EpisomiR, a New Family of miRNAs, and Its Possible Roles in Human Diseases
Abstract
:1. Introduction
2. Regulatory miRNAs Are Implicated in Human Diseases
2.1. Canonical Pathway for Generating miRNAs
2.2. Implication of miRNAs in Human Diseases
2.2.1. Involvement of miRNAs in Human Cancers
2.2.2. Involvement of miRNAs in Human Cardiovascular Diseases
2.2.3. Involvement of miRNAs in Human Neurodegenerative Diseases
2.2.4. Involvement of miRNAs in Human Psychiatric Disorders
2.2.5. Involvement of miRNAs in Human Chronic Inflammatory Diseases
3. Non-Canonical isomiR Synthesis Pathway
3.1. Generation of Multiple Sequences from a Single miRNA Precursor
3.2. Enzymes Involved in isomiR Generation
3.2.1. Enzymes Regulating miRNA Length
Poly(A)-Specific Ribonuclease (PARN)
Nibbler Homolog
Terminal Uridylyltransferase 1 (TUT1/TENT1)
Terminal Uridylyltransferase 7 or Zinc-Finger CCHC Domain-Containing Protein ([TUT7/TENT3B/ZCCHC6 or TUT4/TENT3A/ZCCHC11])
Non-Canonical Poly(A) RNA Polymerase (TUT3/PAPD5/TENT4B)
3.2.2. Enzymes Determining miRNA Sequence
Adenosine Deaminases Acting on RNA (ADAR) Family
APOBEC Family
3.2.3. Enzymes Involved in miRNA Modifications
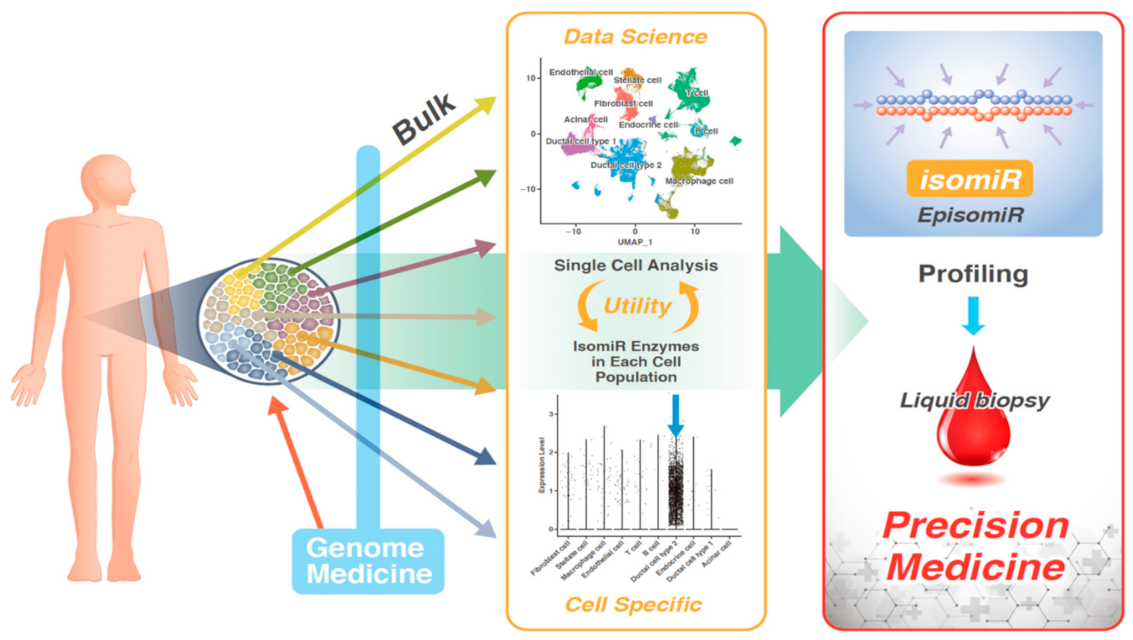
Functions of EpisomiR
Identification of EpisomiR
Sequencing of miRNAs in Single Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa Dos Santos, G., Jr.; Renovato-Martins, M.; de Brito, N.M. The remodel of the “central dogma”: A metabolomics interaction perspective. Metabolomics 2021, 17, 48. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.-L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Cummins, J.M.; He, Y.; Leary, R.J.; Pagliarini, R.; Diaz, L.A., Jr.; Sjoblom, T.; Barad, O.; Bentwich, Z.; Szafranska, A.E.; Labourier, E.; et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 2006, 103, 3687–3692. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, 6, pdb.prot5448. [Google Scholar] [CrossRef]
- Taub, F.E.; DeLeo, J.M.; Thompson, E.B. Sequential comparative hybridizations analyzed by computerized image processing can identify and quantitate regulated RNAs. DNA 1983, 2, 309–327. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Jing, Y.; Londin, E.; Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucl. Acid. Res. 2015, 43, 9158–9175. [Google Scholar] [CrossRef] [Green Version]
- Llorens, F.; Bañez-Coronel, M.; Pantano, L.; del Río, J.A.; Ferrer, I.; Estivill, X.; Martí, E. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genomic. 2013, 14, 104. [Google Scholar] [CrossRef] [Green Version]
- Mercey, O.; Popa, A.; Cavard, A.; Paquet, A.; Chevalier, B.; Pons, N.; Magnone, V.; Zangari, J.; Brest, P.; Zaragosi, L.-E.; et al. Characterizing isomiR variants within the microRNA-34/449 family. FEBS Letter. 2017, 591, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Sulston, J.E.; Horvitz, H.R. Post-Embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977, 56, 110–156. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Auyeung, V.C.; Ulitsky, I.; McGeary, S.E.; Bartel, D.P. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013, 152, 844–858. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Murchison, E.P.; Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004, 16, 223–229. