Enhanced Antibacterial Activity of a Cationic Macromolecule by Its Complexation with a Weakly Active Pyrazole Derivative
Abstract
:1. Introduction
2. Materials and Methods
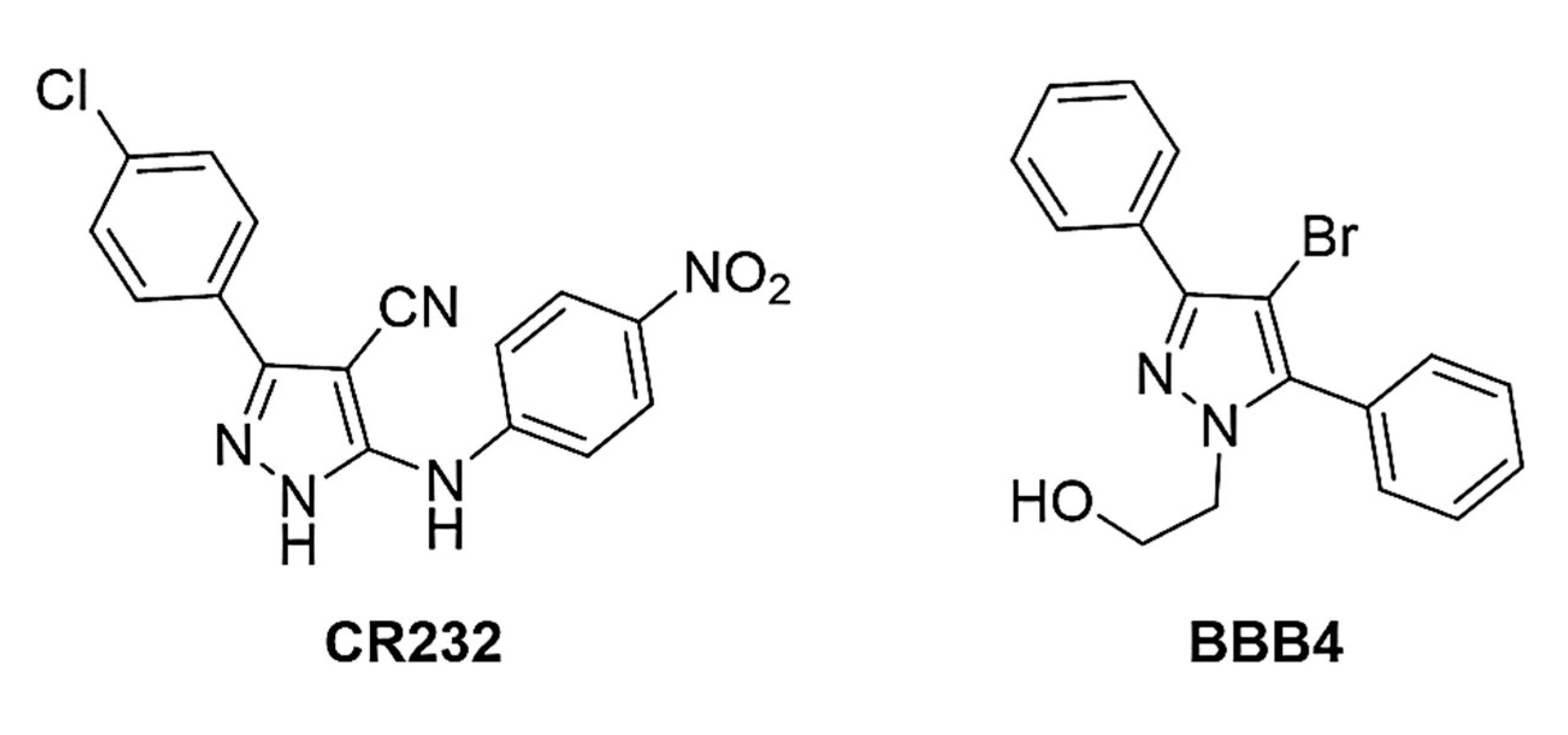
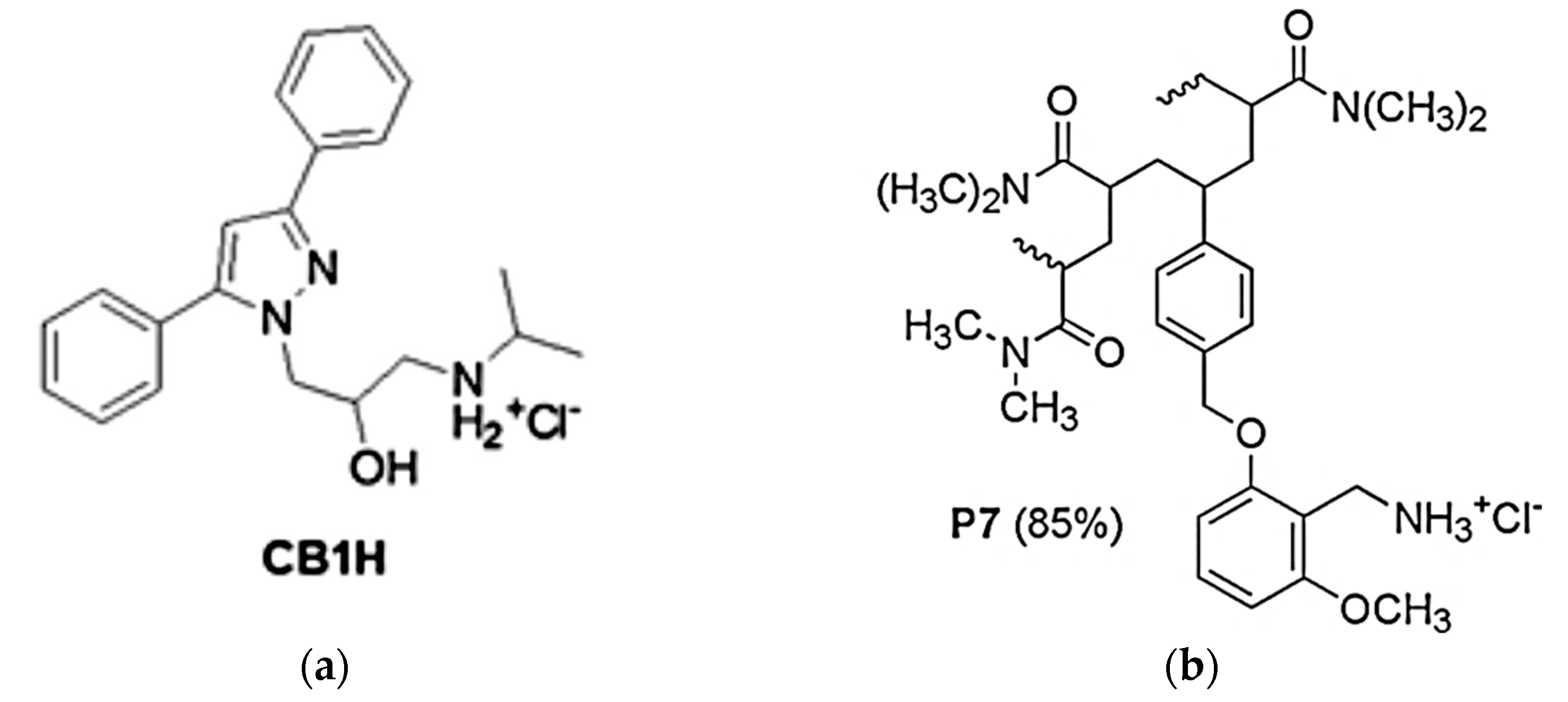
2.1. Synthesis and Characterization of P7, CB1H and CB1H-P7 NPs
2.2. Microbiology
2.2.1. Bacterial Species Considered in This Study
2.2.2. Determination of the Minimal Inhibitory Concentrations (MICs)
