In Vitro Evidence of Statins’ Protective Role against COVID-19 Hallmarks
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Reagents, Treatments, and Antibodies
2.3. Cell Viability Assay
2.4. Western Blot Analysis
2.5. Plasma Membrane-Derived Lipid Rafts Isolation
2.6. Confocal Microscopy
2.7. Analysis of Cytokines and Chemokines Profiles
2.8. Statistical Analysis
3. Results
3.1. Statins Modulate ACE2 Receptor and TMPRSS2 Expression
3.2. Statins Counteract the Effect of Induced Inflammation
3.3. Effect of Statins on Chemokine/Cytokine Profile
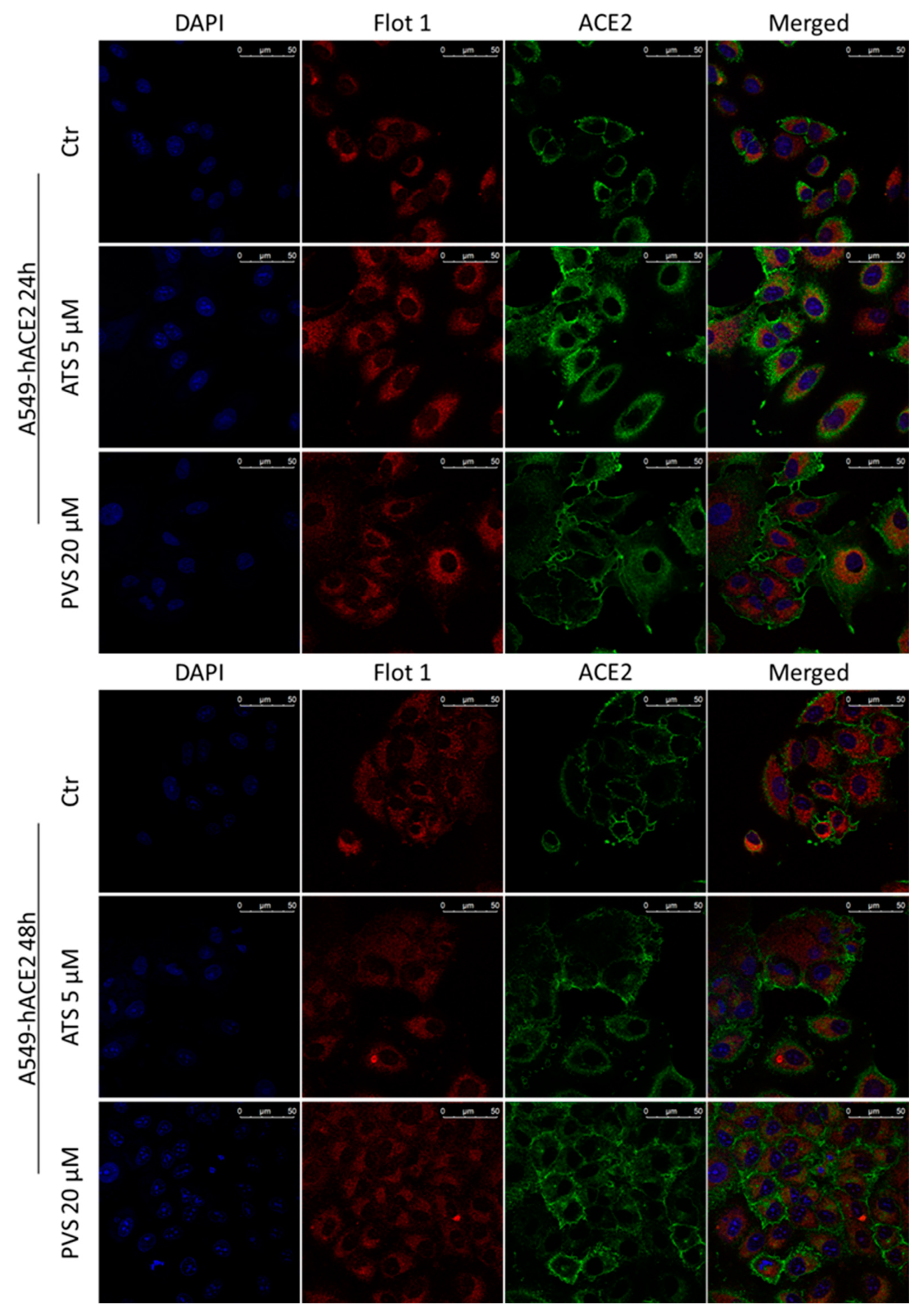
3.4. Hydrophilic and Lipophilic Statins Differentially Affect ACE2 Receptor Localization in Lipid Rafts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| EGF | Epidermal growth factor |
| ENA-78 | Epithelial-neutrophil activating peptide |
| GCSF | granulocyte colony-stimulating factor |
| GM-CSF | Granulocyte-Macrophage Colony-Stimulating Factor |
| GRO | growth-regulated oncogene |
| IFN-γ | Interferon gamma |
| IGF-I | Insulin-like growth factor-1 |
| IL | Interleukin |
| IP-10 | Interferon gamma-induced protein 10 |
| MCP1 | Monocyte chemoattractant protein-1 |
| MCP2 | Monocyte chemoattractant protein-2 |
| MCP3 | Monocyte chemoattractant protein-3 |
| MCSF | macrophage colony-stimulating factor |
| MDC | Macrophage-derived chemokine |
| MIG | Monokine induced by gamma |
| MIP1 | Macrophage Inflammatory Protein-1 |
| PDGF-BB | Platelet-Derived Growth Factor-BB |
| RANTES | Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted |
| SCF | Stem cell factor |
| SDF1 | stromal cell-derived factor 1 |
| TARC | Thymus and activation-regulated chemokine |
| TGF | transforming growth factor |
| TNFβ | Tumor necrosis factor-β |
| TNFα | Tumor necrosis factor-α |
References
- Jiménez, D.; Arias, M.T. Immunouniverse of SARS-CoV-2. Immunol. Med. 2022, 1–39. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Palacios-Rápalo, S.N.; De Jesús-González, L.A.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Osuna-Ramos, J.F.; Martínez-Mier, G.; Quistián-Galván, J.; Muñoz-Pérez, A.; Bernal-Dolores, V.; del Ángel, R.M.; et al. Cholesterol-Rich Lipid Rafts as Platforms for SARS-CoV-2 Entry. Front. Immunol. 2021, 12, 796855. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Irani, S. Immune responses in SARS-CoV-2, SARS-CoV, and MERS-CoV infections: A comparative review. Int. J. Prev. Med. 2022, 13, 45. [Google Scholar] [CrossRef]
- Fauvel, C.; Trimaille, A.; Weizman, O.; Pezel, T.; Mika, D.; Waldmann, V.; Cohen, A.; Bonnet, G. Cardiovascular manifestations secondary to COVID-19: A narrative review. Respir. Med. Res. 2022, 81, 100904. [Google Scholar] [CrossRef]
- Louis, D.W.; Saad, M.; Vijayakumar, S.; Ilyas, S.; Kokkirala, A.; Aronow, H.D. The Cardiovascular Manifestations of COVID-19. Cardiol. Clin. 2022, 40, 277–285. [Google Scholar] [CrossRef]