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-Strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Liu, J.; Elfenbein, S.J.; Ma, Y.; Zhong, M.; Qiu, C.; Ding, Y.; Lu, J. Characterization of the mammalian miRNA turnover landscape. Nucl. Acid. Res. 2015, 43, 2326–2341. [Google Scholar] [CrossRef]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Wakiyama, M.; Takimoto, K.; Ohara, O.; Yokoyama, S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Gene. Dev. 2007, 21, 1857–1862. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Shih, I.-h.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Hibio, N.; Hino, K.; Shimizu, E.; Nagata, Y.; Ui-Tei, K. Stability of miRNA 5′terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci. Rep. 2012, 2, 996. [Google Scholar] [CrossRef]
- Wang, X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics 2014, 30, 1377–1383. [Google Scholar] [CrossRef] [Green Version]
- Wisløff, U.; Najjar, S.M.; Ellingsen, O.; Haram, P.M.; Swoap, S.; Al-Share, Q.; Fernström, M.; Rezaei, K.; Lee, S.J.; Koch, L.G.; et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 2005, 307, 418–420. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, C.; Aoyama, K.; Matsumura, N.; Kikuchi-Utsumi, K.; Watabe, M.; Nakaki, T. Rhythmic oscillations of the microRNA miR-96-5p play a neuroprotective role by indirectly regulating glutathione levels. Nat. Commun. 2014, 5, 3823. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.; Turner, J.A.; Calhoun, V.D.; Perrone-Bizzozero, N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet. 2013, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Buckland, J. Biomarkers: microRNAs under the spotlight in inflammatory arthritis. Nat. Rev. Rheumatol. 2010, 6, 436. [Google Scholar] [CrossRef]
- Tavasolian, F.; Abdollahi, E.; Rezaei, R.; Momtazi-Borojeni, A.A.; Henrotin, Y.; Sahebkar, A. Altered expression of microRNAs in rheumatoid arthritis. J. Cell. Biochem. 2018, 119, 478–487. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Çakmak, H.A.; Demir, M. MicroRNA and cardiovascular diseases. Balkan Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Azad, F.M.; Arabian, M.; Maleki, M.; Malakootian, M. Small molecules with big impacts on cardiovascular diseases. Biochem. Genet. 2020, 58, 359–383. [Google Scholar] [CrossRef]
- Sayed, A.S.; Xia, K.; Salma, U.; Yang, T.; Peng, J. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ. 2014, 23, 503–510. [Google Scholar] [CrossRef]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between microRNAs and oxidative stress in neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef] [Green Version]
- Juźwik, C.A.; Drake, S.S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef]
- Perkovic, M.N.; Paska, A.V.; Konjevod, M.; Kouter, K.; Strac, D.S.; Erjavec, G.N.; Pivac, N. Epigenetics of Alzheimer’s disease. Biomolecules 2021, 11, 195. [Google Scholar] [CrossRef]
- Issler, O.; Chen, A. Determining the role of microRNAs in psychiatric disorders. Nat. Rev. Neurosci. 2015, 16, 201–212. [Google Scholar] [CrossRef]
- Shen, H.; Li, Z. miRNAs in NMDA receptor-dependent synaptic plasticity and psychiatric disorders. Clin. Sci. 2016, 130, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.; Dev, K.; Agarwal, B.; Das, P.; Syed, M.A. Macrophages: Their role, activation and polarization in pulmonary diseases. Immunobiology 2018, 223, 383–396. [Google Scholar] [CrossRef]
