Targeted Protein Degradation to Overcome Resistance in Cancer Therapies: PROTAC and N-Degron Pathway
Abstract
:1. Introduction
2. General Mechanism and Advantages of PROTAC
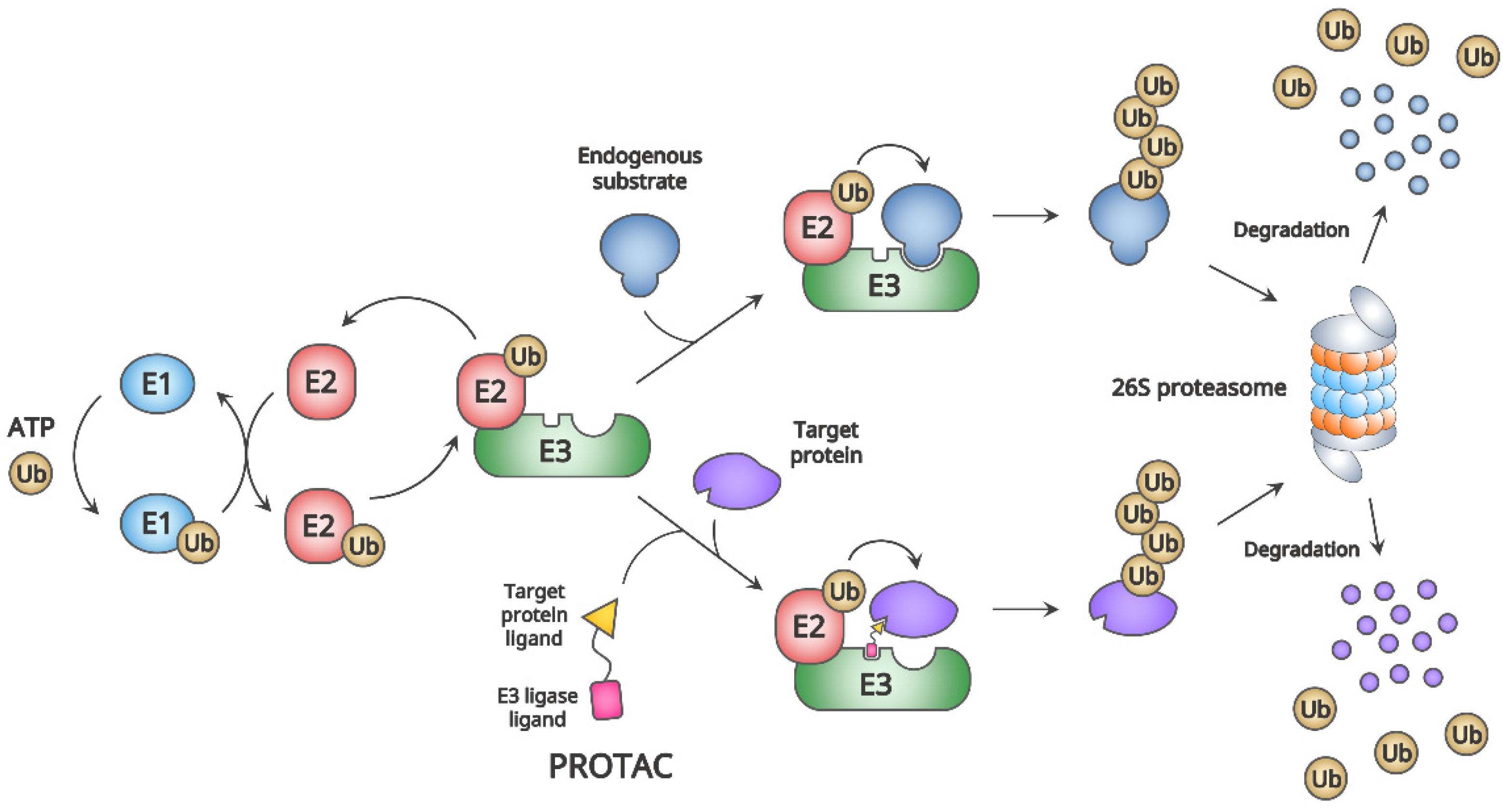
2.1. General Mechanism of PROTAC
2.2. Advantages of PROTAC
3. Small Molecule PROTACs for Molecule-Targeted Cancer Therapies
3.1. CRBN-Based PROTACs
3.2. VHL-Based PROTACs
3.3. MDM2-Based or IAP-Based PROTACs
4. Resistance to PROTACs and Remaining Challenges
5. Expansion of PROTACs beyond CRBN and VHL E3 Ligases
5.1. Possible Alternative E3 Ligases for Novel PROTACs
5.2. Non-Small Molecule PROTACs (NSM-PROTACs)
6. N-degron Pathways as a Possible Novel Strategy for PROTAC
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, N.A.; Stacy, A.E.; Porter, G.M.; Merlot, A.M. Clinical development of targeted and immune based anti-cancer therapies. J. Exp. Clin. Cancer Res. 2019, 38, 156. [Google Scholar] [CrossRef] [PubMed]
- Jana, D.; Zhao, Y. Strategies for enhancing cancer chemodynamic therapy performance. Exploration 2022, 2, 20210238. [Google Scholar] [CrossRef]
- Taefehshokr, N.; Baradaran, B.; Baghbanzadeh, A.; Taefehshokr, S. Promising approaches in cancer immunotherapy. Immunobiology 2020, 225, 151875. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, Y.; Hu, Q. Recent advances in overcoming barriers to cell-based delivery systems for cancer immunotherapy. Exploration 2022, 2, 20210106. [Google Scholar] [CrossRef]
- Asao, T.; Takahashi, F.; Takahashi, K. Resistance to molecularly targeted therapy in non-small-cell lung cancer. Respir. Investig. 2019, 57, 20–26. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef]
- Aldea, M.; Andre, F.; Marabelle, A.; Dogan, S.; Barlesi, F.; Soria, J.C. Overcoming Resistance to Tumor-Targeted and Immune-Targeted Therapies. Cancer Discov. 2021, 11, 874–899. [Google Scholar] [CrossRef]
- Sabnis, A.J.; Bivona, T.G. Principles of Resistance to Targeted Cancer Therapy: Lessons from Basic and Translational Cancer Biology. Trends Mol. Med. 2019, 25, 185–197. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal. Transduct Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Widakowich, C.; De Castro Jr, G.; De Azambuja, E.; Dinh, P.; Awada, A. Review: Side effects of approved molecular targeted therapies in solid cancers. Oncologist 2007, 12, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Braña, I.; Rodon, J.; Taberanero, J. Toxicity as a biomarker of efficacy of molecular targeted therapies: Focus on EGFR and VEGF inhibiting anticancer drugs. Oncologist 2011, 16, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Rudmann, D.G. On-target and off-target-based toxicologic effects. Toxicol. Pathol. 2013, 41, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, T.S.; Wen, P.Y.; Gilbert, M.R.; Schiff, D. Management of treatment-associated toxicites of anti-angiogenic therapy in patients with brain tumors. Neuro-Oncol. 2012, 14, 1203–1214. [Google Scholar] [CrossRef]
- Shoshan, M.C.; Linder, S. Target specificity and off-target effects as determinants of cancer drug efficacy. Expert Opin. Drug Metab. Toxicol. 2008, 4, 273–280. [Google Scholar] [CrossRef]
- Asić, K. Dominant mechanisms of primary resistance differ from dominant mechanisms of secondary resistance to targeted therapies. Crit. Rev. Oncol. Hematol. 2016, 97, 178–196. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018, 834, 188–196. [Google Scholar] [CrossRef]
- Redaelli, S.; Ceccon, M.; Zappa, M.; Sharma, G.G.; Mastini, C.; Mauri, M.; Nigoghossian, M.; Massimino, L.; Cordani, N.; Farina, F.; et al. Lorlatinib treatment elicits multiple on- and off-target mechanisms of resistance in ALK-driven cancer. Cancer Res. 2018, 78, 6866–6880. [Google Scholar] [CrossRef]
- Lyu, N.; Pedersen, B.; Shklovskaya, E.; Rizos, H.; Molloy, M.P.; Wang, Y. SERS characterization of colorectal cancer cell surface markers upon anti-EGFR treatment. Exploration 2022, 2, 20210176. [Google Scholar] [CrossRef]
