Highly Sensitive HBsAg, Anti-HBc and Anti HBsAg Titres in Early Diagnosis of HBV Reactivation in Anti-HBc-Positive Onco-Haematological Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. HBV Laboratory Evaluation
2.2.1. Classical HBV Markers
2.2.2. Innovative HBV Markers
HS-HBs
Quantification of Anti-HBc Titer
Quantification of Serum HBV-RNA
2.3. Population-Based Sequencing of HBV Reverse Transcriptase
2.4. Statistical Methods
3. Results
3.1. Study Population
3.2. Occurrence of HBV-R
3.3. Predictive Role of Serological HBV Markers in the Diagnosis of HBV-R
3.4. Outcome of Patients Experiencing HBV-R
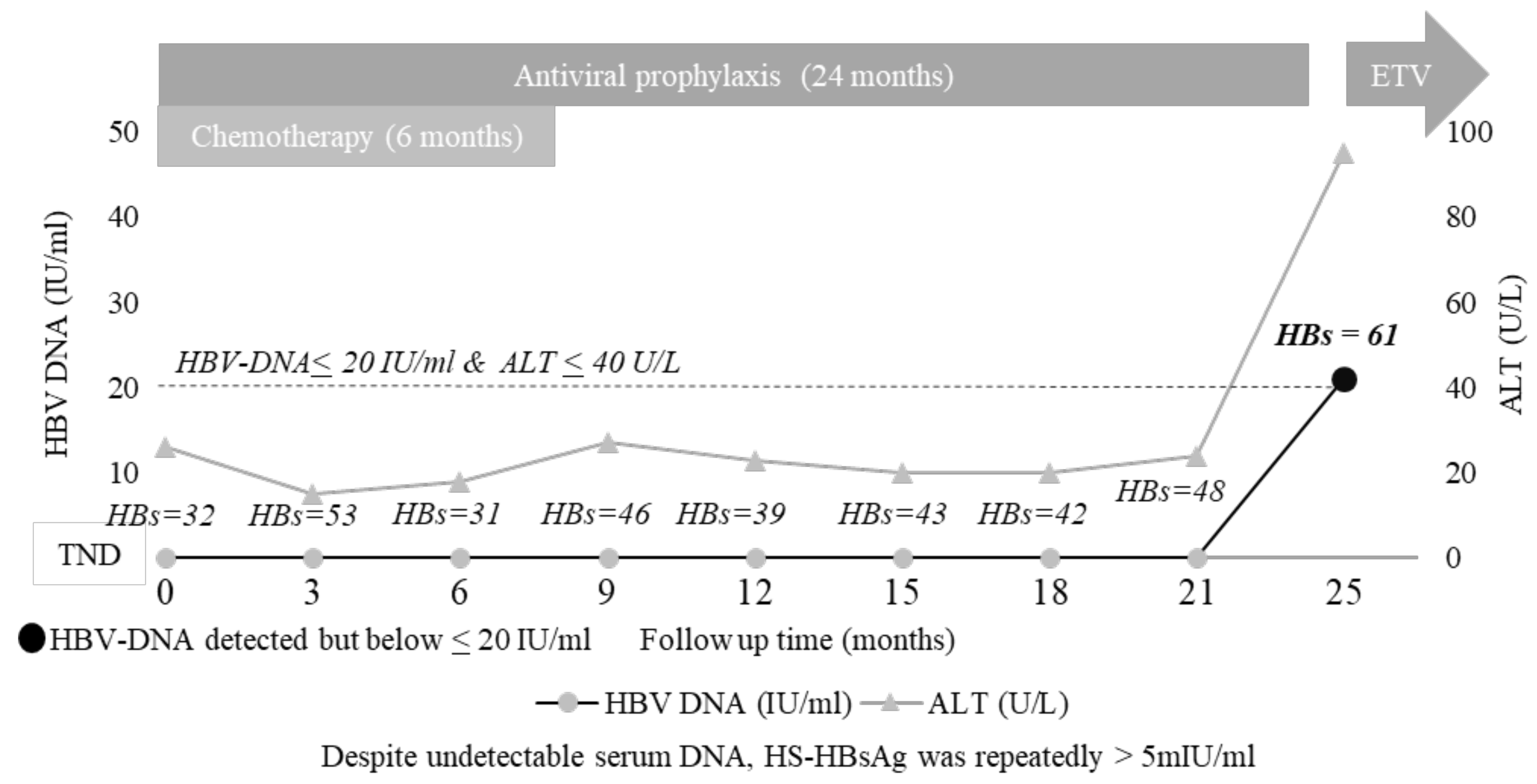
3.5. The Added Value of HS HBsAg Quantification: A Case Report
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sagnelli, C.; Pisaturo, M.; Calò, F.; Martini, S.; Sagnelli, E.; Coppola, N. Reactivation of hepatitis B virus infection in patients with hemo-lymphoproliferative diseases, and its prevention. World J. Gastroenterol. 2019, 14, 3299–3312. [Google Scholar] [CrossRef] [PubMed]
- Myint, A.; Tong, M.J.; Beaven, S.W. Reactivation of Hepatitis B Virus: A Review of Clinical Guidelines. Clin. Liver Dis. 2020, 15, 162–167. [Google Scholar] [CrossRef]
- Terrault, N.A.; Lok, A.S.F.; McMahon, B.J.; Chang, K.-M.; Hwang, J.P.; Jonas, M.M.; Brown, R.S.B., Jr.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef] [PubMed]
- Sarin, S.K.; Kumar, M.P.; Lau, G.K.; Abbas, Z.; Chan, H.L.Y.; Chen, C.J.; Chen, D.-S.; Chen, H.L.; Chien, R.N.; Dokmeci, A.; et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.R.; Beavers, K.L.; Hammond, S.; Lim, J.K.; Falck-Ytter, Y.T. American Gastroenterological Association Institute Guideline on the Prevention and Treatment of Hepatitis B Virus Reactivation During Immunosuppressive Drug Therapy. Gastroenterology 2015, 148, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerva, C.; Colagrossi, L.; Maffongelli, G.; Salpini, R.; Di Carlo, D.; Malagnino, V.; Battisti, A.; Ricciardi, A.; Pollicita, M.; Bianchi, A.; et al. Persistent risk of HBV reactivation despite extensive lamivudine prophylaxis in haematopoietic stem cell transplant recipients who are anti-HBc-positive or HBV-negative recipients with an anti-HBc-positive donor. Clin. Microbiol. Infect. 2016, 22, 946.e1–946.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, T.; Tanaka, Y. Novel biomarkers for the management of chronic hepatitis B. Clin. Mol. Hepatol. 2020, 26, 261–279. [Google Scholar] [CrossRef]
- Charre, C.; Levrero, M.; Zoulim, F.; Scholtès, C. Non-invasive biomarkers for chronic hepatitis B virus infection management. Antivir. Res. 2019, 169, 104553. [Google Scholar] [CrossRef]