- Loboda, A.; Sobczak, M.; Jozkowicz, A.; Dulak, J. TGF-β1/Smads and miR-21 in renal fibrosis and inflammation. Mediator. Inflamm. 2016, 2016, 8319283. [Google Scholar] [CrossRef] [Green Version]
- Cao, B.; Zhou, X.; Ma, J.; Zhou, W.; Yang, W.; Fan, D.; Hong, L. Role of miRNAs in inflammatory bowel disease. Dig. Dis. Sci. 2017, 62, 1426–1438. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, Y.; Shi, C.; Chen, H.; Zhang, H.; Chen, N.; Zhang, P.; Wang, F.; Yang, J.; Yang, J.; et al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem. Biophys. Res. Commun. 2013, 434, 746–752. [Google Scholar] [CrossRef]
- Wu, W.; He, C.; Liu, C.; Cao, A.T.; Xue, X.; Evans-Marin, H.L.; Sun, M.; Fang, L.; Yao, S.; Pinchuk, I.V.; et al. miR-10a inhibits dendritic cell activation and Th1/Th17 cell immune responses in IBD. Gut 2015, 64, 1755–1764. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, C.; Liu, Y.; Tang, L.; Zheng, M.; Xu, C.; Song, J.; Meng, X. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem. Biophys. Res. Commun. 2013, 438, 133–139. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Mizushima, K.; Hirata, I.; Yagi, N.; Tomatsuri, N.; Ando, T.; Oyamada, Y.; Isozaki, Y.; Hongo, H.; et al. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J. Gastroenterol. Hepatol. 2010, 25 (Suppl. S1), S129–S133. [Google Scholar] [CrossRef]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5′ isomiR variation is of functional and evolutionary importance. Nucl. Acid. Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef]
- Ferreira, P.G.; Oti, M.; Barann, M.; Wieland, T.; Ezquina, S.; Friedländer, M.R.; Rivas, M.A.; Esteve-Codina, A.; GEUVADIS Consortium; Rosenstiel, P.; et al. Sequence variation between 462 human individuals fine-tunes functional sites of RNA processing. Sci. Rep. 2016, 6, 32406. [Google Scholar] [CrossRef]
- Buiting, K.; Körner, C.; Ulrich, B.; Wahle, E.; Horsthemke, B. The human gene for the poly(A)-specific ribonuclease (PARN) maps to 16p13 and has a truncated copy in the Prader-Willi/Angelman syndrome region on 15q11-->q13. Cytogenet. Genome Res. 1999, 87, 125–131. [Google Scholar] [CrossRef]
- Shukla, S.; Bjerke, G.A.; Muhlrad, D.; Yi, R.; Parker, R. The RNase PARN controls the levels of specific miRNAs that contribute to p53 regulation. Mol. Cell 2019, 73, 1204–1216.e4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Devany, E.; Murphy, M.R.; Glazman, G.; Persaud, M.; Kleiman, F.E. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucl. Acid. Res. 2015, 43, 10925–10938. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Park, D.; Park, J.H.; Kim, J.H.; Shin, C. Poly(A)-specific ribonuclease sculpts the 3′ ends of microRNAs. RNA 2019, 25, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Katoh, T.; Hojo, H.; Suzuki, T. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucl. Acid Res. 2015, 43, 7521–7534. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, R.; Schnabl, J.; Handler, D.; Mohn, F.; Ameres, S.L.; Brennecke, J. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 2016, 539, 588–592. [Google Scholar] [CrossRef]
- Xie, W.; Sowemimo, I.; Hayashi, R.; Wang, J.; Burkard, T.R.; Brennecke, J.; Ameres, S.L.; Patel, D.J. Structure-function analysis of microRNA 3′-end trimming by Nibbler. Proc. Natl. Acad. Sci. USA 2020, 117, 30370–30379. [Google Scholar] [CrossRef]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-Transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011, 21, 1450–1461. [Google Scholar] [CrossRef] [Green Version]