- Recondo, G.; Bahcall, M.; Spurr, L.F.; Che, J.; Ricciuti, B.; Leonardi, G.C.; Lo, Y.C.; Li, Y.Y.; Lamberti, G.; Nguyen, T.; et al. Molecular Mechanisms of Acquired Resistance to MET Tyrosine Kinase Inhibitors in Patients with MET Exon 14–Mutant NSCLC. Clin. Cancer Res. 2020, 26, 2615–2625. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Kiura, K.; Ueoka, H.; Tabata, M.; Hamasaki, S.; Tanimoto, M. Dramatic effect of ZD1839 (‘Iressa’) in a patient with advanced non-small-cell lung cancer and poor performance status. Lung Cancer 2003, 40, 73–76. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Cheng, Y.; Zhou, C.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.-C.; Majem, M.; Shah, R.; Rukazenkov, Y.; et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann. Oncol. 2018, 29, VIII740. [Google Scholar] [CrossRef]
- Recondo, G.; Facchinetti, F.; Olaussen, K.A.; Besse, B.; Friboulet, L. Making the first move in EGFR-driven or ALK-driven NSCLC: First-generation or next-generation TKI? Nat. Rev. Clin. Oncol. 2018, 15, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.J.; Tan, L.; Lin, J.J.; Wong, S.Q.; Hollizeck, S.; Ebata, K.; Tuch, B.B.; Yoda, S.; Gainor, J.F.; Sequist, L.V.; et al. RET Solvent Front Mutations Mediate Acquired Resistance to Selective RET Inhibition in RET-Driven Malignancies. J. Thorac. Oncol. 2020, 15, 541–549. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Sequist, L.V.; Waltman, B.A.; Dias-Santagata, D.; Digumarthy, S.; Turke, A.B.; Fidias, P.; Bergethon, K.; Shaw, A.T.; Gettinger, S.; Cosper, A.K.; et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011, 3, 75ra26. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.; Park, S.B. Privileged structures: Efficient chemical “navigators” toward unexplored biologically relevant chemical spaces. J. Am. Chem Soc. 2014, 136, 14629–14638. [Google Scholar] [CrossRef]
- Xi, M.; Chen, Y.; Yang, H.; Xu, H.; Du, K.; Wu, C.; Xu, Y.; Deng, L.; Luo, X.; Yu, L.; et al. Small molecule PROTACs in targeted therapy: An emerging strategy to induce protein degradation. Eur. J. Med. Chem. 2019, 174, 159–180. [Google Scholar] [CrossRef]
- Toure, M.; Crews, C.M. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew. Chem. Int. Ed. Engl. 2016, 55, 1966–1973. [Google Scholar] [CrossRef] [PubMed]
- Nero, T.L.; Morton, C.J.; Holien, J.K.; Wielens, J.; Parker, M.W. Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 2014, 14, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 2007, 450, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Buhimschi, A.D.; Armstrong, H.A.; Toure, M.; Jaime-Figueroa, S.; Chen, T.L.; Lehman, A.M.; Woyach, J.A.; Johnson, A.J.; Byrd, J.C.; Crews, C.M. Targeting the C481S Ibrutinib-Resistance Mutation in Bruton’s Tyrosine Kinase Using PROTAC-Mediated Degradation. Biochemistry 2018, 57, 3564–3575. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, N.; Song, Y.; Yang, Z.; Liu, W.; Zhu, J.; Rao, Y. Degradation of Bruton’s tyrosine kinase mutants by PROTACs for potential treatment of ibrutinib-resistant non-Hodgkin lymphomas. Leukemia 2019, 33, 2105–2110. [Google Scholar] [CrossRef]
- Chiron, D.; Di Liberto, M.; Martin, P.; Huang, X.; Sharman, J.; Blecua, P.; Mathew, S.; Vijay, P.; Eng, K.; Ali, S.; et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014, 4, 1022–1035. [Google Scholar] [CrossRef]
- Cheng, S.; Guo, A.; Lu, P.; Ma, J.; Coleman, M.; Wang, Y.L. Functional characterization of BTK(C481S) mutation that confers ibrutinib resistance: Exploration of alternative kinase inhibitors. Leukemia 2015, 29, 895–900. [Google Scholar] [CrossRef]
- Zorba, A.; Nguyen, C.; Xu, Y.; Starr, J.; Borzilleri, K.; Smith, J.; Zhu, H.; Farley, K.A.; Ding, W.; Schiemer, J.; et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc. Natl. Acad. Sci. USA 2018, 115, E7285–E7292. [Google Scholar] [CrossRef]