- Caviglia, G.P.; Abate, M.L.; Tandoi, F.; Ciancio, A.; Amoroso, A.; Salizzoni, M.; Saracco, G.M.; Rizzetto, M.; Romagnoli, R.; Smedile, A. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: A new tool to detect occult infection. J. Hepatol. 2018, 69, 301–307. [Google Scholar] [CrossRef]
- Salpini, R.; Malagnino, V.; Piermatteo, L.; Mulas, T.; Alkhatib, M.; Scutari, R.; Teti, E.; Cerva, C.; La Rosa, K.Y.; Brugneti, M.; et al. Cryptic HBV replicative activity is frequently revealed in anti-HBC-positive/HBsag-negative patients with HIV infection by highly sensitive molecular assays, and can be predicted by integrating classical and novel serological HBV markers. Microorganisms 2020, 8, 1819. [Google Scholar] [CrossRef]
- Anderson, M.; Gersch, J.; Luk, K.-C.; Dawson, G.; Carey, I.; Agarwal, K.; Shah, P.; Dusheiko, G.; Lau, D.; Cloherty, G. Circulating Pregenomic Hepatitis B Virus RNA Is Primarily Full-length in Chronic Hepatitis B Patients Undergoing Nucleos(t)ide Analogue Therapy. Clin. Infect. Dis. 2021, 72, 2029–2031. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, T.; Huang, X.; Kumar, G.R.; Chen, X.; Zeng, Z.; Zhang, R.; Chen, R.; Li, T.; Zhang, T.; et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 2016, 65, 700–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinkai, N.; Kusumoto, S.; Murakami, S.; Ogawa, S.; Ri, M.; Matsui, T.; Tamori, A.; Toyoda, H.; Ishida, T.; Iida, S.; et al. Novel monitoring of hepatitis B reactivation based on ultra-high sensitive hepatitis B surface antigen assay. Liver Int. 2017, 37, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Piermatteo, L.; Scutari, R.; Chirichiello, R.; Alkhatib, M.; Malagnino, V.; Bertoli, A.; Iapadre, N.; Ciotti, M.; Sarmati, L.; Andreoni, M.; et al. Droplet digital PCR assay as an innovative and promising highly sensitive assay to unveil residual and cryptic HBV replication in peripheral compartment. Methods, 2021; in press. [Google Scholar] [CrossRef] [PubMed]
- Lampertico, P.; Agarwal, K.; Berg, T.; Buti, M.; Janssen, H.L.A.; Papatheodoridis, G.V.; Zoulim, F.; Tacke, F. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, W.-K.; Chan, T.S.-Y.; Hwang, Y.-Y.; Wong, D.K.-H.; Fung, J.; Liu, K.S.-H.; Gill, H.; Lam, Y.-F.; Lau, E.; Cheung, K.-S.; et al. Hepatitis B reactivation in occult viral carriers undergoing hematopoietic stem cell transplantation: A prospective study. Hepatology 2017, 65, 1451–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, E.K.; Gersch, J.; McNamara, A.; Luk, K.-C.; Holzmayer, V.; De Medina, M.; Schiff, E.; Kuhns, M.; Cloherty, G.A. Hepatitis B Virus Serum DNA andRNA Levels in Nucleos(t)ide Analog-Treated or Untreated Patients During Chronic and Acute Infection. Hepatology 2018, 68, 2106–2117. [Google Scholar] [CrossRef] [Green Version]
