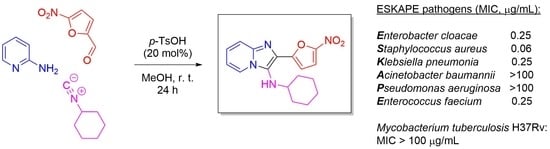
Novel 5-Nitrofuran-Tagged Imidazo-Fused Azines and Azoles Amenable by the Groebke–Blackburn–Bienaymé Multicomponent Reaction: Activity Profile against ESKAPE Pathogens and Mycobacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Organic Synthesis
2.1.1. General Considerations
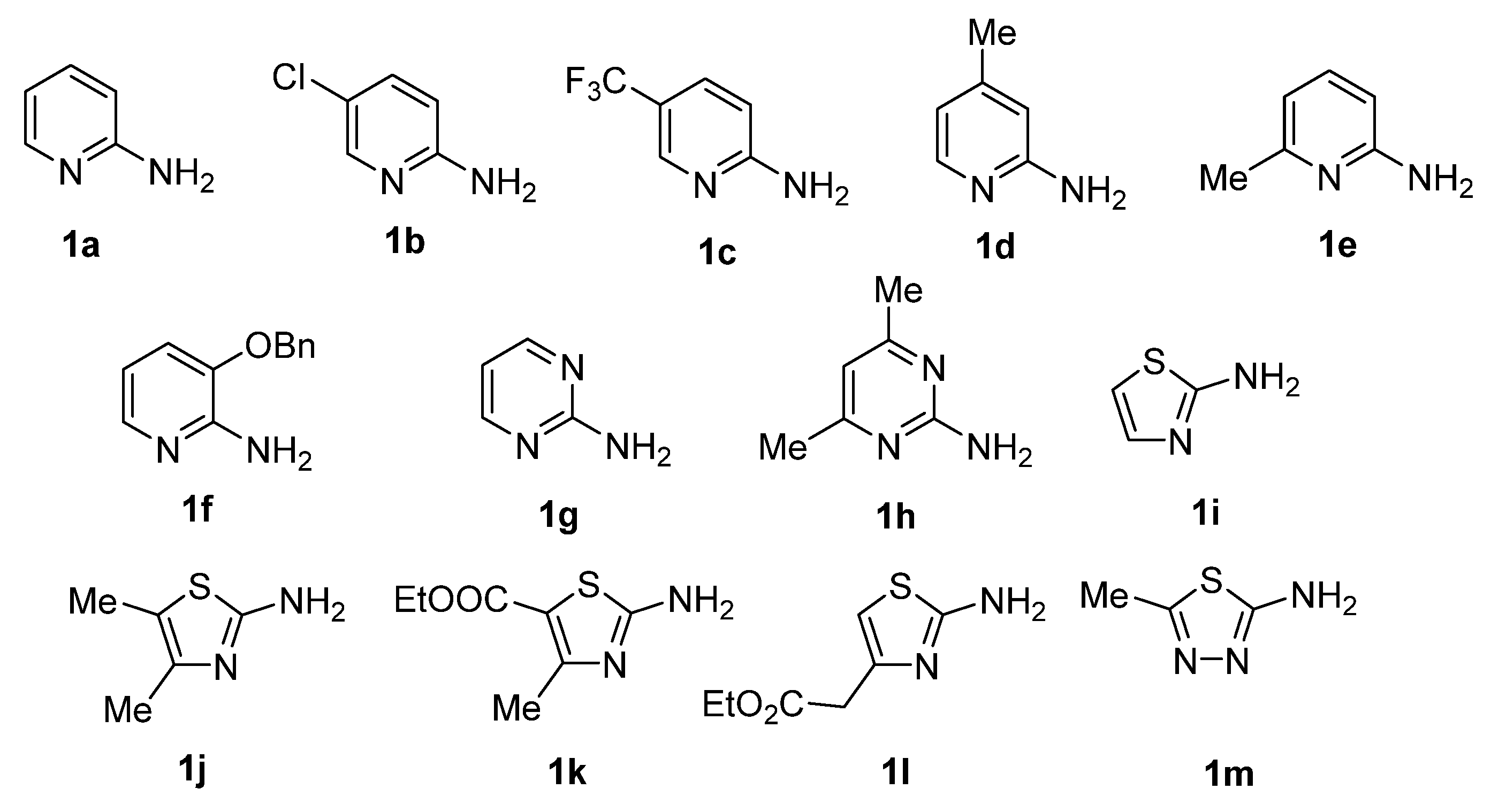
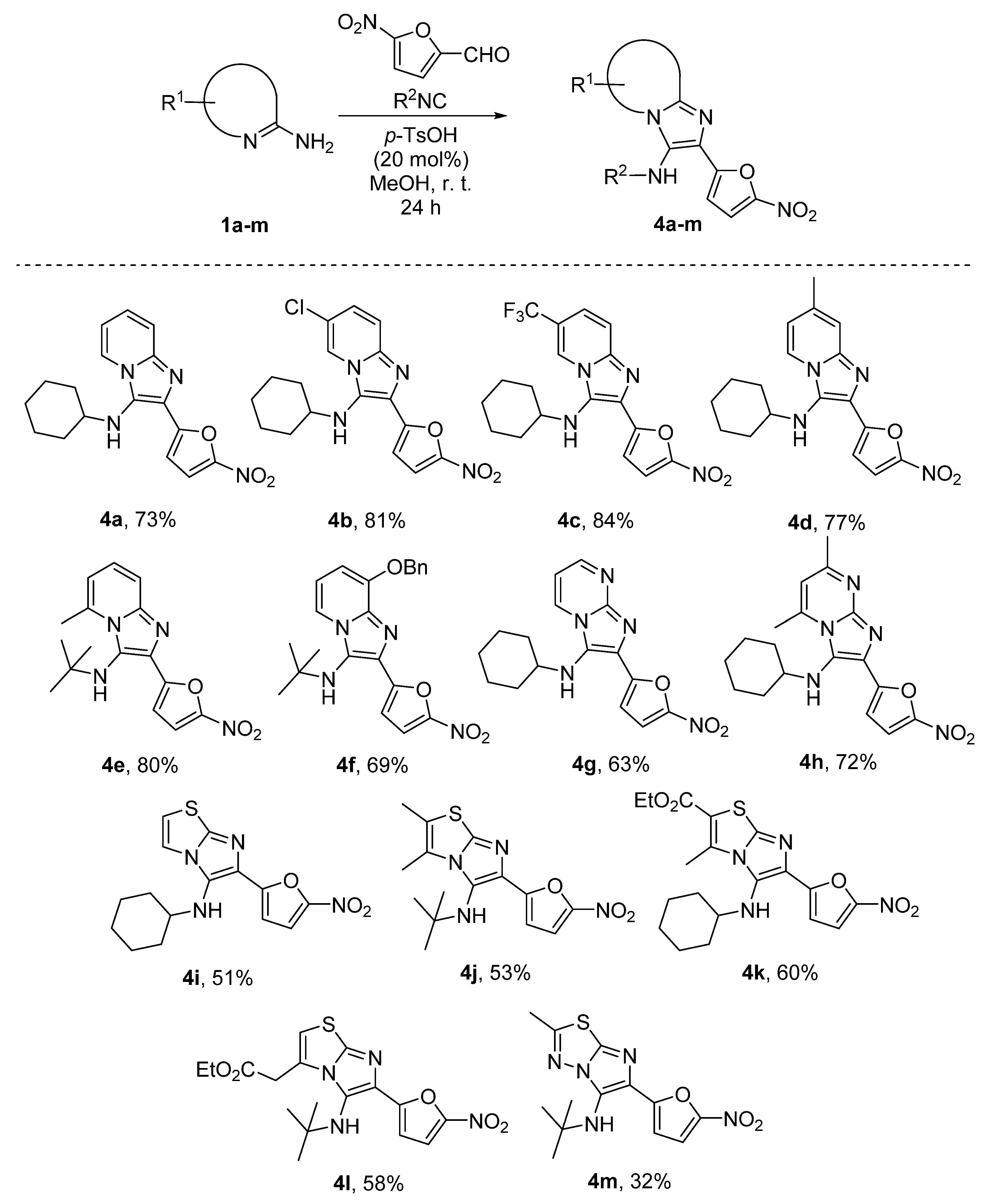
2.1.2. General Procedure for the Synthesis of GBB Reaction Products 4a–m
N-Cyclohexyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyridin-3-amine (4a)
6-Chloro-N-cyclohexyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyridin-3-amine (4b)
N-Cyclohexyl-2-(5-nitrofuran-2-yl)-6-(trifluoromethyl)imidazo[1,2-a]pyridin-3-amine (4c)
