A nop56 Zebrafish Loss-of-Function Model Exhibits a Severe Neurodegenerative Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Care and Maintenance
2.2. Genotyping
2.3. Characterization of General Morphology
2.4. Survival Analysis
2.5. Expression Analysis
2.6. Acridine Orange
2.7. Immunofluorescence
2.8. Behavioral Analysis
2.9. Morpholino Injection
2.10. Statistical Analysis
3. Results
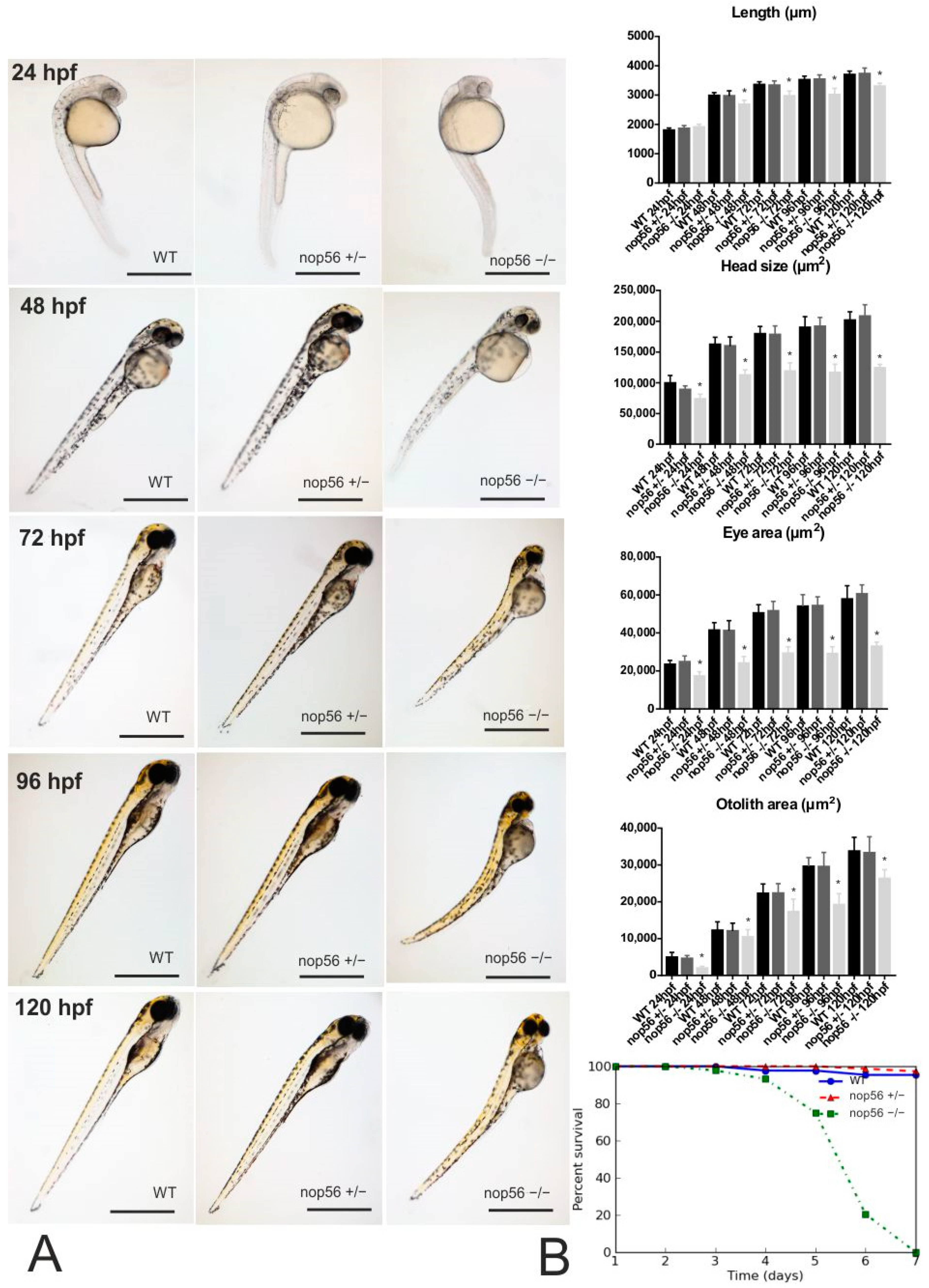
3.1. nop56−/− Embryos Develop an Abnormal Phenotype
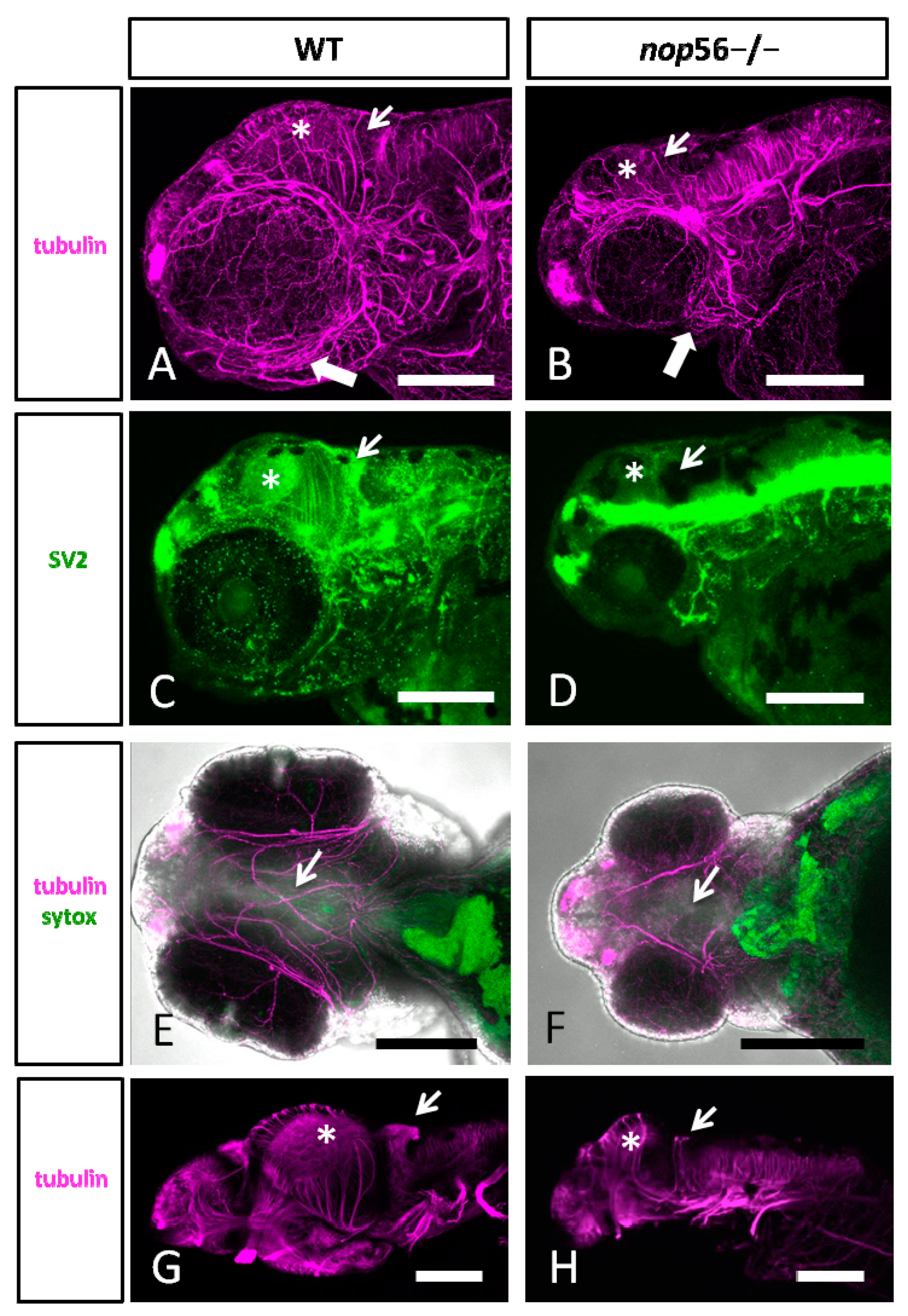
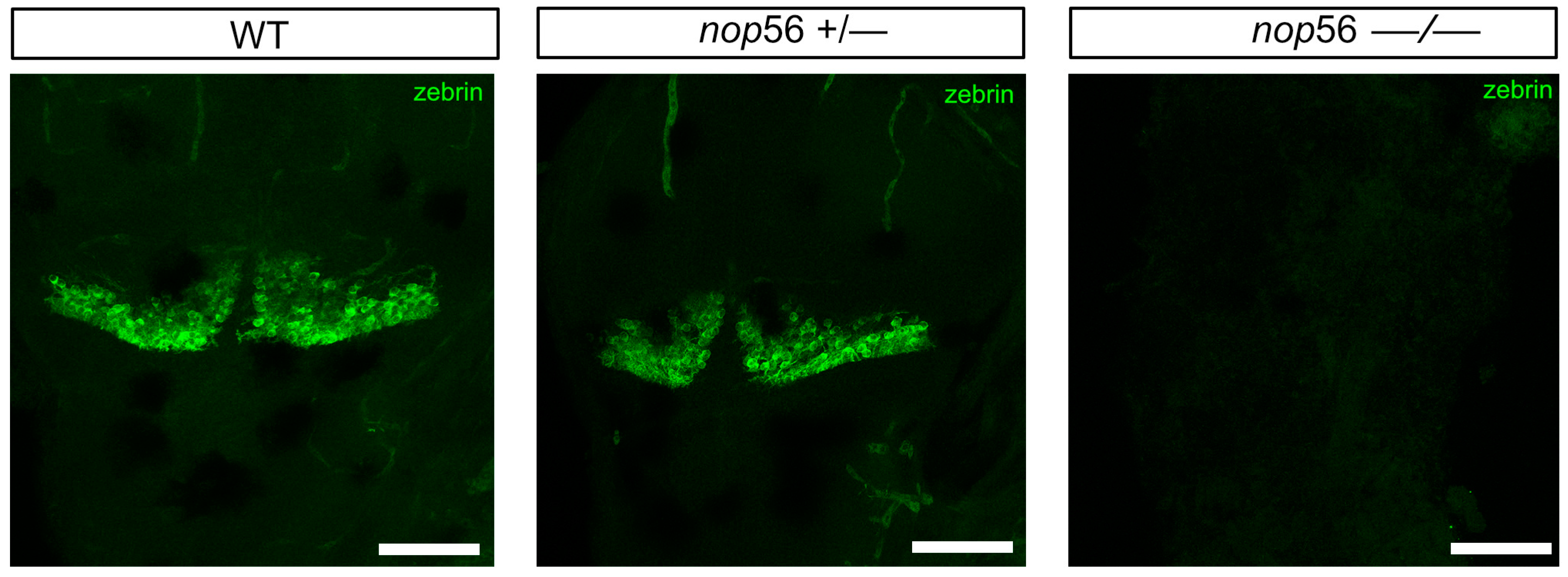
3.2. Malformations in the Central and Peripheral Nervous System of nop56 −/− Larvae
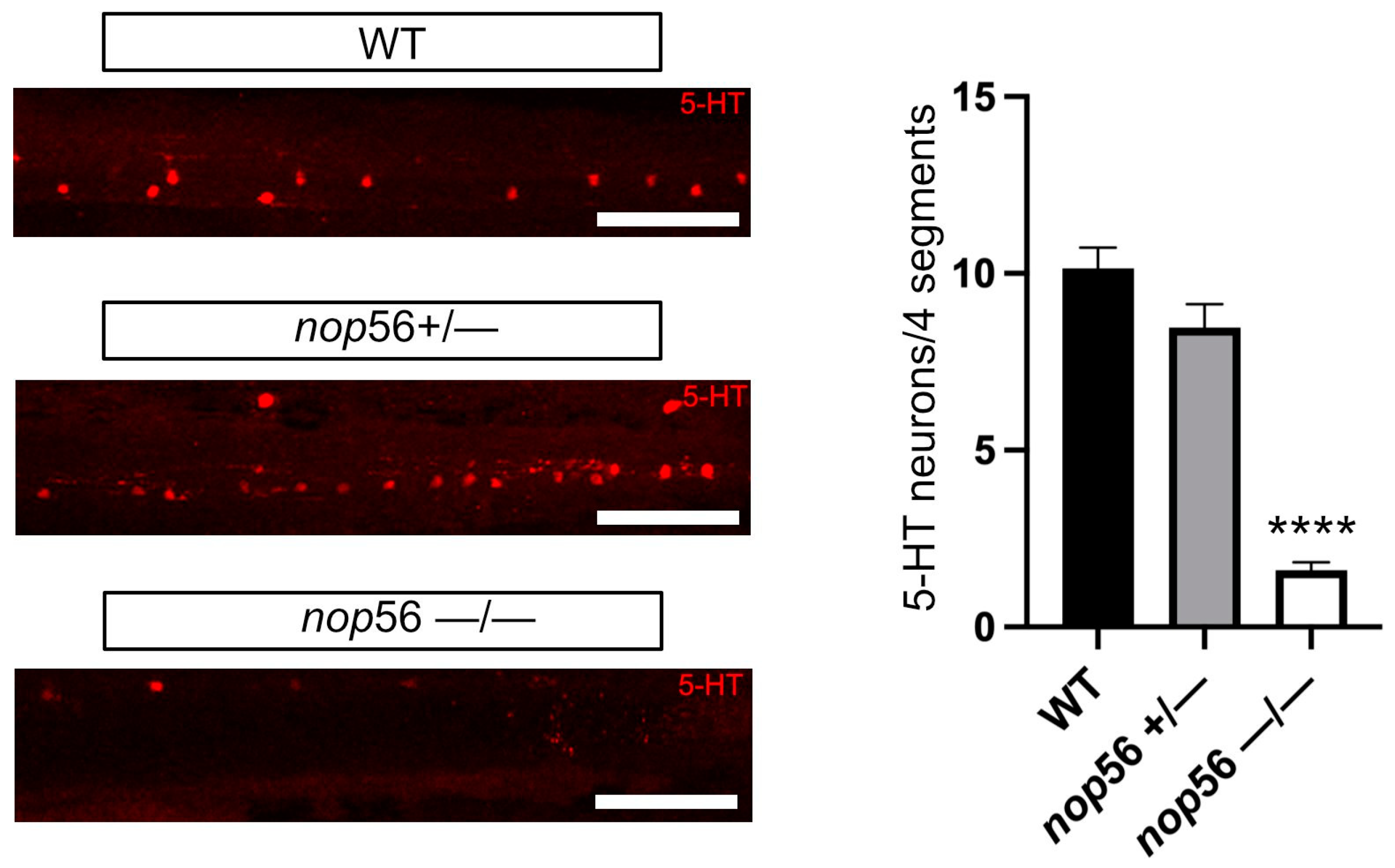
3.3. Neuromuscular Junction and 5-HT Spinal Cord Interneurons
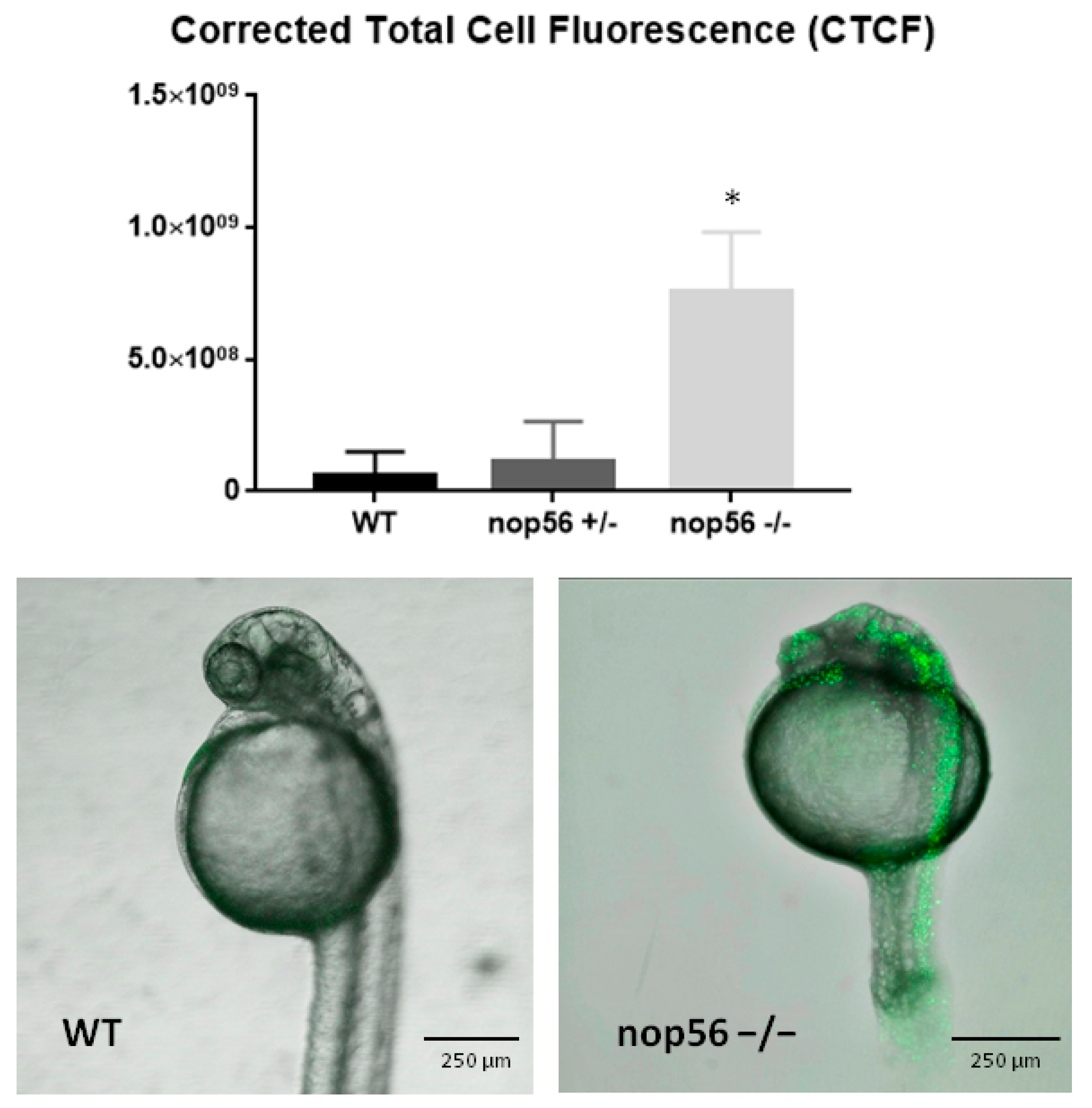
3.4. nop56−/− Larvae Have Increased Apoptosis Mainly in the CNS
3.5. nop56 Knockout Caused Impaired Motor Function
3.6. Reduced Expression of the zpld1 Genes from the Cupula of the Inner Ear
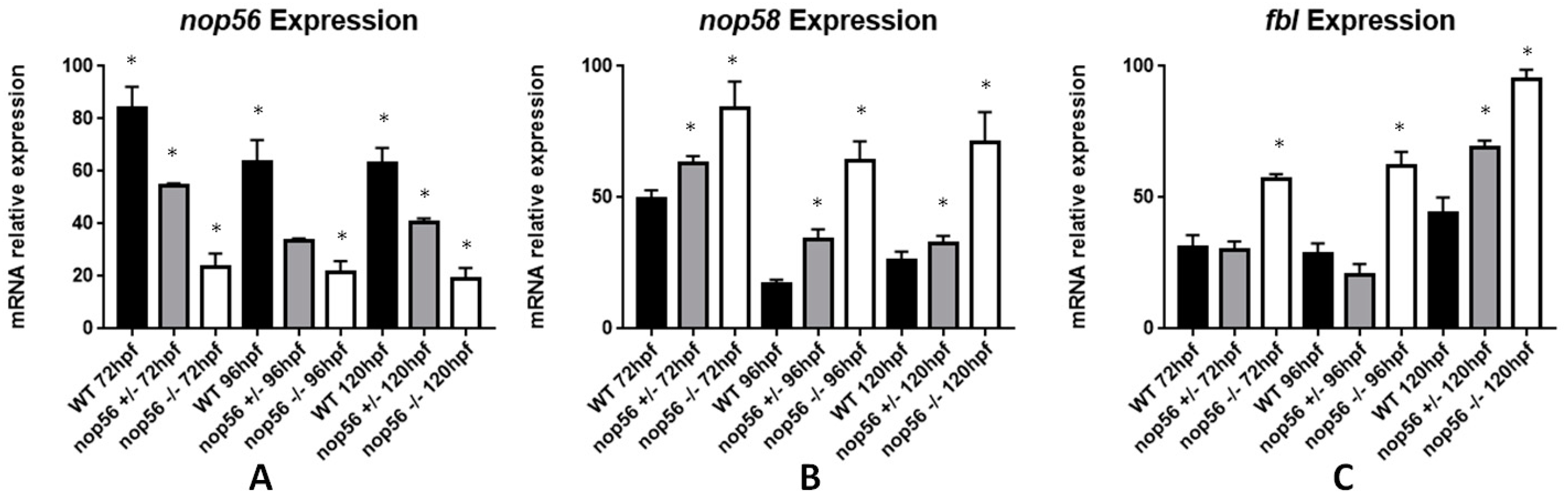
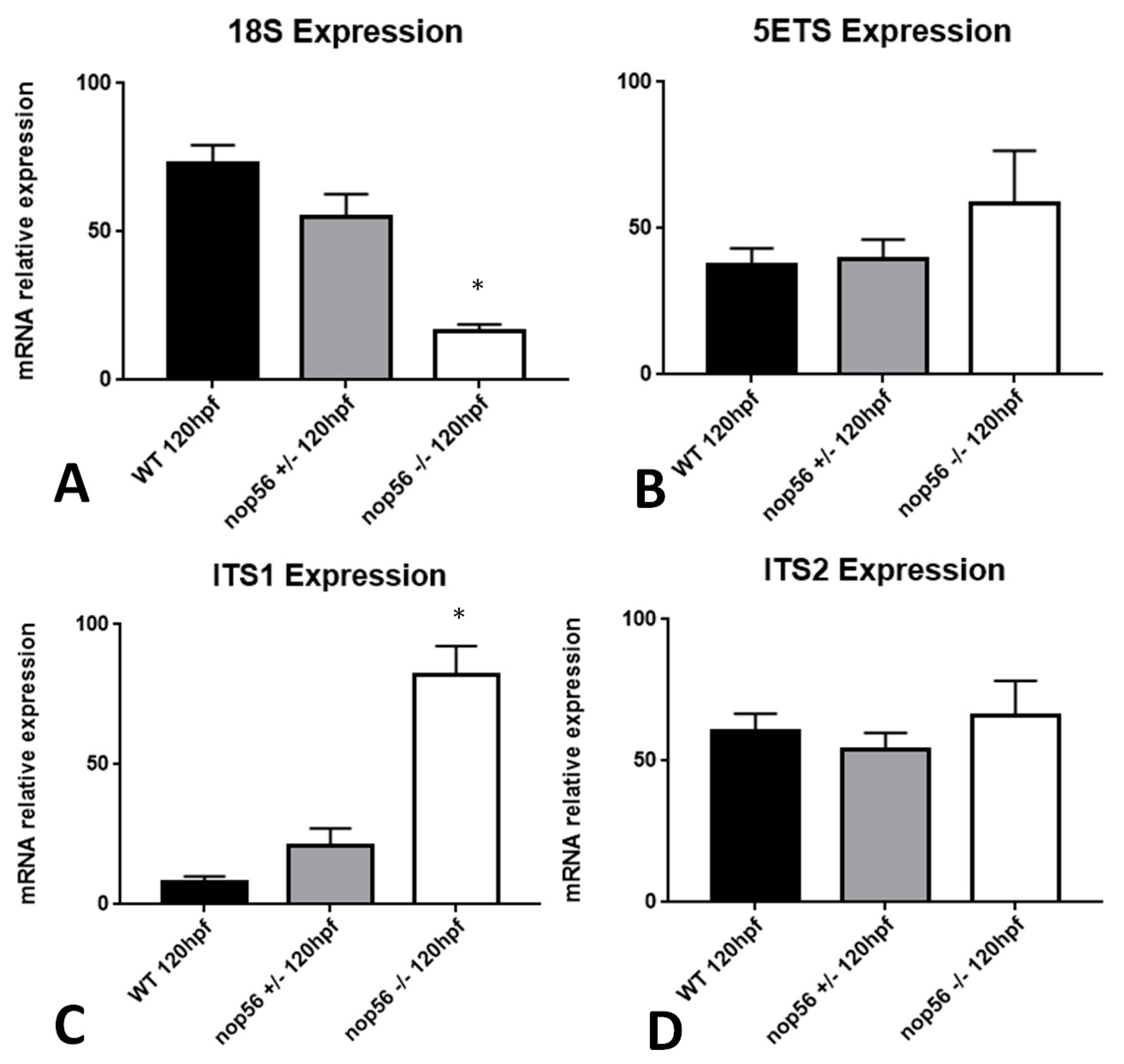
3.7. nop56 Disruption Causes Overexpression in nop58 and fbl Related Genes and Impaired rRNA Processing
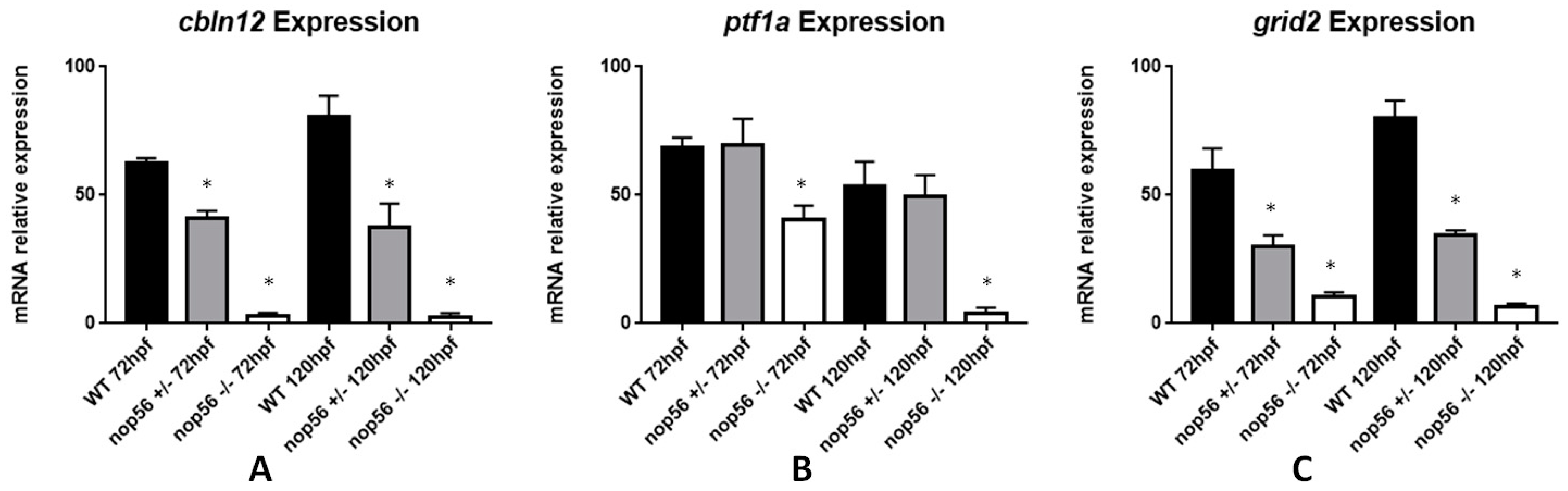
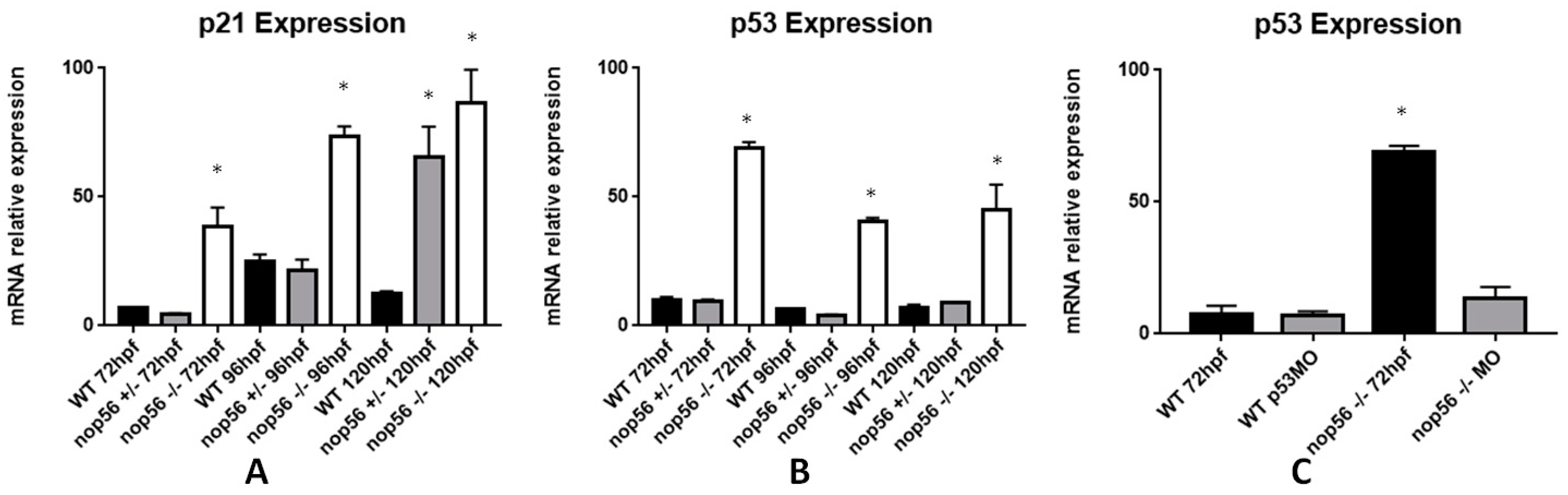
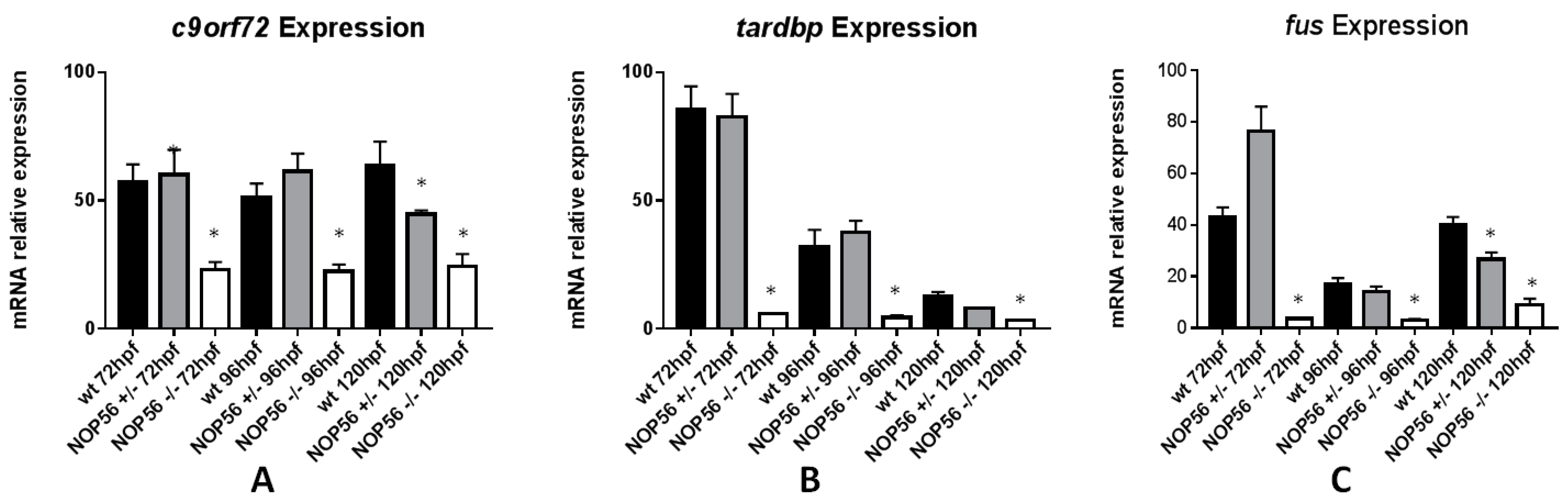
3.8. Expression of Genes Related to ALS Is Also Affected in the nop56 Knockout Embryos
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-Box SnoRNAs Form Methylating and Non-Methylating Ribonucleoprotein Complexes: Old Dogs Show New Tricks. BioEssays 2017, 39, 1600264. [Google Scholar] [CrossRef]