- Salpini, R.; Alteri, C.; Cento, V.; Pollicita, M.; Micheli, V.; Gubertini, G.; De Sanctis, G.; Visca, M.; Romano, S.; Sarrecchia, C.; et al. Snapshot on drug-resistance rate and profiles in patients with chronic hepatitis B receiving nucleos(t)ide analogues in clinical practice. J. Med. Virol. 2013, 85, 996–1004. [Google Scholar] [CrossRef]
- Yang, H.-C.; Tsou, H.-H.; Pei, S.-N.; Chang, C.-S.; Chen, J.-H.; Yao, M.; Lin, S.-J.; Lin, J.; Yuan, Q.; Xia, N.; et al. Quantification of HBV core antibodies may help predict HBV reactivation in patients with lymphoma and resolved HBV infection. J. Hepatol. 2018, 69, 286–292. [Google Scholar] [CrossRef]
- Kusumoto, S.; Tanaka, Y.; Suzuki, R.; Watanabe, T.; Nakata, M.; Sakai, R.; Fukushima, N.; Fukushima, T.; Moriuchi, Y.; Itoh, K.; et al. Ultra-high sensitivity HBsAg assay can diagnose HBV reactivation following rituximab-based therapy in patients with lymphoma. J. Hepatol. 2020, 73, 285–293. [Google Scholar] [CrossRef]
- Yang, D.; Hu, T.; Wu, X.; Li, K.; Zhong, Q.; Liu, W. Droplet-digital polymerase chain reaction for detection of clinical hepatitis B virus DNA samples. J. Med. Virol. 2018, 90, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, Y.; Li, G.; Shen, C.; Meng, Z.; Zheng, J.; Jia, Y.; Chen, S.; Zhang, X.; Zhu, M.; et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. 2018, 68, 16–24. [Google Scholar] [CrossRef] [PubMed]


| Patients’ Characteristics | N = 107 |
|---|---|
| Male/Female, N (%) | 61/46 (57/43) |
| Age (years), median (IQR) | 66 (58–77) |
| Italian origin, N (%) Duration of monitoring, median (IQR) months | 95 (88.8) |
| 44 (31–56) | |
| Oncohaematological Diseases, N (%) | |
| Non-Hodgkin Lymphoma (NHL) | 39 (36.5) |
| Acute Myeloid Leukaemia (AML) | 17 (15.9) |
| Multiple Myeloma (MM) | 15 (14.0) |
| Chronic Lymphocytic Leukaemia (CLL) | 13 (12.1) |
| Hodgkin Lymphoma (HL) | 6 (5.6) |
| Acute Lymphocytic Leukaemia (ALL) | 5 (4.7) |
| Other Diseases a | 12 (11.2) |
| Immunosuppressive Regimens, N (%) | |
| Chemotherapy + Rituximab | 42 (39.2) |
| Allogenic HSCT | 32 (29.9) |
| Autologous HSCT | 8 (7.5) |
| Other Chemotherapies | 25 (23.4) |
| HBV Serological Profiles | |
| Anti-HBc positive anti-HBs negative, N (%) | 22 (20.7%) |
| Anti-HBc positive anti-HBs positive, N (%) | 85 (79.3%) |
| Anti-HBs titre, median (IQR) mIU/mL | 152 (47–976) |
| Antiviral Prophylaxis | |
| Use of antiviral prophylaxis, N (%) | 107 (100) |
| Patient still in antiviral prophylaxis | 39 (36) |
| Patient who interrupted antiviral prophylaxis | 61 (57) |
| Patients’ Characteristics | N = 17 |
|---|---|
| Serum HBV-DNA, median (IQR) IU/mL | 44 (27–40,509) |
| Serum ALT > UNL, N (%) | 7 (44) |
| - Serum ALT, median (IQR) U/L | 81 (49–541) |
| HBV Serological Profiles at HBV-R | |
| HBsAg positive, N (%): | 4 (23.5) |
| HBsAg negative, N (%) | 13 (76.5) |