- Knouf, E.C.; Wyman, S.K.; Tewari, M. The human TUT1 nucleotidyl transferase as a global regulator of microRNA abundance. PLoS ONE 2013, 8, e69630. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.Q.; Lou, Y.F.; He, Z.G.; Ji, M. Nucleotidyl transferase TUT1 inhibits lipogenesis in osteosarcoma cells through regulation of microRNA-24 and microRNA-29a. Tumour Biol. 2014, 35, 11829–11835. [Google Scholar] [CrossRef]
- Lim, J.; Ha, M.; Chang, H.; Kwon, S.C.; Simanshu, D.K.; Patel, D.J.; Kim, V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014, 159, 1365–1376. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.; Ha, M.; Loeff, L.; Chang, H.; Simanshu, D.K.; Li, S.; Fareh, M.; Patel, D.J.; Joo, C.; Kim, V.N. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 2015, 34, 1801–1815. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Chang, H.-M.; Piskounova, E.; Gregory, R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rammelt, C.; Bilen, B.; Zavolan, M.; Keller, W. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 2011, 17, 1737–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.; Kim, D.; Lee, Y.-S.; Ha, M.; Lee, M.; Yeo, J.; Chang, H.; Song, J.; Ahn, K.; Kim, V.M. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 2018, 361, 701–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boele, J.; Persson, H.; Shin, J.W.; Ishizu, Y.; Newie, I.S.; Søkilde, R.; Hawkins, S.M.; Coarfa, C.; Ikeda, K.; Takayama, K.; et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11467–11472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newie, I.; Søkilde, R.; Persson, H.; Jacomasso, T.; Gorbatenko, A.; Borg, Å.; de Hoon, M.; Pedersen, S.F.; Rovira, C. HER2-encoded mir-4728 forms a receptor-independent circuit with miR-21-5p through the non-canonical poly(A) polymerase PAPD5. Sci Rep. 2016, 6, 35664. [Google Scholar] [CrossRef] [Green Version]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs′ Action through miRNA editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [Green Version]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Warf, M.B.; Shepherd, B.A.; Johnson, W.E.; Bass, B.L. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012, 22, 1488–1498. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucl. Acid. Res. 2008, 36, 5270–5280. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Song, Y.; Shi, X.; Liu, J.; Xiong, S.; Chen, W.; Fu, Q.; Huang, Z.; Gu, N.; Zhang, R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018, 28, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Iizasa, H.; Wulff, B.-E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, H.C.; Bennett, R.P.; Kizilyer, A.; McDougall, W.M.; Prohaska, K.M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012, 23, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramatsu, M.; Kinoshita, K.; Fagarasan, S.; Yamada, S.; Shinkai, Y.; Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000, 102, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Hamidane, H.B.; Graumann, J.; Mortazavi, A.; et al. Genome-Wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 30 UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA family gets wider: The isomiRs classification and role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef]