- Lechertier, T.; Grob, A.; Hernandez-Verdun, D.; Roussel, P. Fibrillarin and Nop56 Interact before Being Co-Assembled in Box C/D SnoRNPs. Exp. Cell Res. 2009, 315, 928–942. [Google Scholar] [CrossRef] [PubMed]
- Oruganti, S.; Zhang, Y.; Li, H.; Robinson, H.; Terns, M.P.; Terns, R.M.; Yang, W.; Li, H. Alternative Conformations of the Archaeal Nop56/58-Fibrillarin Complex Imply Flexibility in Box C/D RNPs. J. Mol. Biol. 2007, 371, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Nepomuceno-Mejía, T.; Florencio-Martínez, L.E.; Martínez-Calvillo, S. Nucleolar Division in the Promastigote Stage of Leishmania Major Parasite: A Nop56 Point of View. BioMed Res. Int. 2018, 2018, 1641839. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, K.T.; Biswas, S.; Zhang, X.; Brown, B.A.; Wollenzien, P.; Mattos, C.; Maxwell, E.S. Structurally Conserved Nop56/58 N-Terminal Domain Facilitates Archaeal Box C/D Ribonucleoprotein-Guided Methyltransferase Activity. J. Biol. Chem. 2012, 287, 19418–19428. [Google Scholar] [CrossRef]
- Garcıa-Planells, J.; Paricio, N.; Palau, F.; de Frutos, R. Dnop56, a Drosophila Gene Homologous to the Yeast Nucleolar NOP56 Gene. Genetica 2000, 109, 275–282. [Google Scholar] [CrossRef]
- Gautier, T.; Bergès, T.; Tollervey, D.; Hurt, E. Nucleolar KKE/D Repeat Proteins Nop56p and Nop58p Interact with Nop1p and Are Required for Ribosome Biogenesis. Mol. Cell. Biol. 1997, 17, 7088–7098. [Google Scholar] [CrossRef] [PubMed]