- Niederst, M.J.; Hu, H.; Mulvey, H.E.; Lockerman, E.L.; Garcia, A.R.; Piotrowska, Z.; Sequist, L.V.; Engelman, J.A. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin. Cancer Res. 2015, 21, 3924–3933. [Google Scholar] [CrossRef]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef] [Green Version]
- Chong, C.R.; Janne, P.A. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 2013, 19, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Pao, W. EGFR mutant lung cancer. Curr. Top. Microbiol. Immunol. 2012, 355, 59–81. [Google Scholar] [CrossRef]
- Volpe, G.; Panuzzo, C.; Ulisciani, S.; Cilloni, D. Imatinib resistance in CML. Cancer Lett. 2009, 274, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Niederst, M.J.; Engelman, J.A. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci. Signal. 2013, 6, re6. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef]
- Hafeez, U.; Gan, H.K.; Scott, A.M. Monoclonal antibodies as immunomodulatory therapy against cancer and autoimmune diseases. Curr. Opin. Pharm. 2018, 41, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Gharwan, H.; Groninger, H. Kinase inhibitors and monoclonal antibodies in oncology: Clinical implications. Nat. Rev. Clin. Oncol 2016, 13, 209–227. [Google Scholar] [CrossRef]
- Edmondson, S.D.; Yang, B.; Fallan, C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: Recent progress and future challenges. Bioorg. Med. Chem. Lett. 2019, 29, 1555–1564. [Google Scholar] [CrossRef]
- Coats, S.; Williams, M.; Kebble, B.; Dixit, R.; Tseng, L.; Yao, N.S.; Tice, D.A.; Soria, J.C. Antibody-Drug Conjugates: Future Directions in Clinical and Translational Strategies to Improve the Therapeutic Index. Clin. Cancer Res. 2019, 25, 5441–5448. [Google Scholar] [CrossRef]
- Wolska-Washer, A.; Robak, T. Safety and Tolerability of Antibody-Drug Conjugates in Cancer. Drug Saf. 2019, 42, 295–314. [Google Scholar] [CrossRef] [Green Version]
- Bekes, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Kastl, J.M.; Davies, G.; Godsman, E.; Holdgate, G.A. Small-Molecule Degraders beyond PROTACs-Challenges and Opportunities. SLAS Discov. 2021, 26, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.J.; Testa, A.; Thompson, N.; Churcher, I. The rise and rise of protein degradation: Opportunities and challenges ahead. Drug Discov. Today 2021, 26, 2889–2897. [Google Scholar] [CrossRef] [PubMed]
- Alabi, S.B.; Crews, C.M. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Moriyama, J.; Nakamura, T.; Miki, E.; Takahashi, E.; Sato, A.; Akaike, T.; Itto-Nakama, K.; Arimoto, H. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810.e10. [Google Scholar] [CrossRef]
- Banik, S.M.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.M.; Bertozzi, C.R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef]
- Cotton, A.D.; Nguyen, D.P.; Gramespacher, J.A.; Seiple, I.B.; Wells, J.A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef]
- Sakamoto, K.M. Chimeric molecules to target proteins for ubiquitination and degradation. Methods Enzymol. 2005, 399, 833–847. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Mares, A.; Smith, I.E.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Qian, Y.; Altieri, M.; Dong, H.; Wang, J.; Raina, K.; Hines, J.; Winkler, J.D.; Crew, A.P.; Coleman, K.; et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Swamy, K.B.S.; Schuyler, S.C.; Leu, J.Y. Protein Complexes Form a Basis for Complex Hybrid Incompatibility. Front. Genet. 2021, 12, 609766. [Google Scholar] [CrossRef] [PubMed]
- Gorka, M.; Swart, C.; Siemiatkowska, B.; Martinez-Jaime, S.; Skirycz, A.; Streb, S.; Graf, A. Protein Complex Identification and quantitative complexome by CN-PAGE. Sci. Rep. 2019, 9, 11523. [Google Scholar] [CrossRef]
- Woodsmith, J.; Stelzl, U. Studying post-translational modifications with protein interaction networks. Curr. Opin. Struct. Biol. 2014, 24, 34–44. [Google Scholar] [CrossRef]
- Skanland, S.S.; Mato, A.R. Overcoming resistance to targeted therapies in chronic lymphocytic leukemia. Blood Adv. 2021, 5, 334–343. [Google Scholar] [CrossRef]