- Lateef, O.M.; Akintubosun, M.O.; Olaoba, O.T.; Samson, S.O.; Adamczyk, M. Making sense of “nonsense” and more: Challenges and opportunities in the genetic code expansion, in the world of tRNA modifications. Int. J. Mol. Sci. 2022, 23, 938. [Google Scholar] [CrossRef]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef] [Green Version]
- Ohshiro, T.; Konno, M.; Asai, A.; Komoto, Y.; Yamagata, A.; Doki, Y.; Eguchi, H.; Ofusa, K.; Taniguchi, M.; Ishii, H. Single-molecule RNA sequencing for simultaneous detection of m6A and 5mC. Sci. Rep. 2021, 11, 19304. [Google Scholar] [CrossRef] [PubMed]
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 2019, 74, 1278–1290.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsara, O.; Schneider, R.J. m7G tRNA modification reveals new secrets in the translational regulation of cancer development. Mol. Cell 2021, 81, 3243–3245. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, H.; Liao, J.; Huang, C.; Ren, X.; Zhu, W.; Zhu, S.; Peng, B.; Li, S.; Lai, J.; et al. N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 2021, 81, 3339–3355.e8. [Google Scholar] [CrossRef] [PubMed]
- Van den Homberg, D.A.L.; van der Kwast, R.V.C.T.; Quax, P.H.A.; Nossent, A.Y. N-6-methyladenosine in vasoactive microRNAs during hypoxia; A novel role for METTL4. Int. J. Mol. Sci. 2022, 23, 1057. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Zhang, J.G.; Wu, C.X.; Li, Q. Non-Coding RNA m6A modification in cancer: Mechanisms and therapeutic targets. Front. Cell Dev. Biol. 2021, 9, 778582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, X.; Wang, J.; Jin, Y.; Gong, M.; Ye, Y.; Li, P. METTL3 suppresses neuropathic pain via modulating N6-methyladenosine-dependent primary miR-150 processing. Cell Death Discov. 2022, 8, 80. [Google Scholar] [CrossRef]
- Wang, H.; Song, X.; Song, C.; Wang, X.; Cao, H. m(6)A-seq analysis of microRNAs reveals that the N6-methyladenosine modification of miR-21-5p affects its target expression. Arch. Biochem. Biophys. 2021, 711, 109023. [Google Scholar] [CrossRef]
- Konno, M.; Taniguchi, M.; Ishii, H. Significant epitranscriptomes in heterogeneous cancer. Cancer Sci. 2019, 110, 2318–2327. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D. DART-seq: An antibody-free method for global m6A detection. Nat. Method. 2019, 16, 1275–1280. [Google Scholar] [CrossRef]
- Tegowski, M.; Flamand, M.N.; Meyer, K.D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 2022, 82, 868–878.e10. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L.; et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 2022; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, Q.; Lelyveld, V.S.; Choe, J.; Szostak, J.W.; Gregory, R.I. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 2018, 71, 244–255.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Liu, Q.; Jiang, Y.Z.; Gregory, R.I. Nucleotide resolution profiling ofm7G tRNA modification by TRAC-Seq. Nat. Protoc. 2019, 14, 3220–3242. [Google Scholar] [CrossRef]
- Frasson, I.; Pirota, V.; Richter, S.N.; Doria, F. Multimeric G-quadruplexes: A review on their biological roles and targeting. Int. J. Biol. Macromol. 2022, 204, 89–102. [Google Scholar] [CrossRef] [PubMed]
