De Novo Self-Assembling Peptides Mediate the Conversion of Temozolomide and Delivery of a Model Drug into Glioblastoma Multiforme Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Materials Characterization
2.2.1. Peptide Self-Assembly
2.2.2. Circular Dichroism
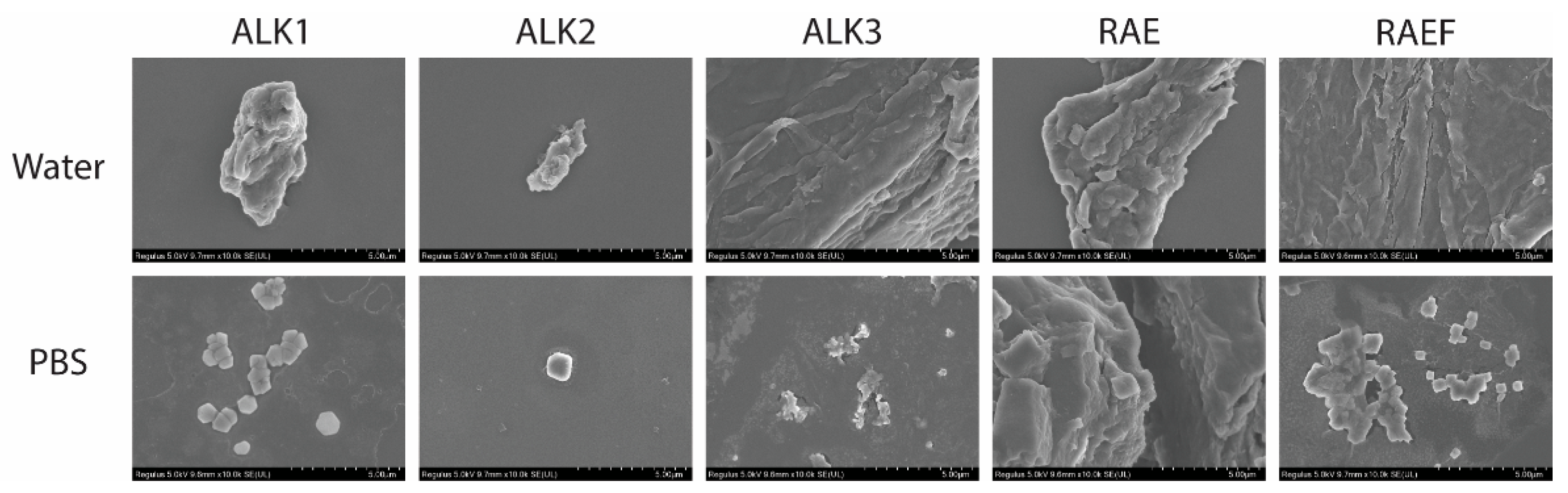
2.2.3. SEM
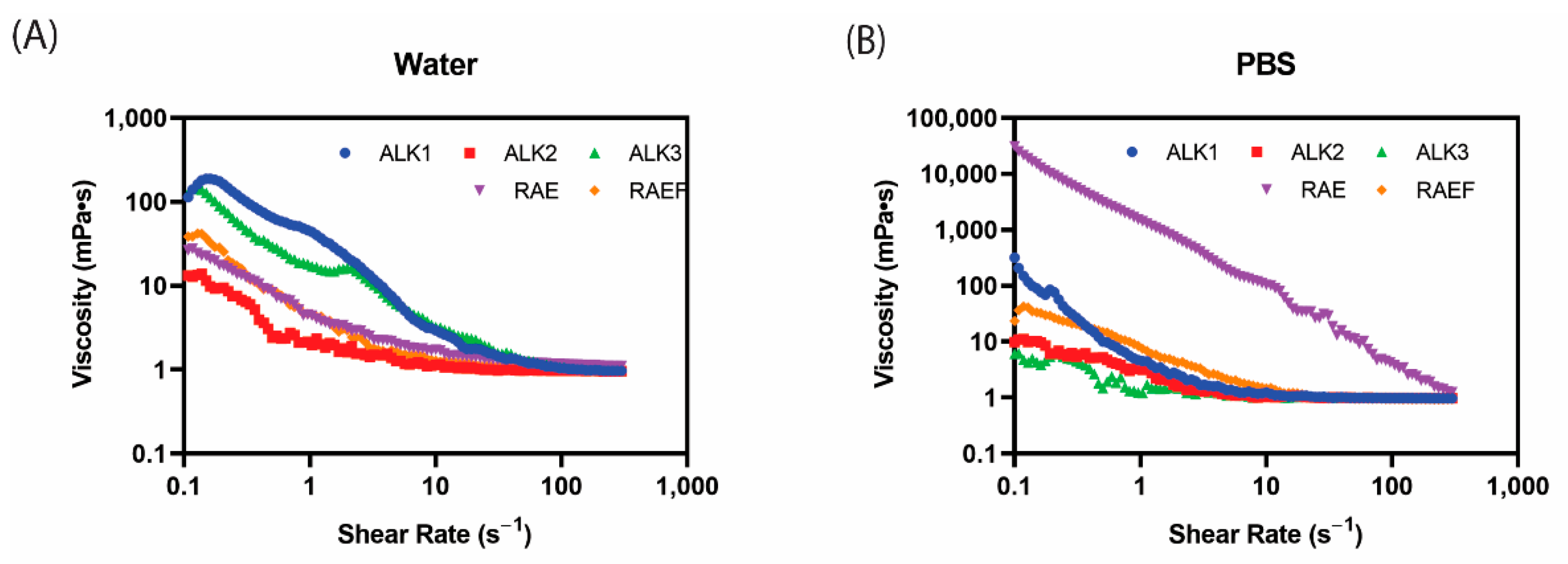
2.2.4. Rheology
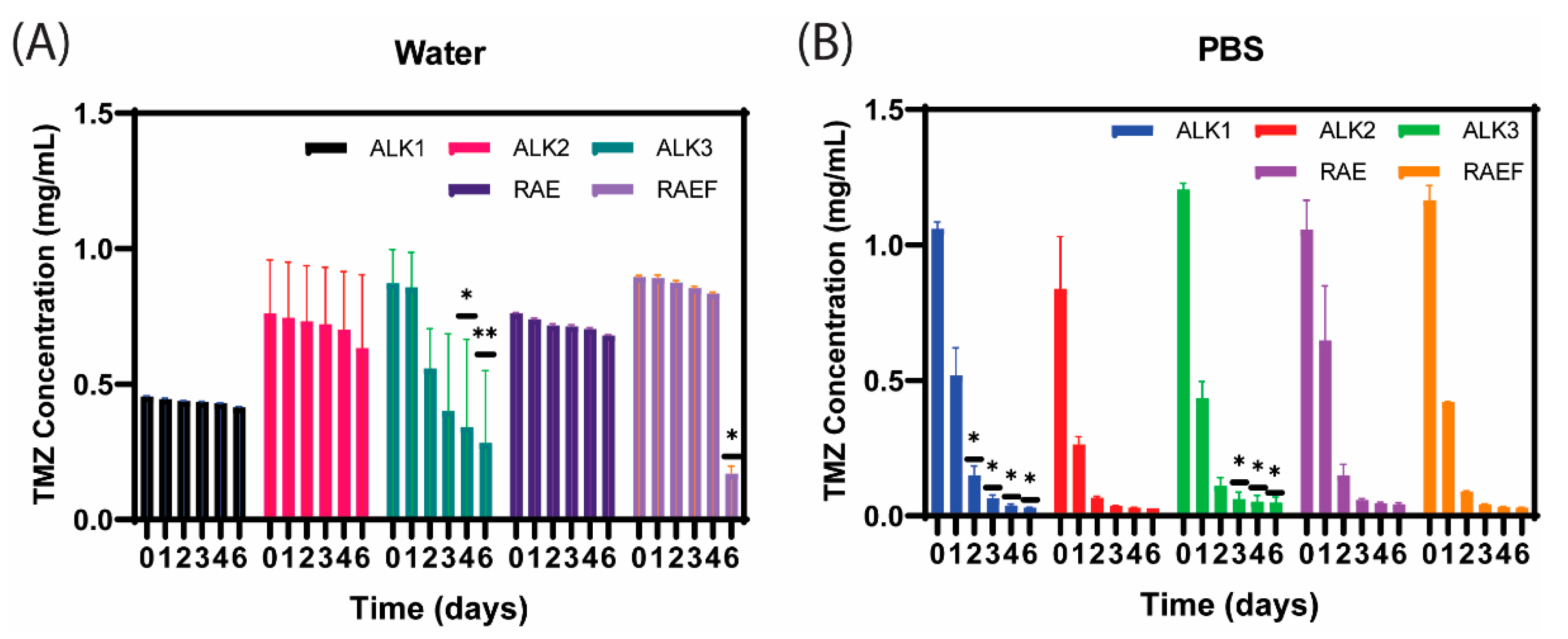
2.2.5. Drug Loading and Conversion
2.3. Cell Studies
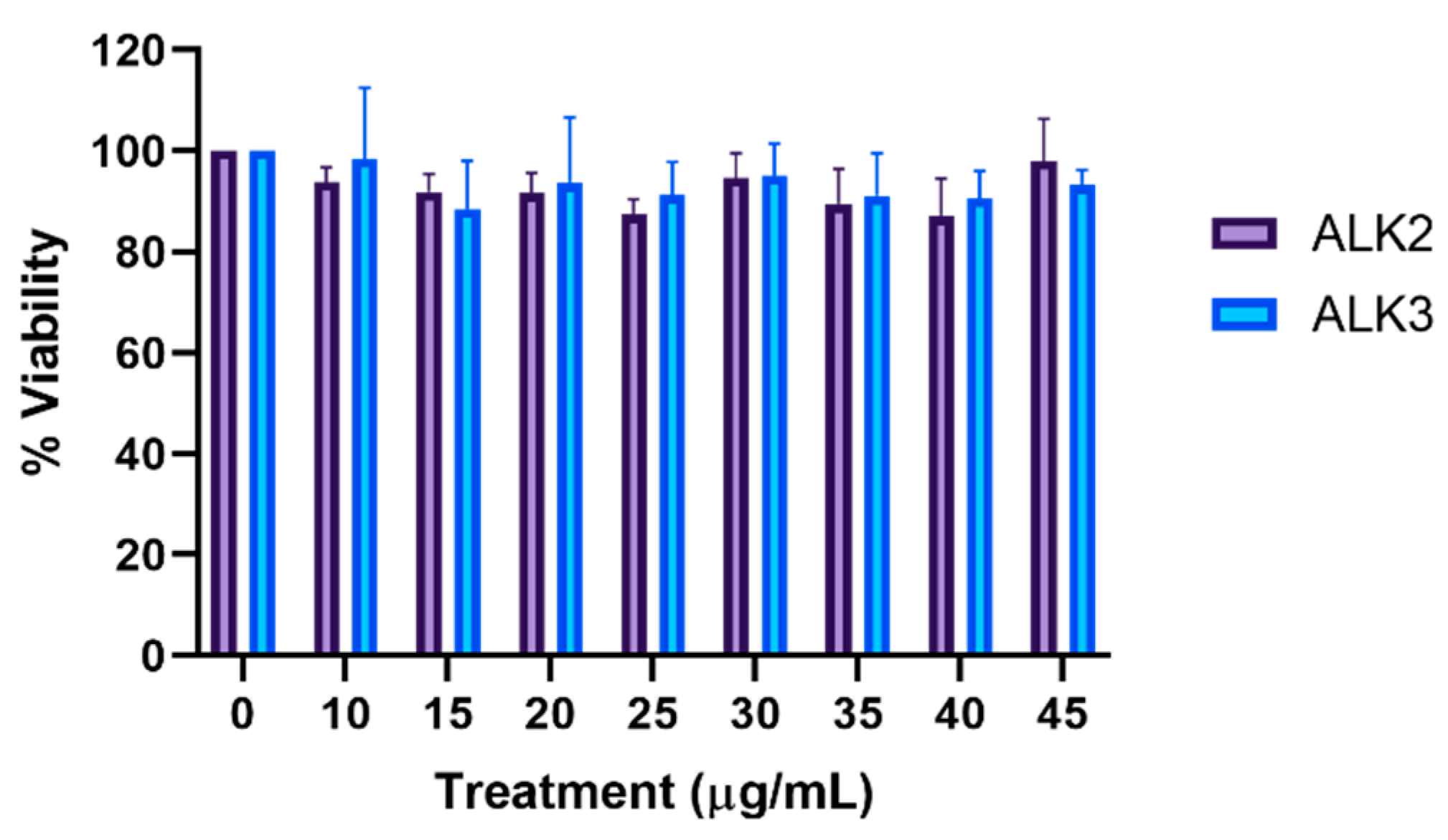
2.3.1. MTS
2.3.2. Cellular Uptake
2.4. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Assemblies Formed via Film Dehydration Method