- Zeng, S.; Huang, W.; Zheng, X.; Liyan, C.; Zhang, Z.; Wang, J.; Shen, Z. Proteolysis targeting chimera (PROTAC) in drug discovery paradigm: Recent progress and future challenges. Eur. J. Med. Chem. 2021, 210, 112981. [Google Scholar] [CrossRef]
- Li, X.; Pu, W.; Zheng, Q.; Ai, M.; Chen, S.; Peng, Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol. Cancer 2022, 21, 99. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Coleman, K.G.; Crews, C.M. Proteolysis-Targeting Chimeras: Harnessing the Ubiquitin-Proteasome System to Induce Degradation of Specific Target Proteins. Annu. Rev. Cancer Biol. 2018, 2, 41–58. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Ishikawa, M.; Naito, M.; Hashimoto, Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: Design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J. Am. Chem Soc. 2010, 132, 5820–5826. [Google Scholar] [CrossRef] [PubMed]
- Okuhira, K.; Ohoka, N.; Sai, K.; Nishimaki-Mogami, T.; Itoh, Y.; Ishikawa, M.; Hashimoto, Y.; Naito, M. Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS Lett. 2011, 585, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Ohoka, N.; Okuhira, K.; Ito, M.; Nagai, K.; Shibata, N.; Hattori, T.; Ujikawa, O.; Shimokawa, K.; Sano, O.; Koyama, R.; et al. In Vivo Knockdown of Pathogenic Proteins via Specific and Nongenetic Inhibitor of Apoptosis Protein (IAP)-dependent Protein Erasers (SNIPERs). J. Biol. Chem. 2017, 292, 4556–4570. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ando, H.; Suzuki, T.; Ogura, T.; Hotta, K.; Imamura, Y.; Yamaguchi, Y.; Handa, H. Identification of a primary target of thalidomide teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef]
- Lopez-Girona, A.; Mendy, D.; Ito, T.; Miller, K.; Gandhi, A.K.; Kang, J.; Karasawa, S.; Carmel, G.; Jackson, P.; Abbasian, M.; et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 2012, 26, 2326–2335. [Google Scholar] [CrossRef]
- Buckley, D.L.; Gustafson, J.L.; Van Molle, I.; Roth, A.G.; Tae, H.S.; Gareiss, P.C.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew. Chem. Int. Ed. Engl. 2012, 51, 11463–11467. [Google Scholar] [CrossRef]
- Buckley, D.L.; Van Molle, I.; Gareiss, P.C.; Tae, H.S.; Michel, J.; Noblin, D.J.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J. Am. Chem Soc. 2012, 134, 4465–4468. [Google Scholar] [CrossRef]
- Fischer, E.S.; Bohm, K.; Lydeard, J.R.; Yang, H.; Stadler, M.B.; Cavadini, S.; Nagel, J.; Serluca, F.; Acker, V.; Lingaraju, G.M.; et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 2014, 512, 49–53. [Google Scholar] [CrossRef]
- Gandhi, A.K.; Kang, J.; Havens, C.G.; Conklin, T.; Ning, Y.; Wu, L.; Ito, T.; Ando, H.; Waldman, M.F.; Thakurta, A.; et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol 2014, 164, 811–821. [Google Scholar] [CrossRef]
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.; Ott, C.J.; Mitsiades, C.S.; Wong, K.K.; Bradner, J.E.; Kaelin, W.G., Jr. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef] [Green Version]
- Kronke, J.; Fink, E.C.; Hollenbach, P.W.; MacBeth, K.J.; Hurst, S.N.; Udeshi, N.D.; Chamberlain, P.P.; Mani, D.R.; Man, H.W.; Gandhi, A.K.; et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 2015, 523, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Kronke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.E.; Buckley, D.L.; Paulk, J.; Roberts, J.M.; Souza, A.; Dhe-Paganon, S.; Bradner, J.E. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Su, H.; Li, Y.; Luo, C.; Xu, H.; Wang, Y.; Sun, H.; Wan, G.; Zhou, B.; Bu, X. Degradation of intracellular TGF-beta1 by PROTACs efficiently reverses M2 macrophage induced malignant pathological events. Chem. Commun. 2020, 56, 2881–2884. [Google Scholar] [CrossRef]
- Sun, D.Y.; Wu, J.Q.; He, Z.H.; He, M.F.; Sun, H.B. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-beta signaling pathway. Life Sci. 2019, 235, 116791. [Google Scholar] [CrossRef]