- Renzi, F.; Filippini, D.; Loreni, F.; Bozzoni, I.; Caffarelli, E. Characterization of the Sequences Encoding for Xenopus Laevis Box C/D SnoRNP Nop56 Protein. Biochim. Et Biophys. Acta (BBA)—Gene Struct. Expr. 2002, 1575, 26–30. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The Box C/D and H/ACA SnoRNPs: Key Players in the Modification, Processing and the Dynamic Folding of Ribosomal RNA: Box C/D and H/ACA SnoRNPs. WIREs RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Ardal, B.K.; Hollensen, A.K.; Damgaard, C.K.; Jensen, T.H. Box C/D SnoRNP Autoregulation by a Cis-Acting SnoRNA in the NOP56 Pre-MRNA. Mol. Cell 2018, 72, 99–111.e5. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L.; et al. A Pan-Cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, K.; Liao, J.; Jiang, F. Small Nucleolar RNAs in Cancer. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2012, 1826, 121–128. [Google Scholar] [CrossRef]
- Williams, G.T.; Farzaneh, F. Are SnoRNAs and SnoRNA Host Genes New Players in Cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef]
- Cowling, V.H.; Turner, S.A.; Cole, M.D. Burkitt’s Lymphoma-Associated c-Myc Mutations Converge on a Dramatically Altered Target Gene Response and Implicate Nol5a/Nop56 in Oncogenesis. Oncogene 2014, 33, 3519–3527. [Google Scholar] [CrossRef]
- He, J.; Yu, J. Long Noncoding RNA FAM83A-AS1 Facilitates Hepatocellular Carcinoma Progression by Binding with NOP58 to Enhance the MRNA Stability of FAM83A. Biosci. Rep. 2019, 39, BSR20192550. [Google Scholar] [CrossRef]
- Marcel, V.; Catez, F.; Diaz, J.-J. P53, a Translational Regulator: Contribution to Its Tumour-Suppressor Activity. Oncogene 2015, 34, 5513–5523. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.-M.; Thompson, I.; Yuan, Z.-M. Elevated SnoRNA Biogenesis Is Essential in Breast Cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef]