2.2.3. Time-Kill Experiments
2.3. Evaluation of Cytotoxicity of CB1H, P7 and CB1H-P7 NPs on Human Keratinocites (HaCaT)
2.3.1. Cell Culture
2.3.2. Viability Assay
2.4. Statistical Analyses
3. Results and Discussion
3.1. Antibacterial Effects of CB1H-P7 NPs
3.1.1. Determination of MIC Values
3.1.2. Time-Killing Curves
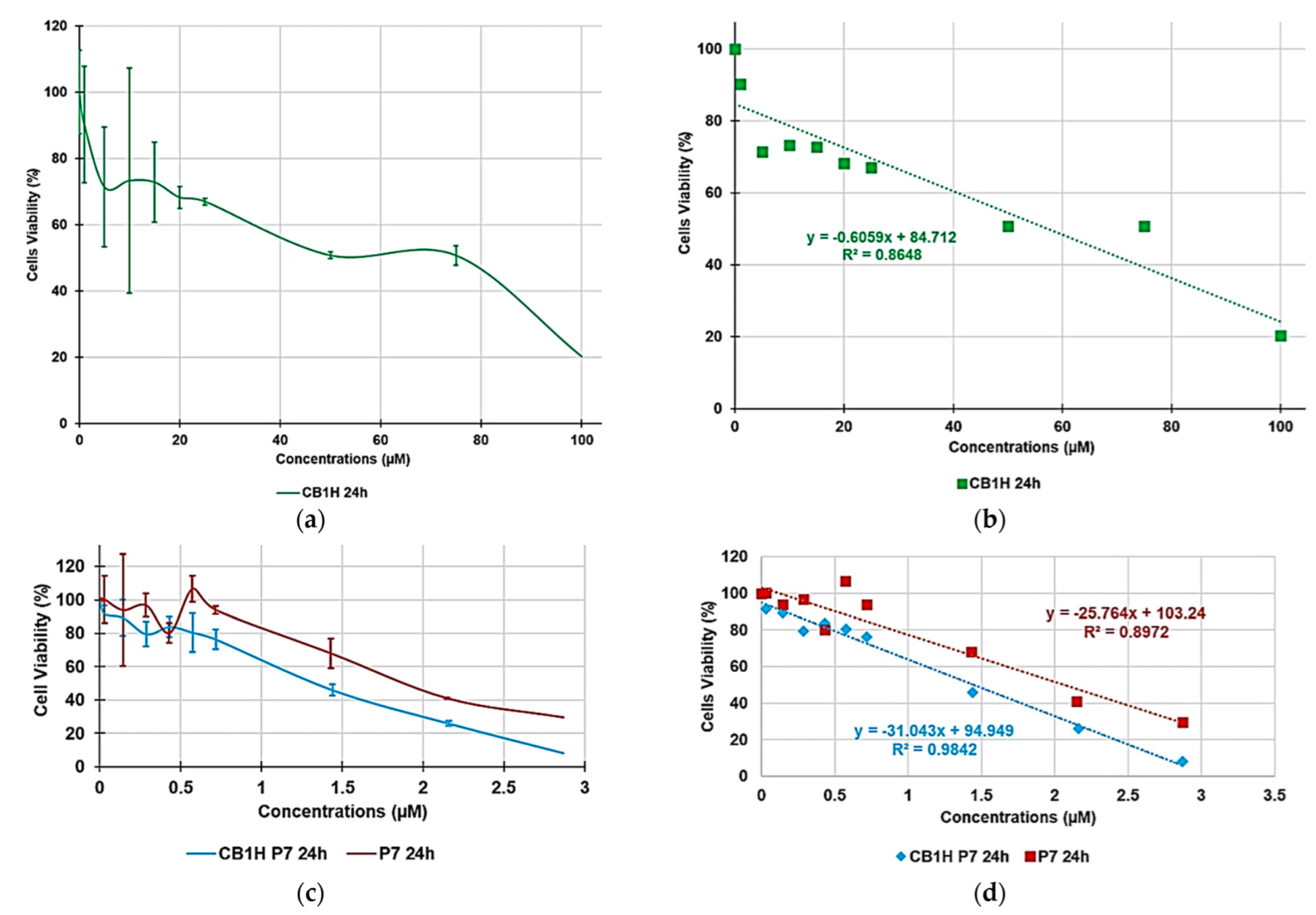
3.2. Cytotoxicity of P7, CB1H and CB1H-P7 NPs on HaCaT Human Keratinocytes Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). No Time to Wait: Securing the Future from Drug-Resistant Infections; Report to the Secretary-General of the United Nations; Interagency Coordination Group on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019. Available online: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_final_report_EN.pdf?ua=1 (accessed on 23 May 2022).