- Dai, G.; Sun, B.; Gong, T.; Pan, Z.; Meng, Q.; Ju, W. Ginsenoside Rb2 inhibits epithelial-mesenchymal transition of colorectal cancer cells by suppressing TGF-beta/Smad signaling. Phytomedicine 2019, 56, 126–135. [Google Scholar] [CrossRef]
- He, M.; Lv, W.; Rao, Y. Opportunities and Challenges of Small Molecule Induced Targeted Protein Degradation. Front. Cell Dev. Biol. 2021, 9, 685106. [Google Scholar] [CrossRef]
- Li, X.; Song, Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020, 13, 50. [Google Scholar] [CrossRef]
- Qi, S.M.; Dong, J.; Xu, Z.Y.; Cheng, X.D.; Zhang, W.D.; Qin, J.J. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front. Pharm. 2021, 12, 692574. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Yang, H.; Fribourgh, J.L.; Wolfe, L.S.; Xiong, Y. Insights into Cullin-RING E3 ubiquitin ligase recruitment: Structure of the VHL-EloBC-Cul2 complex. Structure 2015, 23, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wu, T.; Simon, J.; Takada, M.; Saito, R.; Fan, C.; Liu, X.D.; Jonasch, E.; Xie, L.; Chen, X.; et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 2018, 361, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Q. VHL and Hypoxia Signaling: Beyond HIF in Cancer. Biomedicines 2018, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, A.; Carico, E. Role of HIF-1 in Cancer Progression: Novel Insights. A Review. Curr. Mol. Med. 2018, 18, 343–351. [Google Scholar] [CrossRef]
- Raina, K.; Lu, J.; Qian, Y.; Altieri, M.; Gordon, D.; Rossi, A.M.; Wang, J.; Chen, X.; Dong, H.; Siu, K.; et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 7124–7129. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.J.; Li, H.; Zhou, Z.; Izquierdo-Ferrer, J.; Xue, Y.; Wavelet, C.M.; Schiltz, G.E.; Zhang, B.; Cristofanilli, M.; Lu, X.; et al. A novel strategy to block mitotic progression for targeted therapy. EBioMedicine 2019, 49, 40–54. [Google Scholar] [CrossRef]
- Khan, S.; Zhang, X.; Lv, D.; Zhang, Q.; He, Y.; Zhang, P.; Liu, X.; Thummuri, D.; Yuan, Y.; Wiegand, J.S.; et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019, 25, 1938–1947. [Google Scholar] [CrossRef]
- Han, X.; Zhao, L.; Xiang, W.; Qin, C.; Miao, B.; Xu, T.; Wang, M.; Yang, C.Y.; Chinnaswamy, K.; Stuckey, J.; et al. Discovery of Highly Potent and Efficient PROTAC Degraders of Androgen Receptor (AR) by Employing Weak Binding Affinity VHL E3 Ligase Ligands. J. Med. Chem. 2019, 62, 11218–11231. [Google Scholar] [CrossRef]
- Hou, H.; Sun, D.; Zhang, X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019, 19, 216. [Google Scholar] [CrossRef]
- Wang, W.; Qin, J.J.; Rajaei, M.; Li, X.; Yu, X.; Hunt, C.; Zhang, R. Targeting MDM2 for novel molecular therapy: Beyond oncology. Med. Res. Rev. 2020, 40, 856–880. [Google Scholar] [CrossRef]
- Schneekloth, A.R.; Pucheault, M.; Tae, H.S.; Crews, C.M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008, 18, 5904–5908. [Google Scholar] [CrossRef] [Green Version]
- Hines, J.; Lartigue, S.; Dong, H.; Qian, Y.; Crews, C.M. MDM2-Recruiting PROTAC Offers Superior, Synergistic Antiproliferative Activity via Simultaneous Degradation of BRD4 and Stabilization of p53. Cancer Res. 2019, 79, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Lan, T.; Su, S.; Rao, Y. Induction of apoptosis in MDA-MB-231 breast cancer cells by a PARP1-targeting PROTAC small molecule. Chem. Commun. 2019, 55, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Ohoka, N.; Ujikawa, O.; Shimokawa, K.; Sameshima, T.; Shibata, N.; Hattori, T.; Nara, H.; Cho, N.; Naito, M. Different Degradation Mechanisms of Inhibitor of Apoptosis Proteins (IAPs) by the Specific and Nongenetic IAP-Dependent Protein Eraser (SNIPER). Chem. Pharm Bull. 2019, 67, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, M.; Li, J.; Zhang, B.; Wang, Z.; Shaabani, S.; Ter Brake, F.; Essa, K.; Domling, A. PROTACs- a game-changing technology. Expert Opin. Drug Discov. 2019, 14, 1255–1268. [Google Scholar] [CrossRef]