3.2. TMZ Loading and Conversion in Assemblies Formed via Film Dehydration Method
3.3. Characterization of Assemblies Formed via Incubation Method
3.4. TMZ Loading and Conversion in Assemblies Formed via Incubation Method
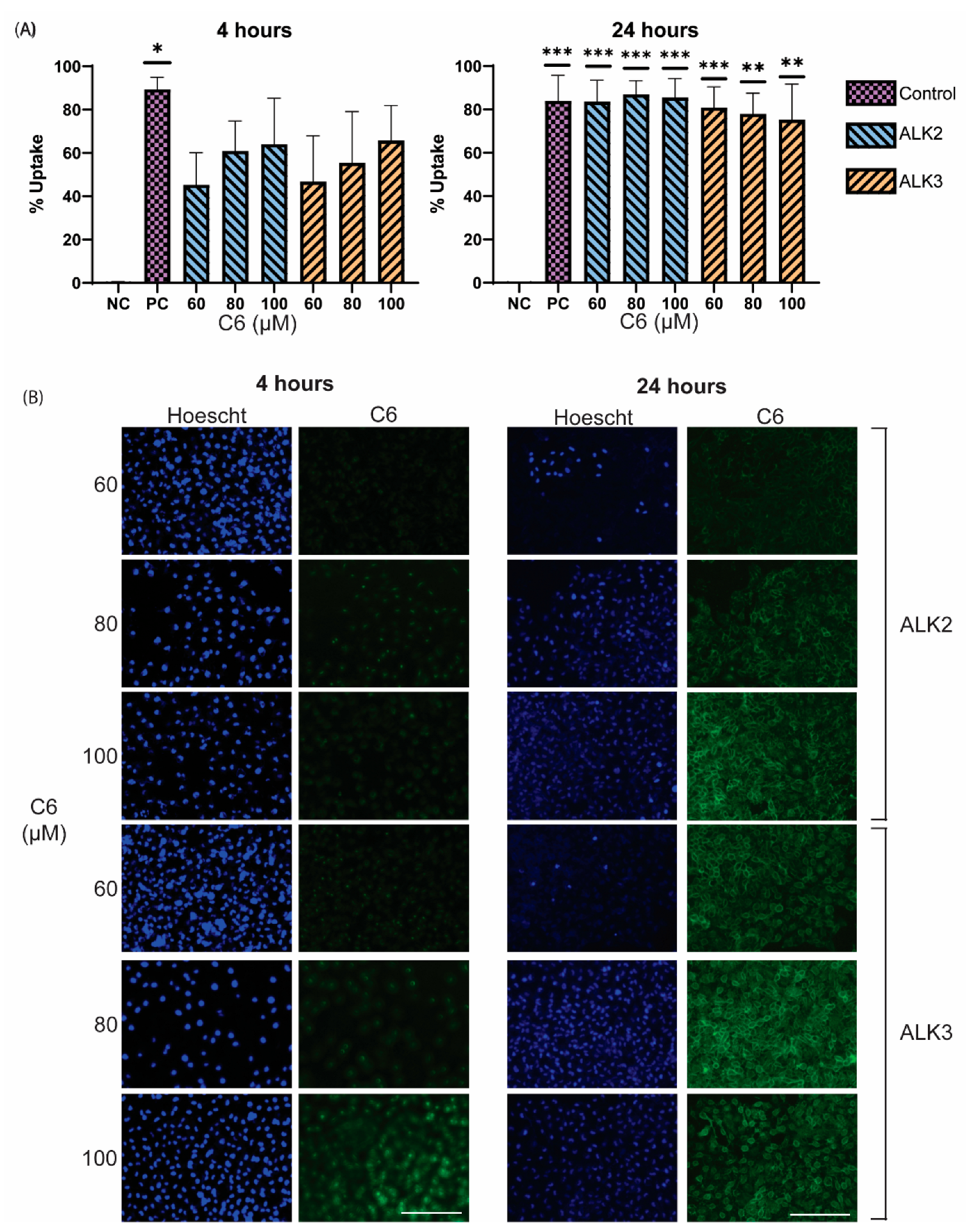
3.5. In Vitro Analysis of Peptide Biocompatiblity and Cellular Uptake
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016, 18, v1–v75. [Google Scholar] [CrossRef]
- Van Tellingen, O.; Yetkin-Arik, B.; De Gooijer, M.C.; Wesseling, P.; Wurdinger, T.; De Vries, H.E. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, M.J.; Sevin, E.; Gosselet, F.; Lima, J.; Coelho, M.; Loureiro, J.; Pereira, M. Receptor-mediated