- Tobler, D.L.; Pruzansky, A.J.; Naderi, S.; Ambrosy, A.P.; Slade, J.J. Long-Term Cardiovascular Effects of COVID-19: Emerging Data Relevant to the Cardiovascular Clinician. Curr. Atheroscler. Rep. 2022, 24, 563–570. [Google Scholar] [CrossRef]
- Pandey, R.; Rai, D.; Tahir, M.; Wahab, A.; Bandyopadhyay, D.; Lesho, E.; Laguio-Vila, M.; Fentanes, E.; Tariq, R.; Naidu, S.; et al. Prevalence of comorbidities and symptoms stratified by severity of illness amongst adult patients with COVID-19: A systematic review. Arch. Med. Sci. Atheroscler. Dis. 2022, 7, 5–23. [Google Scholar] [CrossRef]
- Bifulco, M.; Ciccarelli, M.; Bruzzese, D.; Dipasquale, A.; Lania, A.G.; Mazziotti, G.; Gazzerro, P. The benefit of statins in SARS-CoV-2 patients: Further metabolic and prospective clinical studies are needed. Endocrine 2021, 71, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Bifulco, M.; Gazzerro, P. Statin therapy in COVID-19 infection: Much more than a single pathway. Eur. Heart J. Cardiovasc. Pharmacother. 2020, 6, 410–411. [Google Scholar] [CrossRef] [PubMed]
- Proto, M.C.; Fiore, D.; Piscopo, C.; Pagano, C.; Galgani, M.; Bruzzaniti, S.; Laezza, C.; Gazzerro, P.; Bifulco, M. Lipid homeostasis and mevalonate pathway in COVID-19: Basic concepts and potential therapeutic targets. Prog. Lipid Res. 2021, 82, 101099. [Google Scholar] [CrossRef] [PubMed]
- Torres-Peña, J.D.; Katsiki, N.; Perez-Martinez, P. Could Statin Therapy Be Useful in Patients with Coronavirus Disease 2019 (COVID-19)? Front. Cardiovasc. Med. 2021, 8, 775749. [Google Scholar] [CrossRef]
- Patel, K.K.; Sehgal, V.S.; Kashfi, K. Molecular targets of statins and their potential side effects: Not all the glitter is gold. Eur. J. Pharmacol. 2022, 922, 174906. [Google Scholar] [CrossRef]
- Subir, R.; Jagat, J.M.; Kalyan, K.G. Pros and cons for use of statins in people with coronavirus disease-19 (COVID-19). Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1225–1229. [Google Scholar] [CrossRef]
- Rodrigues-Diez, R.R.; Tejera-Muñoz, A.; Marquez-Exposito, L.; Rayego-Mateos, S.; Sanchez, L.S.; Marchant, V.; Santamaria, L.T.; Ramos, A.M.; Ortiz, A.; Egido, J.; et al. Statins: Could an old friend help in the fight against COVID-19? Br. J. Pharmacol. 2020, 177, 4873–4886. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Poterucha, T.J.; DeFilippis, E.M.; Hennessey, J.A.; Redfors, B.; Eckhardt, C.; Bikdeli, B.; Platt, J.; Nalbandian, A.; et al. Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat. Commun. 2021, 12, 1325. [Google Scholar] [CrossRef]
- Kuno, T.; So, M.; Iwagami, M.; Takahashi, M.; Egorova, N.N. The association of statins use with survival of patients with COVID-19. J. Cardiol. 2022, 79, 494–500. [Google Scholar] [CrossRef]