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Songmee, B.; Jaehoon, L.; Jaehwa, L.; Eunah, K.; Sunhwa, L.; Jaeyon, Y.; Yeonho, K. Antimicrobial Resistance in Haemophilus influenzae Respiratory Tract Isolates in Korea: Results of a Nationwide Acute Respiratory Infections Surveillance. Antimicrob. Agents Chemother. 2010, 54, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Alfei, S.; Brullo, C.; Caviglia, D.; Piatti, G.; Zorzoli, A.; Marimpietri, D.; Zuccari, G.; Schito, A.M. Pyrazole-Based Water-Soluble Dendrimer Nanoparticles as a Potential New Agent against Staphylococci. Biomedicines 2022, 10, 17. [Google Scholar] [CrossRef]
- Khanal, P. Antibiotic resistance¸ causes and consequences. Eur. J. Biomed. Pharm. Sci. 2020, 7, 327–331. [Google Scholar]
- PBS New Hour. We’re Headed towards a ‘Post-Antibiotic Era,’ World Health Organization Warns. Available online: https://www.pbs.org/newshour/health/world-health-organization-warns-headed-post-antibiotic-era#:~:text=%E2%80%9CWithout%20urgent%2C%20coordinated%20action%20by%20many%20stakeholders%2C%20the,Keiji%20Fukuda%2C%20WHO%E2%80%99s%20Assistant%20Director-General%20for%20Health%20Security (accessed on 23 May 2022).
- World Health Organization (WHO). Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 23 May 2022).
- World Economic Forum (WEF). How to Stop Drug-Resistant Superbugs from Causing the Next Pandemic. Available online: https://www.weforum.org/agenda/2020/11/amr-antibiotic-resistance-global-risk-death/#:~:text=Antimicrobial%20resistance%20%28AMR%29%20is%20the%20cause%20of%20death,be%20made%20on%20the%20development%20of%20new%20drugs (accessed on 23 May 2022).
- Conly, J.; Johnston, B. Where are all the new antibiotics? The new antibiotic paradox. Can. J. Infect. Dis. Med. Microbiol. 2005, 16, 159. [Google Scholar] [CrossRef] [Green Version]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Alam, M. Antibacterial Pyrazoles: Tackling Resistant Bacteria. Future Med. Chem. 2022, 14, 343–362. [Google Scholar] [CrossRef]
- Ahmed, W.; Yan, X.; Hu, D.; Adnan, M.; Tang, R.; Cui, Z.-N. Synthesis and fungicidal activity of novel pyrazole derivatives containing 5-Phenyl-2-Furan. Bioorg. Med. Chem. 2019, 27, 115048. [Google Scholar] [CrossRef]
- Ma, H.-J.; Li, Y.-H.; Zhao, Q.-F.; Zhang, T.; Xie, R.-L.; Mei, X.-D.; Ning, J. Synthesis and Herbicidal Activity of Novel N-(2,2,2)-Trifluoroethylpyrazole Derivatives. J. Agric. Food Chem. 2010, 58, 4356–4360. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.-Q.; Liu, J.; Yang, X.-P.; Liu, Z.-J. Stereoselective Synthesis and Antifungal Activities of (E)-α-(Methoxyimino)Benzeneacetate Derivatives Containing 1,3,5-Substituted Pyrazole Ring. J. Agric. Food Chem. 2006, 54, 3636–3640. [Google Scholar] [CrossRef]
- Alfei, S.; Zuccari, G. Recommendations to Synthetize Old and New β-Lactamases Inhibitors: A Review to Encourage Further Production. Pharmaceuticals 2022, 15, 384. [Google Scholar] [CrossRef]