- Schapira, M.; Calabrese, M.F.; Bullock, A.N.; Crews, C.M. Targeted protein degradation: Expanding the toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. [Google Scholar] [CrossRef]
- Zhang, L.; Riley-Gillis, B.; Vijay, P.; Shen, Y. Acquired Resistance to BET-PROTACs (Proteolysis-Targeting Chimeras) Caused by Genomic Alterations in Core Components of E3 Ligase Complexes. Mol. Cancer Ther. 2019, 18, 1302–1311. [Google Scholar] [CrossRef]
- Ottis, P.; Palladino, C.; Thienger, P.; Britschgi, A.; Heichinger, C.; Berrera, M.; Julien-Laferriere, A.; Roudnicky, F.; Kam-Thong, T.; Bischoff, J.R.; et al. Cellular Resistance Mechanisms to Targeted Protein Degradation Converge Toward Impairment of the Engaged Ubiquitin Transfer Pathway. ACS Chem. Biol. 2019, 14, 2215–2223. [Google Scholar] [CrossRef]
- Shirasaki, R.; Matthews, G.M.; Gandolfi, S.; de Matos Simoes, R.; Buckley, D.L.; Raja Vora, J.; Sievers, Q.L.; Bruggenthies, J.B.; Dashevsky, O.; Poarch, H.; et al. Functional Genomics Identify Distinct and Overlapping Genes Mediating Resistance to Different Classes of Heterobifunctional Degraders of Oncoproteins. Cell Rep. 2021, 34, 108532. [Google Scholar] [CrossRef]
- Mogollon, P.; Diaz-Tejedor, A.; Algarin, E.M.; Paino, T.; Garayoa, M.; Ocio, E.M. Biological Background of Resistance to Current Standards of Care in Multiple Myeloma. Cells 2019, 8, 1432. [Google Scholar] [CrossRef]
- Ohtake, F.; Baba, A.; Takada, I.; Okada, M.; Iwasaki, K.; Miki, H.; Takahashi, S.; Kouzmenko, A.; Nohara, K.; Chiba, T.; et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 2007, 446, 562–566. [Google Scholar] [CrossRef]
- Ohoka, N.; Tsuji, G.; Shoda, T.; Fujisato, T.; Kurihara, M.; Demizu, Y.; Naito, M. Development of Small Molecule Chimeras That Recruit AhR E3 Ligase to Target Proteins. ACS Chem. Biol. 2019, 14, 2822–2832. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Liu, T.; Jiao, Q.; Ji, J.; Tao, M.; Liu, Y.; You, Q.; Jiang, Z. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 2018, 146, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Luo, M.; Xie, Y.; Spradlin, J.N.; Tallarico, J.A.; McKenna, J.M.; Schirle, M.; Maimone, T.J.; Nomura, D.K. Bardoxolone conjugation enables targeted protein degradation of BRD4. Sci. Rep. 2020, 10, 15543. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Meng, F.; Park, K.S.; Yim, H.; Velez, J.; Kumar, P.; Wang, L.; Xie, L.; Chen, H.; Shen, Y.; et al. Harnessing the E3 Ligase KEAP1 for Targeted Protein Degradation. J. Am. Chem. Soc. 2021, 143, 15073–15083. [Google Scholar] [CrossRef]
- Li, L.; Mi, D.; Pei, H.; Duan, Q.; Wang, X.; Zhou, W.; Jin, J.; Li, D.; Liu, M.; Chen, Y. In vivo target protein degradation induced by PROTACs based on E3 ligase DCAF15. Signal Transduct. Target. Ther. 2020, 5, 129. [Google Scholar] [CrossRef]
- Ward, C.C.; Kleinman, J.I.; Brittain, S.M.; Lee, P.S.; Chung, C.Y.S.; Kim, K.; Petri, Y.; Thomas, J.R.; Tallarico, J.A.; McKenna, J.M.; et al. Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. ACS Chem. Biol. 2019, 14, 2430–2440. [Google Scholar] [CrossRef]
- Spradlin, J.N.; Hu, X.; Ward, C.C.; Brittain, S.M.; Jones, M.D.; Ou, L.; To, M.; Proudfoot, A.; Ornelas, E.; Woldegiorgis, M.; et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 2019, 15, 747–755. [Google Scholar] [CrossRef]
- Luo, M.; Spradlin, J.N.; Boike, L.; Tong, B.; Brittain, S.M.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Maimone, T.J.; Nomura, D.K. Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem. Biol. 2021, 28, 559–566.e15. [Google Scholar] [CrossRef]
- Henning, N.J.; Manford, A.G.; Spradlin, J.N.; Brittain, S.M.; Zhang, E.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Rape, M.; Nomura, D.K. Discovery of a Covalent FEM1B Recruiter for Targeted Protein Degradation Applications. J. Am. Chem. Soc. 2022, 144, 701–708. [Google Scholar] [CrossRef]