- García-Murias, M.; Quintáns, B.; Arias, M.; Seixas, A.I.; Cacheiro, P.; Tarrío, R.; Pardo, J.; Millán, M.J.; Arias-Rivas, S.; Blanco-Arias, P.; et al. ‘Costa Da Morte’ Ataxia Is Spinocerebellar Ataxia 36: Clinical and Genetic Characterization. Brain 2012, 135, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Abe, K.; Matsuura, T.; Ikeda, Y.; Hitomi, T.; Akechi, Y.; Habu, T.; Liu, W.; Okuda, H.; Koizumi, A. Expansion of Intronic GGCCTG Hexanucleotide Repeat in NOP56 Causes SCA36, a Type of Spinocerebellar Ataxia Accompanied by Motor Neuron Involvement. Am. J. Hum. Genet. 2011, 89, 121–130. [Google Scholar] [CrossRef]
- Arias, M.; García-Murias, M.; Sobrido, M.J. La ataxia espinocerebelosa 36 (SCA36): «Ataxia da Costa da Morte». Neurología 2017, 32, 386–393. [Google Scholar] [CrossRef]
- Ikeda, Y.; Ohta, Y.; Kobayashi, H.; Okamoto, M.; Takamatsu, K.; Ota, T.; Manabe, Y.; Okamoto, K.; Koizumi, A.; Abe, K. Clinical Features of SCA36 A Novel Spinocerebellar Ataxia with Motor Neuron Involvement (Asidan). Neurology 2012, 79, 333–341. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Kouri, N.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Belzil, V.V.; Bauer, P.O.; Prudencio, M.; Gendron, T.F.; Stetler, C.T.; Yan, I.K.; Pregent, L.; Daughrity, L.; Baker, M.C.; Rademakers, R.; et al. Reduced C9orf72 Gene Expression in C9FTD/ALS Is Caused by Histone Trimethylation, an Epigenetic Event Detectable in Blood. Acta Neuropathol. 2013, 126, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Butti, Z.; Pan, Y.E.; Giacomotto, J.; Patten, S.A. Reduced C9orf72 Function Leads to Defective Synaptic Vesicle Release and Neuromuscular Dysfunction in Zebrafish. Commun. Biol. 2021, 4, 792. [Google Scholar] [CrossRef]