PLGA nanoparticles for glioblastoma multiforme treatment. Int. J. Pharm. 2018, 545, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Bikhezar, F.; de Kruijff, R.M.; van der Meer, A.J.G.M.; Villa, G.T.; van der Pol, S.M.A.; Aragon, G.B.; Garcia, A.G.; Narayan, R.S.; de Vries, H.E.; Slotman, B.J.; et al. Preclinical evaluation of binimetinib (MEK162) delivered via polymeric nanocarriers in combination with radiation and temozolomide in glioma. J. Neurooncol. 2019, 146, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Andrasi, M.; Bustos, R.; Gaspar, A.; Gomez, F.A.; Klekner, A. Analysis and stability study of temozolomide using capillary electrophoresis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 1801–1808. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.D.; Wirth, M.; Statkevich, P.; Reidenberg, P.; Alton, K.; Sartorius, S.E.; Dugan, M.; Cutler, D.; Batra, V.; Grochow, L.B.; et al. Absorption, metabolism, and excretion of 14C-temozolomide following oral administration to patients with advanced cancer. Clin. Cancer Res. 1999, 5, 309–317. [Google Scholar]
- Ward, S.M.; Skinner, M.; Saha, B.; Emrick, T. Polymer-Temozolomide Conjugates as Therapeutics for Treating Glioblastoma. Mol. Pharm. 2018, 15, 5263–5276. [Google Scholar] [CrossRef]
- Agarwala, S.S. Temozolomide, a Novel Alkylating Agent with Activity in the Central Nervous System, May Improve the Treatment of Advanced Metastatic Melanoma. Oncologist 2000, 5, 144–151. [Google Scholar] [CrossRef]
- Emamgholizadeh Minaei, S.; Khoei, S.; Khoee, S.; Karimi, M.R. Tri-block copolymer nanoparticles modified with folic acid for temozolomide delivery in glioblastoma. Int. J. Biochem. Cell Biol. 2019, 108, 72–83. [Google Scholar] [CrossRef]
- XELODA ® (Capecitabine). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/020896s037lbl.pdf (accessed on 23 June 2022).
