pires, Carlos E. Iglesias-Aguirre, Adrián Cortés-Martín, María V. Selma, Juan Carlos Espín, Elisabete Nascimento-Gonçalves, and et al. 2022. "An Integrative Approach to Characterize the Early Phases of Dimethylhydrazine-Induced Colorectal Carcinogenesis in the Rat" Biomedicines 10, no. 2: 409. https://doi.org/10.3390/biomedicines10020409
APA StyleSilva-Reis, R., Castro-Ribeiro, C., Gonçalves, M., Ferreira, T., Pires, M. J., Iglesias-Aguirre, C. E., Cortés-Martín, A., Selma, M. V., Espín, J. C., Nascimento-Gonçalves, E., Moreira-Pais, A., Neuparth, M. J., Peixoto, F., Rosa, E., Gama, A., Ferreira, R., Oliveira, P. A., & Faustino-Rocha, A. I. (2022). An Integrative Approach to Characterize the Early Phases of Dimethylhydrazine-Induced Colorectal Carcinogenesis in the Rat. Biomedicines, 10(2), 409. https://doi.org/10.3390/biomedicines10020409