- Alfei, S.; Schito, A.M. β-Lactam Antibiotics and β-Lactamase Enzymes Inhibitors, Part 2: Our Limited Resources. Pharmaceuticals 2022, 15, 476. [Google Scholar] [CrossRef]
- Lusardi, M.; Rotolo, C.; Ponassi, M.; Iervasi, E.; Rosano, C.; Spallarossa, A. One-pot synthesis and antiproliferative activity of highly functionalized pyrazole derivatives. ChemMedChem 2022, 17, e202100670. [Google Scholar] [CrossRef]
- Alfei, S.; Spallarossa, A.; Lusardi, M.; Zuccari, G. Successful Dendrimer and Liposome-Based Strategies to Solubilize an Antiproliferative Pyrazole Otherwise Not Clinically Applicable. Nanomaterials 2022, 12, 233. [Google Scholar] [CrossRef]
- Alfei, S.; Brullo, C.; Caviglia, D.; Zuccari, G. Preparation and Physicochemical Characterization of Water-Soluble Pyrazole-Based Nanoparticles by Dendrimer Encapsulation of an Insoluble Bioactive Pyrazole Derivative. Nanomaterials 2021, 11, 2662. [Google Scholar] [CrossRef]
- Alfei, S.; Caviglia, D.; Zorzoli, A.; Marimpietri, D.; Spallarossa, A.; Lusardi, M.; Zuccari, G.; Schito, A.M. Potent and Broad-Spectrum Bactericidal Activity of a Nanotechnologically Manipulated Novel Pyrazole. Biomedicines 2022, 10, 907. [Google Scholar] [CrossRef]
- Alfei, S.; Zuccari, G.; Caviglia, D.; Brullo, C. Synthesis and Characterization of Pyrazole-Enriched Cationic Nanoparticles as New Promising Antibacterial Agent by Mutual Cooperation. Nanomaterials 2022, 12, 1215. [Google Scholar] [CrossRef]
- Alfei, S.; Marengo, B.; Valenti, G.E.; Domenicotti, C. Synthesis of Polystyrene-Based Cationic Nanomaterials with Pro-Oxidant Cytotoxic Activity on Etoposide-Resistant Neuroblastoma Cells. Nanomaterials 2021, 11, 977. [Google Scholar] [CrossRef]
- Schito, A.M.; Piatti, G.; Caviglia, D.; Zuccari, G.; Alfei, S. Broad-Spectrum Bactericidal Activity of a Synthetic Random Copolymer Based on 2-Methoxy-6-(4-Vinylbenzyloxy)-Benzylammonium Hydrochloride. Int. J. Mol. Sci. 2021, 22, 5021. [Google Scholar] [CrossRef]
- Ganewatta, M.S.; Tang, C. Controlling macromolecular structures towards effective antimicrobial polymers. Polymer 2015, 63, A1–A29. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, G.J.; Som, A.; Madkour, A.E.; Eren, T.; Tew, G.N. Infectious disease: Connecting innate immunity to biocidal polymers. Mater. Sci. Eng. R. Rep. 2007, 57, 28–64. [Google Scholar] [CrossRef] [Green Version]
- Leuschner, C.; Hansel, W. Membrane disrupting lytic peptides for cancer treatments. Curr. Pharm. Des. 2004, 10, 2299–2310. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef]
- Tan, J.; Tay, J.; Hedrick, J.; Yang, Y.Y. Synthetic macromolecules as therapeutics that overcome resistance in cancer and microbial infection. Biomaterials 2020, 252, 120078. [Google Scholar] [CrossRef]
- Schito, A.M.; Piatti, G.; Caviglia, D.; Zuccari, G.; Zorzoli, A.; Marimpietri, D.; Alfei, S. Bactericidal Activity of Non-Cytotoxic Cationic Nanoparticles against Clinically and Environmentally Relevant Pseudomonas spp. Isolates. Pharmaceutics 2021, 13, 1411. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria (accessed on 23 May 2022).
