Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Preparation
2.2. Data Preprocessing and Augmentation
2.3. Endocardial Border Detection and Left Ventricular Functional Assessment
2.4. Metrics
2.4.1. Segmentation Performance
2.4.2. LVEF
2.4.3. GLS and GCS
3. Results
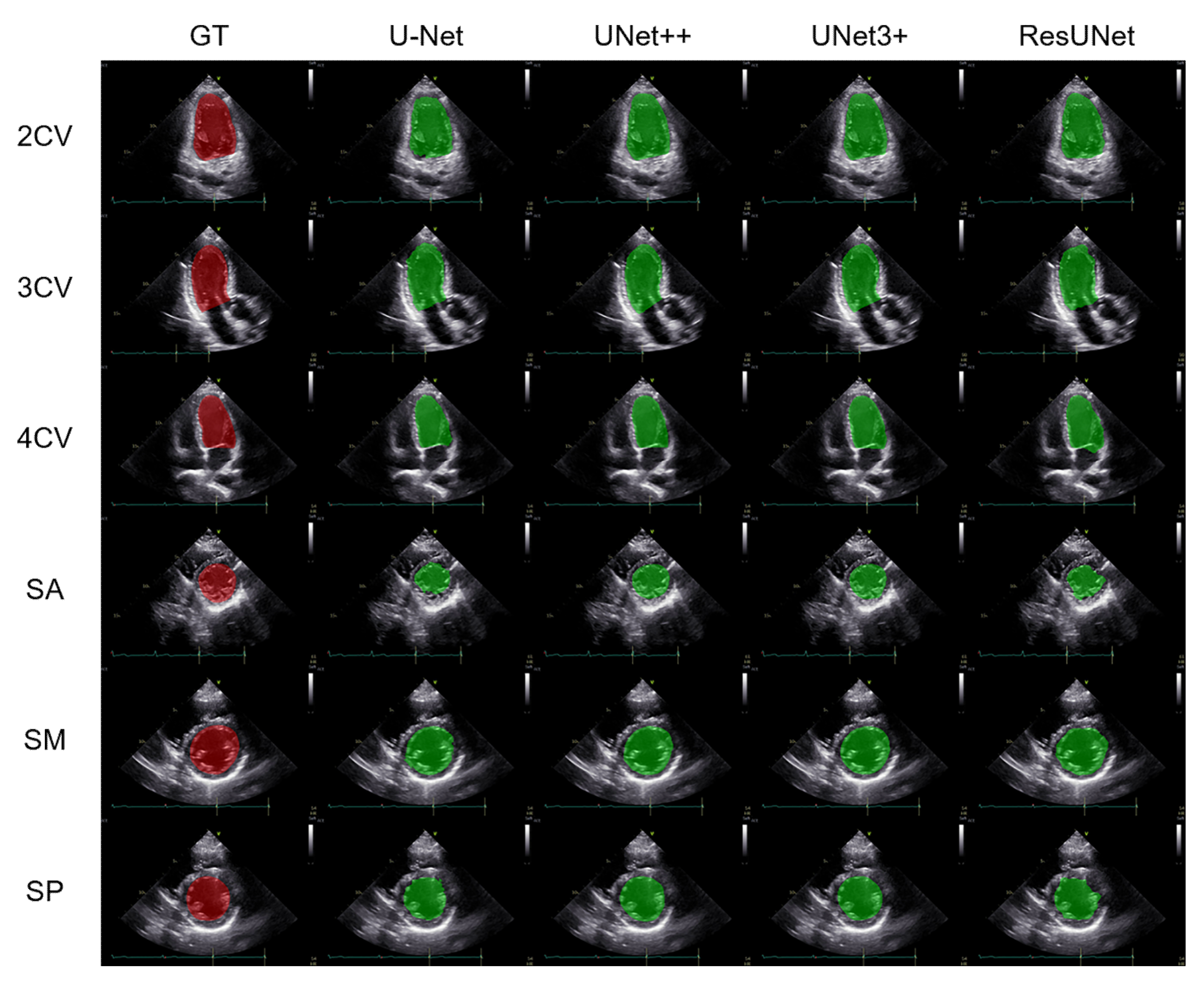
3.1. Performance Comparison of the Segmentation Methods
3.2. Left Ventricular Functional Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lang, R.M.; Addetia, K.; Narang, A.; Mor-Avi, V. 3-dimensional echocardiography: Latest developments and future directions. JACC Cardiovasc. Imaging 2018, 11, 1854–1878. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.M.; Badano, L.P.; Mor-Avi, V.; Afilalo, J.; Armstrong, A.; Ernande, L.; Flachskampf, F.A.; Foster, E.; Goldstein, S.A.; Kuznetsova, T.; et al. Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 2015, 28, 1–39.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, A.C.; Ricketts, E.P.; Cox, C.; Adler, P.; Arynchyn, A.; Liu, K.; Stengel, E.; Sidney, S.; Lewis, C.E.; Schreiner, P.J.; et al. Quality control and reproducibility in M-mode, two-dimensional, and speckle tracking echocardiography acquisition and analysis: The CARDIA study, year 25 examination experience. Echocardiography 2015, 32, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, S.; Dahlslett, T.; Grenne, B.; Sjøli, B.; Smiseth, O.; Edvardsen, T.; Brunvand, H. Global longitudinal strain is a more reproducible measure of left ventricular function than ejection fraction regardless of echocardiographic training. Cardiovasc. Ultrasound 2019, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Riffel, J.H.; Keller, M.G.; Aurich, M.; Sander, Y.; Andre, F.; Giusca, S.; Aus dem Siepen, F.; Seitz, S.; Galuschky, C.; Korosoglou, G.; et al. Assessment of global longitudinal strain using standardized myocardial deformation imaging: A modality independent software approach. Clin. Res. Cardiol. 2015, 104, 591–602. [Google Scholar] [CrossRef]
- Voigt, J.U.; Pedrizzetti, G.; Lysyansky, P.; Marwick, T.H.; Houle, H.; Baumann, R.; Pedri, S.; Ito, Y.; Abe, Y.; Metz, S.; et al. Definitions for a common standard for 2D speckle tracking echocardiography: Consensus document of the EACVI/ASE/Industry Task Force to standardize deformation imaging. Eur. Heart J. Cardiovasc. Imaging 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Farsalinos, K.E.; Daraban, A.M.; Ünlü, S.; Thomas, J.D.; Badano, L.P.; Voigt, J.-U. Head-to-head comparison of global longitudinal strain measurements among nine different vendors: The EACVI/ASE inter-vendor comparison study. J. Am. Soc. Echocardiogr. 2015, 28, 1171–1181.e1172. [Google Scholar] [CrossRef]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of artificial intelligence technology in oncology: Towards the establishment of precision medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef]
- Asada, K.; Takasawa, K.; Machino, H.; Takahashi, S.; Shinkai, N.; Bolatkan, A.; Kobayashi, K.; Komatsu, M.; Kaneko, S.; Okamoto, K.; et al. Single-cell analysis using machine learning techniques and its application to medical research. Biomedicines 2021, 9, 1513. [Google Scholar] [CrossRef]