- Alavijeh, M.S.; Chishty, M.; Qaiser, M.Z.; Palmer, A.M. Drug metabolism and pharmacokinetics, the blood-brain barrier, and central nervous system drug discovery. NeuroRx 2005, 2, 554–571. [Google Scholar] [CrossRef]
- Zhao, M.; Bozzato, E.; Joudiou, N.; Ghiassinejad, S.; Danhier, F.; Gallez, B.; Préat, V. Codelivery of paclitaxel and temozolomide through a photopolymerizable hydrogel prevents glioblastoma recurrence after surgical resection. J. Control. Release 2019, 309, 72–81. [Google Scholar] [CrossRef]
- Sage, W.; Guilfoyle, M.; Luney, C.; Young, A.; Sinha, R.; Sgubin, D.; McAbee, J.H.; Ma, R.; Jefferies, S.; Jena, R.; et al. Local alkylating chemotherapy applied immediately after 5-ALA guided resection of glioblastoma does not provide additional benefit. J. Neurooncol. 2018, 136, 273–280. [Google Scholar] [CrossRef]
- Nagai, Y.; Unsworth, L.D.; Koutsopoulos, S.; Zhang, S. Slow release of molecules in self-assembling peptide nanofiber scaffold. J. Control. Release 2006, 115, 18–25. [Google Scholar] [CrossRef]
- Löwik, D.W.P.M.; Van Hest, J.C.M. Peptide based amphiphiles. Chem. Soc. Rev. 2004, 33, 234–245. [Google Scholar] [CrossRef]
- Pitz, M.E.; Nukovic, A.M.; Elpers, M.A.; Alexander-Bryant, A.A. Factors Affecting Secondary and Supramolecular Structures of Self-Assembling Peptide Nanocarriers. Macromol. Biosci. 2021, 22, e2100347. [Google Scholar] [CrossRef]
- Li, J.; Xing, R.; Bai, S.; Yan, X. Recent advances of self-assembling peptide-based hydrogels for biomedical applications. Soft Matter 2019, 15, 1704–1715. [Google Scholar] [CrossRef]
- Karavasili, C.; Panteris, E.; Vizirianakis, I.S.; Koutsopoulos, S.; Fatouros, D.G. Chemotherapeutic Delivery from a Self-Assembling Peptide Nanofiber Hydrogel for the Management of Glioblastoma. Pharm. Res. 2018, 35, 166. [Google Scholar] [CrossRef]
- Gelain, F.; Bottai, D.; Vescovi, A.; Zhang, S. Designer Self-Assembling Peptide Nanofiber Scaffolds for Adult Mouse Neural Stem Cell 3-Dimensional Cultures. PLoS ONE 2006, 1, e119. [Google Scholar] [CrossRef]
- Wu, Y.; Sadatmousavi, P.; Wang, R.; Lu, S.; Yuan, Y.-F.; Chen, P. Self-assembling peptide-based nanoparticles enhance anticancer effect of ellipticine in vitro and in vivo. Int. J. Nanomed. 2012, 7, 3221–3233. [Google Scholar] [CrossRef]
- Hong, Y.; Legge, R.L.; Zhang, S.; Chen, P. Effect of amino acid sequence and pH on nanofiber formation of self-assembling peptides EAK16-II and EAK16-IV. Biomacromolecules 2003, 4, 1433–1442. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Chacón, P.; Merelo, J.J.; Morán, F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993, 6, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Nikara, S.; Ahmadi, E.; Nia, A.A. Effects of different preparation techniques on the microstructural features of biological materials for scanning electron microscopy. J. Agric. Food Res. 2020, 2, 100036. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. Protein Structure, Neighbor Effect, and a New Index of Amino Acid Dissimilarities. Mol. Biol. Evol. 2002, 19, 58–67. [Google Scholar] [CrossRef]