- Vahedian-Azimi, A.; Mohammadi, S.M.; Banach, M.; Beni, F.H.; Guest, P.C.; Al-Rasadi, K.; Jamialahmadi, T.; Sahebkar, A. Improved COVID-19 Outcomes following Statin Therapy: An Updated Systematic Review and Meta-analysis. BioMed Res. Int. 2021, 2021, 1901772. [Google Scholar] [CrossRef]
- Diaz-Arocutipa, C.; Melgar-Talavera, B.; Alvarado-Yarasca, Á.; Saravia-Bartra, M.M.; Cazorla, P.; Belzusarri, I.; Hernandez, A.V. Statins reduce mortality in patients with COVID-19: An updated meta-analysis of 147 824 patients. Int. J. Infect. Dis. 2021, 110, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Andrews, L.; Goldin, L.; Shen, Y.; Korwek, K.; Kleja, K.; Poland, R.E.; Guy, J.; Sands, K.E.; Perlin, J.B. Discontinuation of atorvastatin use in hospital is associated with increased risk of mortality in COVID-19 patients. J. Hosp. Med. 2022, 17, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Cardona, M.I.; Flórez-Álvarez, L.; Zapata-Builes, W.; Guerra-Sandoval, A.L.; Guerra-Almonacid, C.M.; Hincapié-García, J.; Rugeles, M.T.; Hernandez, J.C. Atorvastatin Effectively Inhibits Ancestral and Two Emerging Variants of SARS-CoV-2 in vitro. Front. Microbiol. 2022, 13, 721103. [Google Scholar] [CrossRef]
- Teixeira, L.; Temerozo, J.R.; Pereira-Dutra, F.S.; Ferreira, A.C.; Mattos, M.; Gonçalves, B.S.; Sacramento, C.Q.; Palhinha, L.; Cunha-Fernandes, T.; Dias, S.S.; et al. Simvastatin Downregulates the SARS-CoV-2-Induced Inflammatory Response and Impairs Viral Infection Through Disruption of Lipid Rafts. Front. Immunol. 2022, 13, 820131. [Google Scholar] [CrossRef] [PubMed]
- Zapatero-Belinchón, F.J.; Moeller, R.; Lasswitz, L.; van Ham, M.; Becker, M.; Brogden, G.; Rosendal, E.; Bi, W.; Carriquí-Madroñal, B.; Islam, K.; et al. Fluvastatin mitigates SARS-CoV-2 infection in human lung cells. iScience 2021, 24, 103469. [Google Scholar] [CrossRef]
- Proto, M.C.; Fiore, D.; Piscopo, C.; Laezza, C.; Bifulco, M.; Gazzerro, P. Modified Adenosines Sensitize Glioblastoma Cells to Temozolomide by Affecting DNA Methyltransferases. Front. Pharmacol. 2022, 13, 815646. [Google Scholar] [CrossRef]
- Fiore, D.; Piscopo, C.; Proto, M.C.; Vasaturo, M.; Dal Piaz, F.; Fusco, B.M.; Pagano, C.; Laezza, C.; Bifulco, M.; Gazzerro, P. N6-Isopentenyladenosine Inhibits Colorectal Cancer and Improves Sensitivity to 5-Fluorouracil-Targeting FBXW7 Tumor Suppressor. Cancers 2019, 11, 1456. [Google Scholar] [CrossRef]
- Fiore, D.; Ramesh, P.; Proto, M.C.; Piscopo, C.; Franceschelli, S.; Anzelmo, S.; Medema, J.P.; Bifulco, M.; Gazzerro, P. Rimonabant Kills Colon Cancer Stem Cells without Inducing Toxicity in Normal Colon Organoids. Front. Pharmacol. 2018, 8, 949. [Google Scholar] [CrossRef]