- Hamamoto, R.; Komatsu, M.; Takasawa, K.; Asada, K.; Kaneko, S. Epigenetics analysis and integrated analysis of multiomics data, including epigenetic data, using artificial intelligence in the era of precision medicine. Biomolecules 2019, 10, 62. [Google Scholar] [CrossRef] [Green Version]
- Asada, K.; Kaneko, S.; Takasawa, K.; Machino, H.; Takahashi, S.; Shinkai, N.; Shimoyama, R.; Komatsu, M.; Hamamoto, R. Integrated Analysis of Whole Genome and Epigenome Data Using Machine Learning Technology: Toward the Establishment of Precision Oncology. Front. Oncol. 2021, 11, 666937. [Google Scholar] [CrossRef]
- Takahashi, S.; Asada, K.; Takasawa, K.; Shimoyama, R.; Sakai, A.; Bolatkan, A.; Shinkai, N.; Kobayashi, K.; Komatsu, M.; Kaneko, S.; et al. Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules 2020, 10, 1460. [Google Scholar] [CrossRef]
- Asada, K.; Komatsu, M.; Shimoyama, R.; Takasawa, K.; Shinkai, N.; Sakai, A.; Bolatkan, A.; Yamada, M.; Takahashi, S.; Machino, H.; et al. Application of artificial intelligence in COVID-19 diagnosis and therapeutics. J. Pers. Med. 2021, 11, 886. [Google Scholar] [CrossRef]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Yamada, M.; Saito, Y.; Imaoka, H.; Saiko, M.; Yamada, S.; Kondo, H.; Takamaru, H.; Sakamoto, T.; Sese, J.; Kuchiba, A.; et al. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci. Rep. 2019, 9, 14465. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Takahashi, M.; Kinoshita, M.; Miyake, M.; Kawaguchi, R.; Shinojima, N.; Mukasa, A.; Saito, K.; Nagane, M.; Otani, R.; et al. Fine-tuning approach for segmentation of gliomas in brain magnetic resonance images with a machine learning method to normalize image differences among facilities. Cancers 2021, 13, 1415. [Google Scholar] [CrossRef]
- Dozen, A.; Komatsu, M.; Sakai, A.; Komatsu, R.; Shozu, K.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Image segmentation of the ventricular septum in fetal cardiac ultrasound videos based on deep learning using time-series information. Biomolecules 2020, 10, 1526. [Google Scholar] [CrossRef]
- Shozu, K.; Komatsu, M.; Sakai, A.; Komatsu, R.; Dozen, A.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Model-agnostic method for thoracic wall segmentation in fetal ultrasound videos. Biomolecules 2020, 10, 1691. [Google Scholar] [CrossRef]
- Yasutomi, S.; Arakaki, T.; Matsuoka, R.; Sakai, A.; Komatsu, R.; Shozu, K.; Dozen, A.; Machino, H.; Asada, K.; Kaneko, S.; et al. Shadow estimation for ultrasound images using auto-encoding structures and synthetic shadows. Appl. Sci. 2021, 11, 1127. [Google Scholar] [CrossRef]
- Komatsu, M.; Sakai, A.; Dozen, A.; Shozu, K.; Yasutomi, S.; Machino, H.; Asada, K.; Kaneko, S.; Hamamoto, R. Towards clinical application of artificial intelligence in ultrasound imaging. Biomedicines 2021, 9, 720. [Google Scholar] [CrossRef]