| Anti-HBs positive, N (%): | 8 (47) |
| - Anti-HBs titre, range mIU/mL | 13–505 |
| Immunosuppressive Regimen, N (%) | |
| Chemotherapy + Rituximab 3 (17.6) | |
| Allogeneic HSCT 8 (47.2) | |
| Autologous HSCT 3 (17.6) | |
| Other Chemotherapies 3 (17.6) | |
| HBV reactivation occurrence | |
| - During prophylaxis, N (%) | 6 (35.3) |
| - After prophylaxis completion, N (%) | 11 (64.7) |
| Months after prophylaxis completion, median (IQR) | 4 (2–13) |
| Variables a | Univariable Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| Crude OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value | |
| Gender (male vs. female) | 2.0 (0.7–6.2) | 0.224 | - | - |
| Age (for 1 year increase) | 1.0 (1.0–1.1) | 0.101 | 1.1 (1.0–1.1) | 0.435 |
| HSCT vs. RTX and other chemotherapies | 3.9 (1.3–11.5) | 0.015 | 3.0 (0.5–17.2) | 0.018 |
| Combination of anti-HBc > 3 COI + anti-HBs < 50 mIU/mL | 9.1 (2.7–30.2) | <0.001 | 7.2 (1.4–39.2) | 0.020 |
| Detection to HS-HBs and/or to serum HBV-DNA < 20 IU/mL | 13.8 (3.6–52.6) | <0.001 | 5.3 (1.0–27.8) | 0.049 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerva, C.; Salpini, R.; Alkhatib, M.; Malagnino, V.; Piermatteo, L.; Battisti, A.; Bertoli, A.; Gersch, J.; Holzmayer, V.; Kuhns, M.; et al. Highly Sensitive HBsAg, Anti-HBc and Anti HBsAg Titres in Early Diagnosis of HBV Reactivation in Anti-HBc-Positive Onco-Haematological Patients. Biomedicines 2022, 10, 443. https://doi.org/10.3390/biomedicines10020443
Cerva C, Salpini R, Alkhatib M, Malagnino V, Piermatteo L, Battisti A, Bertoli A, Gersch J, Holzmayer V, Kuhns M, et al. Highly Sensitive HBsAg, Anti-HBc and Anti HBsAg Titres in Early Diagnosis of HBV Reactivation in Anti-HBc-Positive Onco-Haematological Patients. Biomedicines. 2022; 10(2):443. https://doi.org/10.3390/biomedicines10020443
Chicago/Turabian StyleCerva, Carlotta, Romina Salpini, Mohammad Alkhatib, Vincenzo Malagnino, Lorenzo Piermatteo, Arianna Battisti, Ada Bertoli, Jeff Gersch, Vera Holzmayer, Mary Kuhns, and et al. 2022. "Highly Sensitive HBsAg, Anti-HBc and Anti HBsAg Titres in Early Diagnosis of HBV Reactivation in Anti-HBc-Positive Onco-Haematological Patients" Biomedicines 10, no. 2: 443. https://doi.org/10.3390/biomedicines10020443
APA StyleCerva, C., Salpini, R., Alkhatib, M., Malagnino, V., Piermatteo, L., Battisti, A., Bertoli, A., Gersch, J., Holzmayer, V., Kuhns, M., Cloherty, G., Ferrari, L., Laura, C., Teti, E., Cantonetti, M., Arcese, W., Ceccherini-Silberstein, F., Perno, C.-F., Andreoni, M., ... Sarmati, L. (2022). Highly Sensitive HBsAg, Anti-HBc and Anti HBsAg Titres in Early Diagnosis of HBV Reactivation in Anti-HBc-Positive Onco-Haematological Patients. Biomedicines, 10(2), 443. https://doi.org/10.3390/biomedicines10020443