- Zhang, X.; Crowley, V.M.; Wucherpfennig, T.G.; Dix, M.M.; Cravatt, B.F. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol. 2019, 15, 737–746. [Google Scholar] [CrossRef]
- Zhang, X.; Luukkonen, L.M.; Eissler, C.L.; Crowley, V.M.; Yamashita, Y.; Schafroth, M.A.; Kikuchi, S.; Weinstein, D.S.; Symons, K.T.; Nordin, B.E.; et al. DCAF11 Supports Targeted Protein Degradation by Electrophilic Proteolysis-Targeting Chimeras. J. Am. Chem. Soc. 2021, 143, 5141–5149. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ciulli, A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discov. 2021, 26, 484–502. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted protein degradation: Mechanisms, strategies and application. Signal. Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Banik, S.M.; Miller, C.L.; Riley, N.M.; Cochran, J.R.; Bertozzi, C.R. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol. 2021, 17, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Gao, Q.; Mao, M.; Zhang, C.; Yang, L.; Yang, Y.; Han, D. Bispecific Aptamer Chimeras Enable Targeted Protein Degradation on Cell Membranes. Angew. Chem. Int. Ed. 2021, 60, 11267–11271. [Google Scholar] [CrossRef]
- Zebisch, M.; Xu, Y.; Krastev, C.; Macdonald, B.T.; Chen, M.; Gilbert, R.J.C.; He, X.; Jones, E.Y. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun. 2013, 4, 2787. [Google Scholar] [CrossRef]
- Verdurmen, W.P.R.; Mazlami, M.; Plückthun, A. A quantitative comparison of cytosolic delivery via different protein uptake systems. Sci. Rep. 2017, 7, 13194. [Google Scholar] [CrossRef]
- Han, Y.; Da, Y.; Yu, M.; Cheng, Y.; Wang, X.; Xiong, J.; Guo, G.; Li, Y.; Jiang, X.; Cai, X. Protein labeling approach to improve lysosomal targeting and efficacy of antibody-drug conjugates. Org. Biomol. Chem. 2020, 18, 3229–3233. [Google Scholar] [CrossRef]
- Zhang, H.; Han, Y.; Yang, Y.; Lin, F.; Li, K.; Kong, L.; Liu, H.; Dang, Y.; Lin, J.; Chen, P.R. Covalently Engineered Nanobody Chimeras for Targeted Membrane Protein Degradation. J. Am. Chem. Soc. 2021, 143, 16377–16382. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Grumati, P.; Dikic, I. Ubiquitin signaling and autophagy. J. Biol. Chem. 2018, 293, 5404–5413. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.H.; Kwon, Y.T. Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.M.; Choi, H.R.; Sung, K.W.; Lee, Y.J.; Kim, S.T.; Kim, D.; Mun, S.R.; Hwang, J.; Cha-Molstad, H.; Ciechanover, A.; et al. The endoplasmic reticulum-residing chaperone BiP is short-lived and metabolized through N-terminal arginylation. Sci. Signal. 2018, 11, eaan0630. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.D.; Mun, S.R.; Ji, C.H.; Sung, K.W.; Kang, K.Y.; Heo, A.J.; Lee, S.H.; An, J.Y.; Hwang, J.; Xie, X.Q.; et al. N-terminal arginylation generates a bimodal degron that modulates autophagic proteolysis. Proc. Natl. Acad. Sci. USA 2018, 115, E2716–E2724. [Google Scholar] [CrossRef]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-End rule pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell. Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef]
- Dissmeyer, N.; Rivas, S.; Graciet, E. Life and death of proteins after protease cleavage: Protein degradation by the N-end rule pathway. New Phytol. 2018, 218, 929–935. [Google Scholar] [CrossRef]
- Eldeeb, M.A.; Leitao, L.C.A.; Fahlman, R.P. Emerging branches of the N-end rule pathways are revealing the sequence complexities of N-termini dependent protein degradation. Biochem. Cell Biol. 2018, 96, 289–294. [Google Scholar] [CrossRef]