- Schito, A.M.; Piatti, G.; Stauder, M.; Bisio, A.; Giacomelli, E.; Romussi, G.; Pruzzo, C. Effects of demethylfruticuline A and fruticuline A from Salvia corrugata Vahl. on biofilm production in vitro by multiresistant strains of Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2011, 37, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Di Paolo, D.; Pastorino, F.; Zuccari, G.; Caffa, I.; Loi, M.; Marimpietri, D.; Brignole, C.; Perri, P.; Cilli, M.; Nico, B.; et al. Enhanced Anti-Tumor and Anti-Angiogenic Efficacy of a Novel Liposomal Fenretinide on Human Neuroblastoma. J. Control. Rel. 2013, 170, 445–451. [Google Scholar] [CrossRef]
- Schito, A.M.; Caviglia, D.; Piatti, G.; Zorzoli, A.; Marimpietri, D.; Zuccari, G.; Schito, G.C.; Alfei, S. Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci. Pharmaceutics 2021, 13, 1976. [Google Scholar] [CrossRef]
- Alnufaie, R.; Raj, K.C.H.; Alsup, N.; Whitt, J.; Andrew Chambers, S.; Gilmore, D.; Alam, M.A. Synthesis and Antimicrobial Studies of Coumarin-Substituted Pyrazole Derivatives as Potent Anti-Staphylococcus aureus Agents. Molecules 2020, 25, 2758. [Google Scholar] [CrossRef]
- Sharma, S.; Verma, K.K. Skin and soft tissue infection. Indian J. Pediatrics 2001, 68 (Suppl. S3), S46–S50. [Google Scholar]


| 36 Bacterial Species | ||||
|---|---|---|---|---|
| 12 Gram-Positive | Resistance Information | 24 Gram-Negative | Resistance Information | |
| 6 Enterococci | 3 E. faecalis | VRE * | 9 MDR P. aeruginosa # | 1 colistin-resistant, 1 resistant to CAZ-AVI, 3 pyomelanin-producing, 4 from fibrosis cystic patients |
| 3 E. faecium | VRE * | |||
| 6 Staphylococci | 3 S. aureus | MRSA | 3 E. coli | 2 KPC-producing, 1 NDM carbapenemase-producing |
| 1 E. aerogenes | Carbapenems-resistant | |||
| MRSE ** | 4 MDR S. maltophilia | 2 co-trimoxazole-resistant, 2 co-trimoxazole intermediates | ||
| 3 S. epidermidis | 4 K. pneumoniae | KPC-producing K. pneumoniae § | ||
| 3 MDR A. baumannii | 1 co-trimoxazole-resistant | |||
| Samples | Concentrations (µM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CB1H | 0 | 1 | 5 | 10 | 15 | 20 | 25 | 50 | 75 | 100 |
| CB1H-P7 | 0 | 0.02874 | 0.1437 | 0.2874 | 0.4311 | 0.5748 | 0.7185 | 1.4370 | 2.1555 | 2.8740 |
| P7 | 0 | 0.02867 | 0.1433 | 0.2867 | 0.4300 | 0.5734 | 0.7167 | 1.4334 | 2.1501 | 2.8668 |
| Original P7 (13,719) 1 | Original CB1H (371.9) 2 | CB1H-P7 NPs (26,624) 1 | Complexed P7 (13,719) 1 | Complexed CB1H (371.9) 2 | ||
|---|---|---|---|---|---|---|
| Strains | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | Selectivity Indices 3 |
| Enterococcus genus | ||||||
| E. faecalis 1 *,# | 2.3 (32) | 344.2 (128) | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 1.2 |
| E. faecalis 365 * | 2.3 (32) | 344.2 (128) | 2.4 (64) | 2.4 (32.9) | 83.6 (31.1) | 0.6 |
| E. faecalis 450 * | 2.3 (32) | 344.2 (128) | 2.4 (64) | 2.4 (32.9) | 83.6 (31.1) | 0.6 |
| E. faecium 21 * | 1.2 (16) | 344.2 (128) | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 1.2 |
| E. faecium 325 * | 1.2 (16) | 344.2 (128) | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 2.4 |
| E. faecium 341 *,# | 1.2 (16) | 344.2 (128) | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 2.4 |
| Staphylococcus genus | ||||||
| S. aureus 18 ** | 4.6 (64) | 344.2 (128) | 2.4 (64) | 2.4 (32.9) | 83.6 (31.1) | 0.6 |
| S. aureus 187 ** | 4.6 (64) | 344.2 (128) | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 0.3 |
| S. aureus 195 ** | 4.6 (64) | 344.2 (128) | 2.4 (64) | 2.4 (32.9) | 83.6 (31.1) | 0.6 |
| S. epidermidis 22 ** | 1.2 (16) | 344.2 (128) | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 2.4 |
| S. epidermidis 180 *** | 1.2 (16) | 344.2 (128) | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 1.2 |