- Furuta, N.; Tsukagoshi, S.; Hirayanagi, K.; Ikeda, Y. Suppression of the Yeast Elongation Factor Spt4 Ortholog Reduces Expanded SCA36 GGCCUG Repeat Aggregation and Cytotoxicity. Brain Res. 2019, 1711, 29–40. [Google Scholar] [CrossRef]
- Matsuzono, K.; Imamura, K.; Murakami, N.; Tsukita, K.; Yamamoto, T.; Izumi, Y.; Kaji, R.; Ohta, Y.; Yamashita, T.; Abe, K.; et al. Antisense Oligonucleotides Reduce RNA Foci in Spinocerebellar Ataxia 36 Patient IPSCs. Mol. Ther. Nucleic Acids 2017, 8, 211–219. [Google Scholar] [CrossRef]
- McEachin, Z.T.; Gendron, T.F.; Raj, N.; García-Murias, M.; Banerjee, A.; Purcell, R.H.; Ward, P.J.; Todd, T.W.; Merritt-Garza, M.E.; Jansen-West, K.; et al. Chimeric Peptide Species Contribute to Divergent Dipeptide Repeat Pathology in C9ALS/FTD and SCA36. Neuron 2020, 107, 292–305.e6. [Google Scholar] [CrossRef]
- Wang, H.; Chen, X.; He, T.; Zhou, Y.; Luo, H. Evidence for Tissue-Specific JAK/STAT Target Genes in Drosophila Optic Lobe Development. Genetics 2013, 195, 1291–1306. [Google Scholar] [CrossRef]
- Todd, T.W.; McEachin, Z.T.; Chew, J.; Burch, A.R.; Jansen-West, K.; Tong, J.; Yue, M.; Song, Y.; Castanedes-Casey, M.; Kurti, A.; et al. Hexanucleotide Repeat Expansions in C9FTD/ALS and SCA36 Confer Selective Patterns of Neurodegeneration In Vivo. Cell Rep. 2020, 31, 107616. [Google Scholar] [CrossRef]