- Uemura, K.; Sasaki, M.; Sanaki, T.; Toba, S.; Takahashi, Y.; Orba, Y.; Hall, W.W.; Maenaka, K.; Sawa, H.; Sato, A. MRC5 cells engineered to express ACE2 serve as a model system for the discovery of antivirals targeting SARS-CoV-2. Sci. Rep. 2021, 11, 5376. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.; Greer, B.; Hooper, J.; Zijlstra, A.; Walker, B.; Quigley, J.; Hawthorne, S. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 2005, 388, 967–972. [Google Scholar] [CrossRef]
- Schneider, M.A.; Richtmann, S.; Gründing, A.R.; Wrenger, S.; Welte, T.; Meister, M.; Kriegsmann, M.; Winter, H.; Muley, T.; Janciauskiene, S. Transmembrane serine protease 2 is a prognostic factor for lung adenocarcinoma. Int. J. Oncol. 2022, 60, 39. [Google Scholar] [CrossRef] [PubMed]
- Attiq, A.; Yao, L.J.; Afzal, S.; Khan, M.A. The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int. Immunopharmacol. 2021, 101, 108255. [Google Scholar] [CrossRef] [PubMed]
- David, A.; Parkinson, N.; Peacock, T.P.; Pairo-Castineira, E.; Khanna, T.; Cobat, A.; Tenesa, A.; Sancho-Shimizu, V.; Casanova, J.-L.; Abel, L.; et al. A common TMPRSS2 variant has a protective effect against severe COVID-19. Curr. Res. Transl. Med. 2022, 70, 103333. [Google Scholar] [CrossRef]
- Riganti, C.; Pinto, H.; Bolli, E.; Belisario, D.C.; Calogero, R.A.; Bosia, A.; Cavallo, F. Atorvastatin modulates anti-proliferative and pro-proliferative signals in Her2/neu-positive mammary cancer. Biochem. Pharmacol. 2011, 82, 1079–1089. [Google Scholar] [CrossRef][Green Version]
- Gazzerro, P.; Proto, M.C.; Gangemi, G.; Malfitano, A.M.; Ciaglia, E.; Pisanti, S.; Santoro, A.; Laezza, C.; Bifulco, M. Pharmacological Actions of Statins: A Critical Appraisal in the Management of Cancer. Pharmacol. Rev. 2012, 64, 102–146. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Qin, J.-J.; Cheng, X.; Shen, L.; Zhao, Y.-C.; Yuan, Y.; Lei, F.; Chen, M.-M.; Yang, H.; Bai, L.; et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020, 32, 176–187.e4. [Google Scholar] [CrossRef]
- Daniels, L.B.; Kumar, K. The role of statin therapy in COVID-19. Expert Rev. Cardiovasc. Ther. 2022, 20, 415–417. [Google Scholar] [CrossRef]
- Pal, R.; Banerjee, M.; Yadav, U.; Bhattacharjee, S. Statin use and clinical outcomes in patients with COVID-19: An updated systematic review and meta-analysis. Postgrad. Med. J. 2022, 98, 354–359. [Google Scholar] [CrossRef]
- Kow, C.S.; Hasan, S.S. The Association Between the Use of Statins and Clinical Outcomes in Patients with COVID-19: A Systematic Review and Meta-analysis. Am. J. Cardiovasc. Drugs 2022, 22, 167–181. [Google Scholar] [CrossRef]
- Zein, A.F.M.Z.; Sulistiyana, C.S.; Khasanah, U.; Wibowo, A.; Lim, M.A.; Pranata, R. Statin and mortality in COVID-19: A systematic review and meta-analysis of pooled adjusted effect estimates from propensity-matched cohorts. Postgrad. Med. J. 2021, 98, 503–508. [Google Scholar] [CrossRef]