- Narang, A.; Mor-Avi, V.; Prado, A.; Volpato, V.; Prater, D.; Tamborini, G.; Fusini, L.; Pepi, M.; Goyal, N.; Addetia, K.; et al. Machine learning based automated dynamic quantification of left heart chamber volumes. Eur. Heart J. Cardiovasc. Imaging 2019, 20, 541–549. [Google Scholar] [CrossRef]
- Fiorito, A.M.; Østvik, A.; Smistad, E.; Leclerc, S.; Bernard, O.; Lovstakken, L. Detection of cardiac events in echocardiography using 3D convolutional recurrent neural networks. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS), Kobe, Japan, 22–25 October 2018; pp. 1–4. [Google Scholar]
- Salte, I.M.; Østvik, A.; Smistad, E.; Melichova, D.; Nguyen, T.M.; Karlsen, S.; Brunvand, H.; Haugaa, K.H.; Edvardsen, T.; Lovstakken, L.; et al. Artificial intelligence for automatic measurement of left ventricular strain in echocardiography. JACC Cardiovasc. Imaging 2021, 14, 1918–1928. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A nested U-Net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A full-scale connected UNet for medical image segmentation. In Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Sakai, A.; Komatsu, R.; Matsuoka, R.; Yasutomi, S.; Shozu, K.; Dozen, A.; Machino, H.; Hidaka, H.; Arakaki, T.; et al. Detection of cardiac structural abnormalities in fetal ultrasound videos using deep learning. Appl. Sci. 2021, 11, 371. [Google Scholar] [CrossRef]
- Sakai, A.; Komatsu, M.; Komatsu, R.; Matsuoka, R.; Yasutomi, S.; Dozen, A.; Shozu, K.; Arakaki, T.; Machino, H.; Asada, K.; et al. Medical professional enhancement using explainable artificial intelligence in fetal cardiac ultrasound screening. Biomedicines 2022, 10, 551. [Google Scholar] [CrossRef]
- Zyuzin, V.; Sergey, P.; Mukhtarov, A.; Chumarnaya, T.; Solovyova, O.; Bobkova, A.; Myasnikov, V. Identification of the left ventricle endocardial border on two-dimensional ultrasound images using the convolutional neural network Unet. In Proceedings of the 2018 Ural Symposium on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 7–8 May 2018; pp. 76–78. [Google Scholar]
- Ouyang, D.; He, B.; Ghorbani, A.; Yuan, N.; Ebinger, J.; Langlotz, C.P.; Heidenreich, P.A.; Harrington, R.A.; Liang, D.H.; Ashley, E.A.; et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020, 580, 252–256. [Google Scholar] [CrossRef]
- Wu, H.; Liu, J.; Xiao, F.; Wen, Z.; Cheng, L.; Qin, J. Semi-supervised segmentation of echocardiography videos via noise-resilient spatiotemporal semantic calibration and fusion. Med. Image Anal. 2022, 78, 102397. [Google Scholar] [CrossRef]
- Kim, T.; Hedayat, M.; Vaitkus, V.V.; Belohlavek, M.; Krishnamurthy, V.; Borazjani, I. Automatic segmentation of the left ventricle in echocardiographic images using convolutional neural networks. Quant. Imaging Med. Surg. 2021, 11, 1763–1781. [Google Scholar] [CrossRef]
- Chuang, M.L.; Hibberd, M.G.; Salton, C.J.; Beaudin, R.A.; Riley, M.F.; Parker, R.A.; Douglas, P.S.; Manning, W.J. Importance of imaging method over imaging modality in noninvasive determination of left ventricular volumes and ejection fraction: Assessment by two- and three-dimensional echocardiography and magnetic resonance imaging. J. Am. Coll. Cardiol. 2000, 35, 477–484. [Google Scholar] [CrossRef]