- Kozol, R.A.; Abrams, A.J.; James, D.M.; Buglo, E.; Yan, Q.; Dallman, J.E. Function Over Form: Modeling Groups of Inherited Neurological Conditions in Zebrafish. Front. Mol. Neurosci. 2016, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Kishi, S. Molecular Cloning and Functional Characterization of Zebrafish ATM. Int. J. Biochem. Cell Biol. 2005, 37, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Issa, F.A.; Mazzochi, C.; Mock, A.F.; Papazian, D.M. Spinocerebellar Ataxia Type 13 Mutant Potassium Channel Alters Neuronal Excitability and Causes Locomotor Deficits in Zebrafish. J. Neurosci. 2011, 31, 6831–6841. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, X.; Ning, G.; Zhu, S.; Ma, X.; Liu, X.; Liu, C.; Huang, M.; Schmitt, I.; Wüllner, U.; et al. The Machado–Joseph Disease Deubiquitinase Ataxin-3 Regulates the Stability and Apoptotic Function of P53. PLoS Biol. 2016, 14, e2000733. [Google Scholar] [CrossRef]
- Quelle-Regaldie, A.; Sobrido-Cameán, D.; Barreiro-Iglesias, A.; Sobrido, M.J.; Sánchez, L. Zebrafish Models of Autosomal Dominant Ataxias. Cells 2021, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Quelle-Regaldie, A.; Sobrido-Cameán, D.; Barreiro-Iglesias, A.; Sobrido, M.J.; Sánchez, L. Zebrafish Models of Autosomal Recessive Ataxias. Cells 2021, 10, 836. [Google Scholar] [CrossRef]
- Yanicostas, C.; Barbieri, E.; Hibi, M.; Brice, A.; Stevanin, G.; Soussi-Yanicostas, N. Requirement for Zebrafish Ataxin-7 in Differentiation of Photoreceptors and Cerebellar Neurons. PLoS ONE 2012, 7, e50705. [Google Scholar] [CrossRef]
- Ciura, S.; Lattante, S.; Le Ber, I.; Latouche, M.; Tostivint, H.; Brice, A.; Kabashi, E. Loss of Function of C9orf72 Causes Motor Deficits in a Zebrafish Model of Amyotrophic Lateral Sclerosis. Ann. Neurol. 2013, 74, 180–187. [Google Scholar] [CrossRef]