- Saad, M.; Kennedy, K.F.; Louis, D.W.; Imran, H.; Sherrod, C.F.; Aspry, K.; Mentias, A.; Poppas, A.; Abbott, J.D.; Aronow, H.D. Preadmission Statin Treatment and Outcome in Patients Hospitalized With COVID-19. Am. J. Cardiol. 2022, 177, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, K.; Baricault, B.; Semenzato, L.; Botton, J.; Bertrand, M.; Drouin, J.; Dray-Spira, R.; Weill, A.; Zureik, M. Association of Statins for Primary Prevention of Cardiovascular Diseases With Hospitalization for COVID-19: A Nationwide Matched Population-Based Cohort Study. J. Am. Heart Assoc. 2022, 11, e023357. [Google Scholar] [CrossRef] [PubMed]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e16. [Google Scholar] [CrossRef] [PubMed]
- Arefieva, T.I.; Filatova, A.Y.; Potekhina, A.V.; Shchinova, A.M. Immunotropic Effects and Proposed Mechanism of Action for 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase Inhibitors (Statins). Biochemistry 2018, 83, 874–889. [Google Scholar] [CrossRef]
- Silhol, F.; Sarlon, G.; Deharo, J.-C.; Vaïsse, B. Downregulation of ACE2 induces overstimulation of the renin–angiotensin system in COVID-19: Should we block the renin–angiotensin system? Hypertens. Res. 2020, 43, 854–856. [Google Scholar] [CrossRef]
- Mehrabadi, M.E.; Hemmati, R.; Tashakor, A.; Homaei, A.; Yousefzadeh, M.; Hemati, K.; Hosseinkhani, S. Induced dysregulation of ACE2 by SARS-CoV-2 plays a key role in COVID-19 severity. Biomed. Pharmacother. 2021, 137, 111363. [Google Scholar] [CrossRef]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef]
- Klann, K.; Bojkova, D.; Tascher, G.; Ciesek, S.; Münch, C.; Cinatl, J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol. Cell 2020, 80, 164–174.e4. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiore, D.; Proto, M.C.; Franceschelli, S.; Pascale, M.; Bifulco, M.; Gazzerro, P. In Vitro Evidence of Statins’ Protective Role against COVID-19 Hallmarks. Biomedicines 2022, 10, 2123. https://doi.org/10.3390/biomedicines10092123
Fiore D, Proto MC, Franceschelli S, Pascale M, Bifulco M, Gazzerro P. In Vitro Evidence of Statins’ Protective Role against COVID-19 Hallmarks. Biomedicines. 2022; 10(9):2123. https://doi.org/10.3390/biomedicines10092123
Chicago/Turabian StyleFiore, Donatella, Maria Chiara Proto, Silvia Franceschelli, Maria Pascale, Maurizio Bifulco, and Patrizia Gazzerro. 2022. "In Vitro Evidence of Statins’ Protective Role against COVID-19 Hallmarks" Biomedicines 10, no. 9: 2123. https://doi.org/10.3390/biomedicines10092123
APA StyleFiore, D., Proto, M. C., Franceschelli, S., Pascale, M., Bifulco, M., & Gazzerro, P. (2022). In Vitro Evidence of Statins’ Protective Role against COVID-19 Hallmarks. Biomedicines, 10(9), 2123. https://doi.org/10.3390/biomedicines10092123