- Cole, G.D.; Dhutia, N.M.; Shun-Shin, M.J.; Willson, K.; Harrison, J.; Raphael, C.E.; Zolgharni, M.; Mayet, J.; Francis, D.P. Defining the real-world reproducibility of visual grading of left ventricular function and visual estimation of left ventricular ejection fraction: Impact of image quality, experience and accreditation. Int. J. Cardiovasc. Imaging 2015, 31, 1303–1314. [Google Scholar] [CrossRef] [Green Version]


| Method | Projection | mIoU | mDice |
|---|---|---|---|
| U-Net | 2CV | 0.855 ± 0.068 | 0.920 ± 0.041 |
| 3CV | 0.752 ± 0.137 | 0.851 ± 0.097 | |
| 4CV | 0.816 ± 0.100 | 0.895 ± 0.063 | |
| SA | 0.670 ± 0.153 | 0.791 ± 0.125 | |
| SM | 0.841 ± 0.090 | 0.911 ± 0.057 | |
| SP | 0.813 ± 0.093 | 0.893 ± 0.062 | |
| UNet++ | 2CV | 0.890 ± 0.042 | 0.941 ± 0.024 |
| 3CV | 0.886 ± 0.034 | 0.939 ± 0.019 | |
| 4CV | 0.871 ± 0.067 | 0.929 ± 0.040 | |
| SA | 0.808 ± 0.125 | 0.887 ± 0.099 | |
| SM | 0.887 ± 0.066 | 0.939 ± 0.039 | |
| SP | 0.888 ± 0.064 | 0.939 ± 0.040 | |
| UNet3+ | 2CV | 0.891 ± 0.039 | 0.942 ± 0.022 |
| 3CV | 0.901 ± 0.028 | 0.948 ± 0.016 | |
| 4CV | 0.864 ± 0.063 | 0.926 ± 0.039 | |
| SA | 0.817 ± 0.116 | 0.893 ± 0.095 | |
| SM | 0.887 ± 0.079 | 0.938 ± 0.047 | |
| SP | 0.873 ± 0.084 | 0.930 ± 0.056 | |
| ResUNet | 2CV | 0.851 ± 0.056 | 0.919 ± 0.034 |
| 3CV | 0.837 ± 0.063 | 0.910 ± 0.038 | |
| 4CV | 0.822 ± 0.088 | 0.900 ± 0.057 | |
| SA | 0.732 ± 0.155 | 0.834 ± 0.130 | |
| SM | 0.834 ± 0.090 | 0.907 ± 0.057 | |
| SP | 0.814 ± 0.082 | 0.895 ± 0.056 |
| Method | LVEF | GLS | GCS | |||
|---|---|---|---|---|---|---|
| Mean | Median | Mean | Median | Mean | Median | |
| U-Net | 24.3 | 23.3 | 36.4 | 37.7 | 17.7 | 14.7 |
| UNet++ | 10.8 | 7.8 | 8.5 | 8.7 | 5.8 | 5.2 |
| UNet3+ | 11.7 | 10.7 | 14.6 | 16.0 | 6.4 | 5.2 |
| ResUNet | 12.5 | 13.9 | 13.0 | 15.7 | 16.2 | 22.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ono, S.; Komatsu, M.; Sakai, A.; Arima, H.; Ochida, M.; Aoyama, R.; Yasutomi, S.; Asada, K.; Kaneko, S.; Sasano, T.; et al. Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning. Biomedicines 2022, 10, 1082. https://doi.org/10.3390/biomedicines10051082
Ono S, Komatsu M, Sakai A, Arima H, Ochida M, Aoyama R, Yasutomi S, Asada K, Kaneko S, Sasano T, et al. Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning. Biomedicines. 2022; 10(5):1082. https://doi.org/10.3390/biomedicines10051082
Chicago/Turabian StyleOno, Shunzaburo, Masaaki Komatsu, Akira Sakai, Hideki Arima, Mie Ochida, Rina Aoyama, Suguru Yasutomi, Ken Asada, Syuzo Kaneko, Tetsuo Sasano, and et al. 2022. "Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning" Biomedicines 10, no. 5: 1082. https://doi.org/10.3390/biomedicines10051082
APA StyleOno, S., Komatsu, M., Sakai, A., Arima, H., Ochida, M., Aoyama, R., Yasutomi, S., Asada, K., Kaneko, S., Sasano, T., & Hamamoto, R. (2022). Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning. Biomedicines, 10(5), 1082. https://doi.org/10.3390/biomedicines10051082