- Swaminathan, A.; Bouffard, M.; Liao, M.; Ryan, S.; Callister, J.B.; Pickering-Brown, S.M.; Armstrong, G.A.B.; Drapeau, P. Expression of C9orf72-Related Dipeptides Impairs Motor Function in a Vertebrate Model. Hum. Mol. Genet. 2018, 27, 1754–1762. [Google Scholar] [CrossRef]
- Swinnen, B.; Bento-Abreu, A.; Gendron, T.F.; Boeynaems, S.; Bogaert, E.; Nuyts, R.; Timmers, M.; Scheveneels, W.; Hersmus, N.; Wang, J.; et al. A Zebrafish Model for C9orf72 ALS Reveals RNA Toxicity as a Pathogenic Mechanism. Acta Neuropathol. 2018, 135, 427–443. [Google Scholar] [CrossRef]
- Yeh, T.-H.; Liu, H.-F.; Li, Y.-W.; Lu, C.-S.; Shih, H.-Y.; Chiu, C.-C.; Lin, S.-J.; Huang, Y.-C.; Cheng, Y.-C. C9orf72 Is Essential for Neurodevelopment and Motility Mediated by Cyclin G1. Exp. Neurol. 2018, 304, 114–124. [Google Scholar] [CrossRef]
- Busch-Nentwich, E.; Kettleborough, R.; Dooley, C.M.; Scahill, C.; Sealy, I.; White, R.; Stemple, D.L. Sanger Institute Zebrafish Mutation Project Mutant Data Submission. ZFIN Direct Data Submiss. 2013. Available online: http://zfin.org (accessed on 1 July 2022).
- Draper, B.W.; McCallum, C.M.; Stout, J.L.; Slade, A.J.; Moens, C.B. A High-Throughput Method for Identifying N-Ethyl-N-Nitrosourea (ENU)-Induced Point Mutations in Zebrafish. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 77, pp. 91–112. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and Husbandry Recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. 2000. Available online: http://zfin.org/zf_info/zfbook/zfbk.html (accessed on 1 July 2022).