| S. epidermidis 181 *** | 1.2 (16) | 344.2 (128) | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 2.4 |
| Original P7 (13,719) 1 | Original CB1H (371.9) 2 | CB1H-P7 NPs (26,624) 1 | Complexed P7 (13,719) 1 | Complexed CB1H (371.9) 2 | ||
|---|---|---|---|---|---|---|
| Strains | MIC µM (µg/mL) | MIC (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | Selectivity Indices 3 |
| Enterobacteriaceae family | ||||||
| E. coli 238 # | 4.6 (64) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| E. coli 477 # | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| E. coli 462 § | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| K. pneumoniae 375 # | 4.6 (64) | >688.4 (>256) | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 0.3 |
| K. pneumoniae 376 # | 9.3 (128) | >688.4 (>256) | 9.6 (256) | 9.1 (124.4) | 35.4 (131.6) | 0.2 |
| K. pneumoniae 377 # | 9.3 (128) | >688.4 (>256) | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 0.3 |
| K. pneumoniae 490 CR # | 4.6 (64) | >688.4 (>256) | 2.4 (64) | 2.3 (32.9) | 83.6 (31.1) | 0.6 |
| E. aerogenes 484 ## | 4.6 (64) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6)) | 1.3 |
| Non-fermenting species | ||||||
| A. baumannii 257 | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| A. baumannii 279 | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.2 |
| A. baumannii 245 COR | 9.3 (128) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 1V ## | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 5V ## | 4.6 (64) | >688.4 (>256) | 2.4 (64) | 2.3 (32.9) | 83.6 (31.1) | 0.6 |
| P. aeruginosa 6V ## | 4.6 (64) | >688.4 (>256) | 2.4 (64) | 2.3 (32.9) | 83.6 (31.1) | 0.6 |
| P. aeruginosa 7G ## | 4.6 (64) | >688.4 (>256) | 2.4 (64) | 2.3 (32.9) | 83.6 (31.1) | 0.6 |
| P. aeruginosa 265 CR ## | ≥18.6 (≥256) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 432 py,## | 1.2 (16) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 447 py,## | 4.6 (64) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 244 py,## | 4.6 (64) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| P. aeruginosa 259 *,## | 9.3 (128) | >688.4 (>256) | 9.6 (256) | 9.1 (124.4) | 35.4 (131.6) | 0.2 |
| S. maltophilia 2 COI | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| S. maltophilia 280 COI | 4.6 (64) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| S. maltophilia 384 COR | 2.3 (32) | >688.4 (>256) | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 1.3 |
| S. maltophilia 390 COR | 2.3 (32) | >688.4 (>256) | 2.4 (64) | 2.3 (32.9) | 83.6 (31.1) | 0.6 |
| Strains | CB1H-P7 NPs (26,624) 1 | Complexed P7 (13,719) 1 | Complexed CB1H (371.9) 2 | Reference Antibiotics |
|---|---|---|---|---|
| MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | |
| E. faecalis 1 *,# | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 366.3 (128) 3 |
| E. faecalis 365 * | 2.4 (64) | 2.4 (32.9) | 83.9 (31.1) | 366.3 (128) 3 |
| E. faecalis 450 * | 2.4 (64) | 2.4 (32.9) | 83.9 (31.1) | 366.3 (128) 3 |
| E. faecium 21 | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 366.3 (128) 3 |
| E. faecium 325 * | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 366.3 (128) 3 |
| E. faecium 341 *,# | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 366.3 (128) 3 |
| S. aureus 18 ** | 2.4 (64) | 2.4 (32.9) | 83.9 (31.1) | 386.4 (128) 4, 1275.5 (512) 5 |
| S. aureus 187 ** | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 386.4 (128) 4, 1275.5 (512) 5 |
| S. aureus 195 ** | 2.4 (64) | 2.4 (32.9) | 83.9 (31.1) | 386.4 (128) 4, 1275.5 (512) 5 |
| S. epidermidis 22 ** | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 193.2 (64) 4, 637.8 (256) 5 |
| S. epidermidis 180 *** | 1.2 (32) | 1.2 (16.4) | 41.9 (15.6) | 193.2 (64) 4, 637.8 (256) 5 |
| S. epidermidis 181 *** | 0.6 (16) | 0.6 (8.2) | 21.0 (7.8) | 193.2 (64) 4, 637.8 (256) 5 |