- Bachmair, A.; Varshavsky, A. The degradation signal in a short-lived protein. Cell 1989, 56, 1019–1032. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, R.R.; Oh, J.H.; Cho, H.; Varshavsky, A.; Hwang, C.S. The N-terminal methionine of cellular proteins as a degradation signal. Cell 2014, 156, 158–169. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.S.; Shemorry, A.; Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 2010, 327, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Park, S.E.; Kim, J.M.; Seok, O.H.; Cho, H.; Wadas, B.; Kim, S.Y.; Varshavsky, A.; Hwang, C.S. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 2015, 347, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Wu, X.; Wadas, B.; Oh, J.H.; Varshavsky, A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 2017, 355, eaal3655. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zhang, H.; Li, L.; Tempel, W.; Loppnau, P.; Min, J. Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway. Nat. Chem. Biol. 2018, 14, 466–473. [Google Scholar] [CrossRef]
- Kim, J.M.; Seok, O.H.; Ju, S.; Heo, J.E.; Yeom, J.; Kim, D.S.; Yoo, J.Y.; Varshavsky, A.; Lee, C.; Hwang, C.S. Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway. Science 2018, 362, eaat0174. [Google Scholar] [CrossRef]
- Kim, J.M. N-terminal formylmethionine as a novel initiator and N-degron of eukaryotic proteins. BMB Rep. 2019, 52, 163–164. [Google Scholar] [CrossRef]
- Bachmair, A.; Finley, D.; Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 1986, 234, 179–186. [Google Scholar] [CrossRef]
- Eldeeb, M.A. N-Terminal-Dependent Protein Degradation and Targeting Cancer Cells. Anticancer Agents Med. Chem. 2021, 21, 231–236. [Google Scholar] [CrossRef]
- Eldeeb, M.A.; Zorca, C.E.; Fahlman, R.P. Targeting Cancer Cells via N-degron-based PROTACs. Endocrinology 2020, 161, bqaa185. [Google Scholar] [CrossRef]
- Shanmugasundaram, K.; Shao, P.; Chen, H.; Campos, B.; McHardy, S.F.; Luo, T.; Rao, H. A modular PROTAC design for target destruction using a degradation signal based on a single amino acid. J. Biol. Chem. 2019, 294, 15172–15175. [Google Scholar] [CrossRef]
- Lee, Y.; Heo, J.; Jeong, H.; Hong, K.T.; Kwon, D.H.; Shin, M.H.; Oh, M.; Sable, G.A.; Ahn, G.O.; Lee, J.S.; et al. Targeted Degradation of Transcription Coactivator SRC-1 through the N-Degron Pathway. Angew. Chem. Int. Ed. Engl. 2020, 59, 17548–17555. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Liu, Z.; Chen, H.; Xu, J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009, 69, 3819–3827. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.A.; Qin, L.; Tien, J.C.; Young, L.S.; Xu, J. The function of steroid receptor coactivator-1 in normal tissues and cancer. Int. J. Biol. Sci. 2012, 8, 470–485. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Yeh, C.W.; Chen, Y.F.; Lee, T.T.; Hsieh, P.Y.; Rusnac, D.V.; Lin, S.Y.; Elledge, S.J.; Zheng, N.; Yen, H.C.S. C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol. Cell 2018, 70, 602–613.e3. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.W.; Huang, W.C.; Hsu, P.H.; Yeh, K.H.; Wang, L.C.; Hsu, P.W.C.; Lin, H.C.; Chen, Y.N.; Chen, S.C.; Yeang, C.H.; et al. The C-degron pathway eliminates mislocalized proteins and products of deubiquitinating enzymes. EMBO J. 2021, 40, e105846. [Google Scholar] [CrossRef] [PubMed]
- Timms, R.T.; Koren, I. Tying up loose ends: The N-degron and C-degron pathways of protein degradation. Biochem. Soc. Trans. 2020, 48, 1557–1567. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Park, J.; Kim, J.-M. Targeted Protein Degradation to Overcome Resistance in Cancer Therapies: PROTAC and N-Degron Pathway. Biomedicines 2022, 10, 2100. https://doi.org/10.3390/biomedicines10092100
Kim H, Park J, Kim J-M. Targeted Protein Degradation to Overcome Resistance in Cancer Therapies: PROTAC and N-Degron Pathway. Biomedicines. 2022; 10(9):2100. https://doi.org/10.3390/biomedicines10092100
Chicago/Turabian StyleKim, Hanbyeol, Jeongbae Park, and Jeong-Mok Kim. 2022. "Targeted Protein Degradation to Overcome Resistance in Cancer Therapies: PROTAC and N-Degron Pathway" Biomedicines 10, no. 9: 2100. https://doi.org/10.3390/biomedicines10092100
APA StyleKim, H., Park, J., & Kim, J.-M. (2022). Targeted Protein Degradation to Overcome Resistance in Cancer Therapies: PROTAC and N-Degron Pathway. Biomedicines, 10(9), 2100. https://doi.org/10.3390/biomedicines10092100