- González, I.A.; Stewart, D.R.; Schultz, K.A.P.; Field, A.P.; Hill, D.A.; Dehner, L.P. DICER1 tumor predisposition syndrome: An evolving story initiated with the pleuropulmonary blastoma. Modern Pathol. 2022, 35, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008, 359, 2641–2650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Tsuruta, F.; Okajima, T.; Yano, S.; Sato, B.; Chiba, T. KLHL7 promotes TUT1 ubiquitination associated with nucleolar integrity: Implications for retinitis pigmentosa. Biochem. Biophys. Res. Commun. 2017, 494, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Łabno, A.; Warkocki, Z.; Kuliński, T.; Krawczyk, P.S.; Bijata, K.; Tomecki, R.; Dziembowski, A. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acid. Res. 2016, 44, 10437–10453. [Google Scholar] [CrossRef] [Green Version]
- Tummala, H.; Walne, A.; Collopy, L.; Cardoso, S.; de la Fuente, J.; Lawson, S.; Powell, J.; Cooper, N.; Foster, A.; Mohammed, S.; et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Investig. 2015, 125, 2151–2160. [Google Scholar] [CrossRef]
- Kono, M.; Suganuma, M.; Dutta, A.; Ghosh, S.K.; Takeichi, T.; Muro, Y.; Akiyama, M. Bilateral striatal necrosis and dyschromatosis symmetrica hereditaria: A-I editing efficiency of ADAR1 mutants and phenotype expression. Br. J. Dermatol. 2018, 179, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.-J.; Meng, X.Y.; Fontugne, J.; Chen, C.L.; Radvanyi, F.; Bernard-Pierrot, I. Identification of new driver and passenger mutations within APOBEC-induced hotspot mutations in bladder cancer. Genome Med. 2020, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]



| Technology | Target (RNA) | Single-Cell Level | Single Molecule Level | Throughput | Reference |
|---|---|---|---|---|---|
| Cap-MS | m6A, 5mC, and others (miR) | n.d. | possible | low | [90] |
| Tunnel current sequencing | m6A, 5mC, and others (miR) | n.d. | applicable | moderate or high | [91] |
| TRAC-seq | 7mG (tRNA; the technology can be possibly applied to miR) | possible | possible | high | [94] |
| BoRed-seq | 7mG (miR) | possible | possible | high | [92] |
| scDART-seq | m6A (mRNA, possible to miR) | applicable | possible | high | [101] |
| M6A-SAC-seq | m6A (mRNA, possible to miR) | applicable | applicable | high | [102] |
| isomiR-Related Genes | Human Diseases | Reference |
|---|---|---|
| DICER1 | pleuropulmonary blastoma familial tumor predisposition syndrome (G) | [106] |
| DROSHA | ovarian cancer (S) | [107] |
| TUT1 | retinitis pigmentosa (U) | [108] |
| TUT4, TUT7 | Perlman syndrome (U) | [109] |
| PARN | dyskeratosis congenita (G) | [110] |
| ADAR | dyschromatosis symmetrica hereditaria (G) | [111] |
| APOBEC | bladder cancer (U) | [112] |
| METTL3 | acute myeloid leukemia (S) | [113] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arao, Y.; Nakayama, M.; Tsuji, Y.; Hamano, Y.; Otsuka, C.; Vecchione, A.; Ofusa, K.; Ishii, H. EpisomiR, a New Family of miRNAs, and Its Possible Roles in Human Diseases. Biomedicines 2022, 10, 1280. https://doi.org/10.3390/biomedicines10061280
Arao Y, Nakayama M, Tsuji Y, Hamano Y, Otsuka C, Vecchione A, Ofusa K, Ishii H. EpisomiR, a New Family of miRNAs, and Its Possible Roles in Human Diseases. Biomedicines. 2022; 10(6):1280. https://doi.org/10.3390/biomedicines10061280
Chicago/Turabian StyleArao, Yasuko, Mika Nakayama, Yoshiko Tsuji, Yumiko Hamano, Chihiro Otsuka, Andrea Vecchione, Ken Ofusa, and Hideshi Ishii. 2022. "EpisomiR, a New Family of miRNAs, and Its Possible Roles in Human Diseases" Biomedicines 10, no. 6: 1280. https://doi.org/10.3390/biomedicines10061280