| Strains | CB1H-P7 NPs (26,624) 1 | Complexed P7 (13,719) 1 | Complexed CB1H (371.9) 2 | Reference Antibiotics |
|---|---|---|---|---|
| MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | MIC µM (µg/mL) | |
| E. coli 238 # | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 96.6 (32) 3 |
| E. coli 477 # | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 96.6 (32) 3 |
| E. coli 462 § | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 96.6 (32) 3 |
| K. pneumoniae 375 # | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 96.6 (32) 3 |
| K. pneumoniae 376 # | 9.3 (256) | 9.6 (131.6) | 334.5 (124.4) | 96.6 (32) 3 |
| K. pneumoniae 377 # | 4.8 (128) | 4.8 (65.8) | 167.2 (62.2) | 96.6 (32) 3 |
| K. pneumoniae 490 CR # | 2.4 (64) | 2.3 (32.9) | 83.9 (31.1) | 18.5 (16) 4 |
| E. aerogenes 484 ## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 96.6 (32) 3 |
| A. baumannii 257 | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 193.2 (64) 3 |
| A. baumannii 279 | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 193.2 (64) 3 |
| A. baumannii 245 COR | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 193.2 (64) 3 |
| P. aeruginosa 1V ## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 76.2 (64) 5 |
| P. aeruginosa 5V ## | 2.4 (64) | 2.3 (32.9) | 83.9 (31.1) | 76.2 (64) 5 |
| P. aeruginosa 6V ## | 2.4 (64) | 2.3 (32.9) | 83.9 (31.1) | 76.2 (64) 5 |
| P. aeruginosa 7G ## | 2.4 (64) | 2.3 (32.9) | 83.9 (31.1) | 76.2 (64) 5 |
| P. aeruginosa 265 CR ## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 18.5 (16) 4 |
| P. aeruginosa 432 py,## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 76.2 (64) 5 |
| P. aeruginosa 447 py,## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 76.2 (64) 5 |
| P. aeruginosa 244 py,## | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 76.2 (64) 5 |
| P. aeruginosa 259 *,## | 9.6 (256) | 9.6 (131.6) | 334.5 (124.4) | 76.2 (64) 5 |
| S. maltophilia 2 COI | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 117.7 (64) 6 |
| S. maltophilia 280 COI | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 117.7 (64) 6 |
| S. maltophilia 384 COR | 1.2 (32) | 1.2 (16.5) | 41.9 (15.6) | 117.7 (64) 6 |
| S. maltophilia 390 COR | 2.4 (64) | 2.3 (32.9) | 83.9 (31.1) | 117.7 (64) 6 |
| Sample | Equations | R2 | LD50 (µM) | SI | |
|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | ||||
| P7 | y = −25.7640x + 103.4 | 0.8972 | 2.1 | 0.5–1.8 | 0.1–1.8 |
| CB1H | y = −0.6059x + 84.712 | 0.8648 | 57.3 | ≤0.4. | ≤0.2. |
| CB1H-P7 NPs | y = −31.0430x + 94.949 | 0.9842 | 1.5 | 0.3–2.4 | 0.2–1.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schito, A.M.; Caviglia, D.; Brullo, C.; Zorzoli, A.; Marimpietri, D.; Alfei, S. Enhanced Antibacterial Activity of a Cationic Macromolecule by Its Complexation with a Weakly Active Pyrazole Derivative. Biomedicines 2022, 10, 1607. https://doi.org/10.3390/biomedicines10071607
Schito AM, Caviglia D, Brullo C, Zorzoli A, Marimpietri D, Alfei S. Enhanced Antibacterial Activity of a Cationic Macromolecule by Its Complexation with a Weakly Active Pyrazole Derivative. Biomedicines. 2022; 10(7):1607. https://doi.org/10.3390/biomedicines10071607
Chicago/Turabian StyleSchito, Anna Maria, Debora Caviglia, Chiara Brullo, Alessia Zorzoli, Danilo Marimpietri, and Silvana Alfei. 2022. "Enhanced Antibacterial Activity of a Cationic Macromolecule by Its Complexation with a Weakly Active Pyrazole Derivative" Biomedicines 10, no. 7: 1607. https://doi.org/10.3390/biomedicines10071607
APA StyleSchito, A. M., Caviglia, D., Brullo, C., Zorzoli, A., Marimpietri, D., & Alfei, S. (2022). Enhanced Antibacterial Activity of a Cationic Macromolecule by Its Complexation with a Weakly Active Pyrazole Derivative. Biomedicines, 10(7), 1607. https://doi.org/10.3390/biomedicines10071607