- Zhang, S.; Lockshin, C.; Cook, R.; Rich, A. Unusually stable β-sheet formation in an ionic self-complementary oligopeptide. Biopolymers 1994, 34, 663–672. [Google Scholar] [CrossRef]
- Yokoi, H.; Kinoshita, T.; Zhang, S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl. Acad. Sci. USA 2005, 102, 8414–8419. [Google Scholar] [CrossRef]
- Pavani, P.; Kumar, K.; Rani, A.; Venkatesu, P.; Lee, M.J. The influence of sodium phosphate buffer on the stability of various proteins: Insights into protein-buffer interactions. J. Mol. Liq. 2021, 331, 115753. [Google Scholar] [CrossRef]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577. [Google Scholar] [CrossRef] [Green Version]





| Peptide Name | Sequence |
|---|---|
| ALK1 | AEAEAEAEAEAEKKKK |
| ALK2 | AEAEAEAEKKKKKKKK |
| ALK3 | AEAEKKKKKKKKKKKK |
| RAE | RARAEARARAEARARAEA |
| RAEF | RRAEARRAFARRAEA |
| Peptide | Film Dehydration | |||||
|---|---|---|---|---|---|---|
| Water | 1×PBS | |||||
| Alpha Helix | Beta Sheet | Irregular | Alpha Helix | Beta Sheet | Irregular | |
| ALK1 | 0.12 | 0.34 | 0.54 | 0.10 | 0.41 | 0.49 |
| ALK2 | 0.04 | 0.48 | 0.48 | 0.09 | 0.37 | 0.54 |
| ALK3 | 0.09 | 0.47 | 0.44 | 0.05 | 0.35 | 0.60 |
| RAE | 0.05 | 0.47 | 0.48 | 0.04 | 0.48 | 0.48 |
| RAEF | 0.05 | 0.47 | 0.48 | 0.05 | 0.47 | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitz, M.; Elpers, M.; Nukovic, A.; Wilde, S.; Gregory, A.J.; Alexander-Bryant, A. De Novo Self-Assembling Peptides Mediate the Conversion of Temozolomide and Delivery of a Model Drug into Glioblastoma Multiforme Cells. Biomedicines 2022, 10, 2164. https://doi.org/10.3390/biomedicines10092164
Pitz M, Elpers M, Nukovic A, Wilde S, Gregory AJ, Alexander-Bryant A. De Novo Self-Assembling Peptides Mediate the Conversion of Temozolomide and Delivery of a Model Drug into Glioblastoma Multiforme Cells. Biomedicines. 2022; 10(9):2164. https://doi.org/10.3390/biomedicines10092164
Chicago/Turabian StylePitz, Megan, Margaret Elpers, Alexandra Nukovic, Sarah Wilde, Arica Jordan Gregory, and Angela Alexander-Bryant. 2022. "De Novo Self-Assembling Peptides Mediate the Conversion of Temozolomide and Delivery of a Model Drug into Glioblastoma Multiforme Cells" Biomedicines 10, no. 9: 2164. https://doi.org/10.3390/biomedicines10092164
APA StylePitz, M., Elpers, M., Nukovic, A., Wilde, S., Gregory, A. J., & Alexander-Bryant, A. (2022). De Novo Self-Assembling Peptides Mediate the Conversion of Temozolomide and Delivery of a Model Drug into Glioblastoma Multiforme Cells. Biomedicines, 10(9), 2164. https://doi.org/10.3390/biomedicines10092164