- Bouffard, S.; Dambroise, E.; Brombin, A.; Lempereur, S.; Hatin, I.; Simion, M.; Corre, R.; Bourrat, F.; Joly, J.-S.; Jamen, F. Fibrillarin Is Essential for S-Phase Progression and Neuronal Differentiation in Zebrafish Dorsal Midbrain and Retina. Dev. Biol. 2018, 437, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kani, S.; Bae, Y.-K.; Shimizu, T.; Tanabe, K.; Satou, C.; Parsons, M.J.; Scott, E.; Higashijima, S.; Hibi, M. Proneural Gene-Linked Neurogenesis in Zebrafish Cerebellum. Dev. Biol. 2010, 343, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Yamaguchi, S.; Sakakibara, Y.; Hayashi, T.; Matsuda, K.; Hara, Y.; Tanegashima, C.; Shimizu, T.; Kuraku, S.; Hibi, M. Gene Expression Profiling of Granule Cells and Purkinje Cells in the Zebrafish Cerebellum: Transcriptomes In Zebrafish Cerebellar Neurons. J. Comp. Neurol. 2017, 525, 1558–1585. [Google Scholar] [CrossRef]
- Gomez-Manzano, C.; Fueyo, J.; Kyritsis, A.P.; Steck, P.A.; Levin, V.A.; Alfred Yung, W.K.; McDonnell, T.J. Characterization of P53 and P21 Functional Interactions in Glioma Cells En Route to Apoptosis. JNCI J. Natl. Cancer Inst. 1997, 89, 1036–1044. [Google Scholar] [CrossRef]
- Moulton, J.D. Using Morpholinos to Control Gene Expression. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 4–30. [Google Scholar] [CrossRef]
- Dernedde, J.; Weise, C.; Müller, E.-C.; Hagiwara, A.; Bachmann, S.; Suzuki, M.; Reutter, W.; Tauber, R.; Scherer, H. Cupulin Is a Zona Pellucida-Like Domain Protein and Major Component of the Cupula from the Inner Ear. PLoS ONE 2014, 9, e111917. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lin, S.; Staats, K.A.; Li, Y.; Chang, W.-H.; Hung, S.-T.; Hendricks, E.; Linares, G.R.; Wang, Y.; Son, E.Y.; et al. Haploinsufficiency Leads to Neurodegeneration in C9ORF72 ALS/FTD Human Induced Motor Neurons. Nat. Med. 2018, 24, 313–325. [Google Scholar] [CrossRef]
- Mitra, J.; Guerrero, E.N.; Hegde, P.M.; Liachko, N.F.; Wang, H.; Vasquez, V.; Gao, J.; Pandey, A.; Taylor, J.P.; Kraemer, B.C.; et al. Motor Neuron Disease-Associated Loss of Nuclear TDP-43 Is Linked to DNA Double-Strand Break Repair Defects. Proc. Natl. Acad. Sci. USA 2019, 116, 4696–4705. [Google Scholar] [CrossRef]
- Wang, W.-Y.; Pan, L.; Su, S.C.; Quinn, E.J.; Sasaki, M.; Jimenez, J.C.; Mackenzie, I.R.A.; Huang, E.J.; Tsai, L.-H. Interaction of FUS and HDAC1 Regulates DNA Damage Response and Repair in Neurons. Nat. Neurosci. 2013, 16, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Naumann, M.; Pal, A.; Goswami, A.; Lojewski, X.; Japtok, J.; Vehlow, A.; Naujock, M.; Günther, R.; Jin, M.; Stanslowsky, N.; et al. Impaired DNA Damage Response Signaling by FUS-NLS Mutations Leads to Neurodegeneration and FUS Aggregate Formation. Nat. Commun. 2018, 9, 335. [Google Scholar] [CrossRef]
- Recher, G.; Jouralet, J.; Brombin, A.; Heuzé, A.; Mugniery, E.; Hermel, J.-M.; Desnoulez, S.; Savy, T.; Herbomel, P.; Bourrat, F.; et al. Zebrafish Midbrain Slow-Amplifying Progenitors Exhibit High Levels of Transcripts for Nucleotide and Ribosome Biogenesis. Development 2013, 140, 4860–4869. [Google Scholar] [CrossRef]
- Trouillas, P.; Xie, J.; Adeleine, P. Treatment of Cerebellar Ataxia with Buspirone: A Double-Blind Study. Lancet. Neurol. 1996, 348, 759. [Google Scholar] [CrossRef]
- Trouillas, P. The Cerebellar Serotoninergic System and Its Possible Involvement in Cerebellar Ataxia. Can. J. Neurol. Sci. 1993, 20, S78–S82. [Google Scholar] [CrossRef]
- Takei, A.; Hamada, T.; Yabe, I.; Sasaki, H. Treatment of Cerebellar Ataxia with 5-HT1A Agonist. Cerebellum 2005, 4, 211–215. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Hong, Z.-X.; Lin, S.-Z.; Harn, H.-J. Identifying Therapeutic Targets for Spinocerebellar Ataxia Type 3/Machado–Joseph Disease through Integration of Pathological Biomarkers and Therapeutic Strategies. Int. J. Mol. Sci. 2020, 21, 3063. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Jones, S.M.; Jones, T.A.; Tian, C.; Johnson, K.R. Spontaneous Mutations of the Zpld1 Gene in Mice Cause Semicircular Canal Dysfunction but Do Not Impair Gravity Receptor or Hearing Functions. Sci. Rep. 2019, 9, 12430. [Google Scholar] [CrossRef]
- Gianfrancesco, F.; Esposito, T.; Penco, S.; Maglione, V.; Liquori, C.L.; Patrosso, M.C.; Zuffardi, O.; Ciccodicola, A.; Marchuk, D.A.; Squitieri, F. ZPLD1 Gene Is Disrupted in a Patient with Balanced Translocation That Exhibits Cerebral Cavernous Malformations. Neuroscience 2008, 155, 345–349. [Google Scholar] [CrossRef]
- Miyazaki, K.; Yamashita, T.; Morimoto, N.; Sato, K.; Mimoto, T.; Kurata, T.; Ikeda, Y.; Abe, K. Early and Selective Reduction of NOP56 (Asidan) and RNA Processing Proteins in the Motor Neuron of ALS Model Mice. Neurol. Res. 2013, 35, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Sivadasan, R.; Hornburg, D.; Drepper, C.; Frank, N.; Jablonka, S.; Hansel, A.; Lojewski, X.; Sterneckert, J.; Hermann, A.; Shaw, P.J.; et al. C9ORF72 Interaction with Cofilin Modulates Actin Dynamics in Motor Neurons. Nat. Neurosci. 2016, 19, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Jiang, J.; Gendron, T.F.; McAlonis-Downes, M.; Jiang, L.; Taylor, A.; Diaz Garcia, S.; Ghosh Dastidar, S.; Rodriguez, M.J.; King, P.; et al. Reduced C9ORF72 Function Exacerbates Gain of Toxicity from ALS/FTD-Causing Repeat Expansion in C9orf72. Nat. Neurosci. 2020, 23, 615–624. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quelle-Regaldie, A.; Folgueira, M.; Yáñez, J.; Sobrido-Cameán, D.; Alba-González, A.; Barreiro-Iglesias, A.; Sobrido, M.-J.; Sánchez, L. A nop56 Zebrafish Loss-of-Function Model Exhibits a Severe Neurodegenerative Phenotype. Biomedicines 2022, 10, 1814. https://doi.org/10.3390/biomedicines10081814
Quelle-Regaldie A, Folgueira M, Yáñez J, Sobrido-Cameán D, Alba-González A, Barreiro-Iglesias A, Sobrido M-J, Sánchez L. A nop56 Zebrafish Loss-of-Function Model Exhibits a Severe Neurodegenerative Phenotype. Biomedicines. 2022; 10(8):1814. https://doi.org/10.3390/biomedicines10081814
Chicago/Turabian StyleQuelle-Regaldie, Ana, Mónica Folgueira, Julián Yáñez, Daniel Sobrido-Cameán, Anabel Alba-González, Antón Barreiro-Iglesias, María-Jesús Sobrido, and Laura Sánchez. 2022. "A nop56 Zebrafish Loss-of-Function Model Exhibits a Severe Neurodegenerative Phenotype" Biomedicines 10, no. 8: 1814. https://doi.org/10.3390/biomedicines10081814
APA StyleQuelle-Regaldie, A., Folgueira, M., Yáñez, J., Sobrido-Cameán, D., Alba-González, A., Barreiro-Iglesias, A., Sobrido, M.-J., & Sánchez, L. (2022). A nop56 Zebrafish Loss-of-Function Model Exhibits a Severe Neurodegenerative Phenotype. Biomedicines, 10(8), 1814. https://doi.org/10.3390/biomedicines10081814