N-Cyclohexyl-7-methyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyridin-3-amine (4d)
N-(tert-Butyl)-5-methyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyridin-3-amine (4e)
8-(Benzyloxy)-N-(tert-butyl)-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyridin-3-amine (4f)
N-Cyclohexyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyrimidin-3-amine (4g)
N-Cyclohexyl-5,7-dimethyl-2-(5-nitrofuran-2-yl)imidazo[1,2-a]pyrimidin-3-amine (4h)
N-Cyclohexyl-6-(5-nitrofuran-2-yl)imidazo[2,1-b]thiazol-5-amine (4i)
N-(tert-Butyl)-2,3-dimethyl-6-(5-nitrofuran-2-yl)imidazo[2,1-b]thiazol-5-amine (4j)
Ethyl 5-(cyclohexylamino)-3-methyl-6-(5-nitrofuran-2-yl)imidazo[2,1-b]thiazole-2-carboxylate (4k)
Ethyl 2-(5-(tert-butylamino)-6-(5-nitrofuran-2-yl)imidazo[2,1-b]thiazol-3-yl)acetate (4l)
N-(tert-Butyl)-2-methyl-6-(5-nitrofuran-2-yl)imidazo[2,1-b][1,3,4]thiadiazol-5-amine (4m)
2.2. ESKAPE Pathogen Susceptibility Testing
2.3. Testing against Mycobacterium tuberculosis
2.4. Molecular Modeling
2.4.1. Target Selection
2.4.2. Protein and Ligand Structure Preparation
2.4.3. Molecular Docking
2.4.4. Molecular Mechanics with Generalized Born and Surface Area Solvation (MM-GBSA)
3. Results
3.1. Compound Synthesis
3.2. Activity against ESKAPE Pathogens
3.3. Activity against Mycobacterium tuberculosis
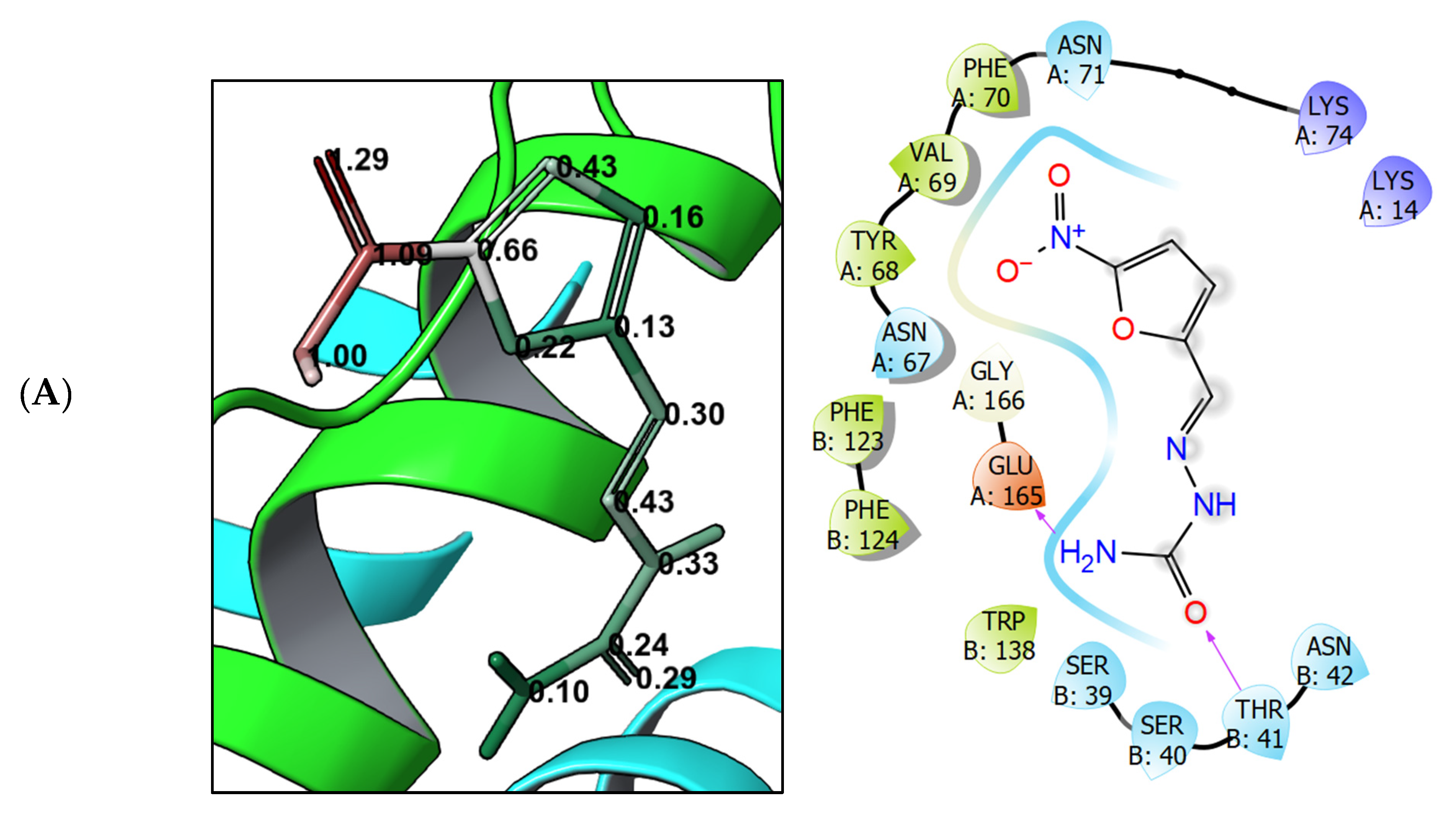
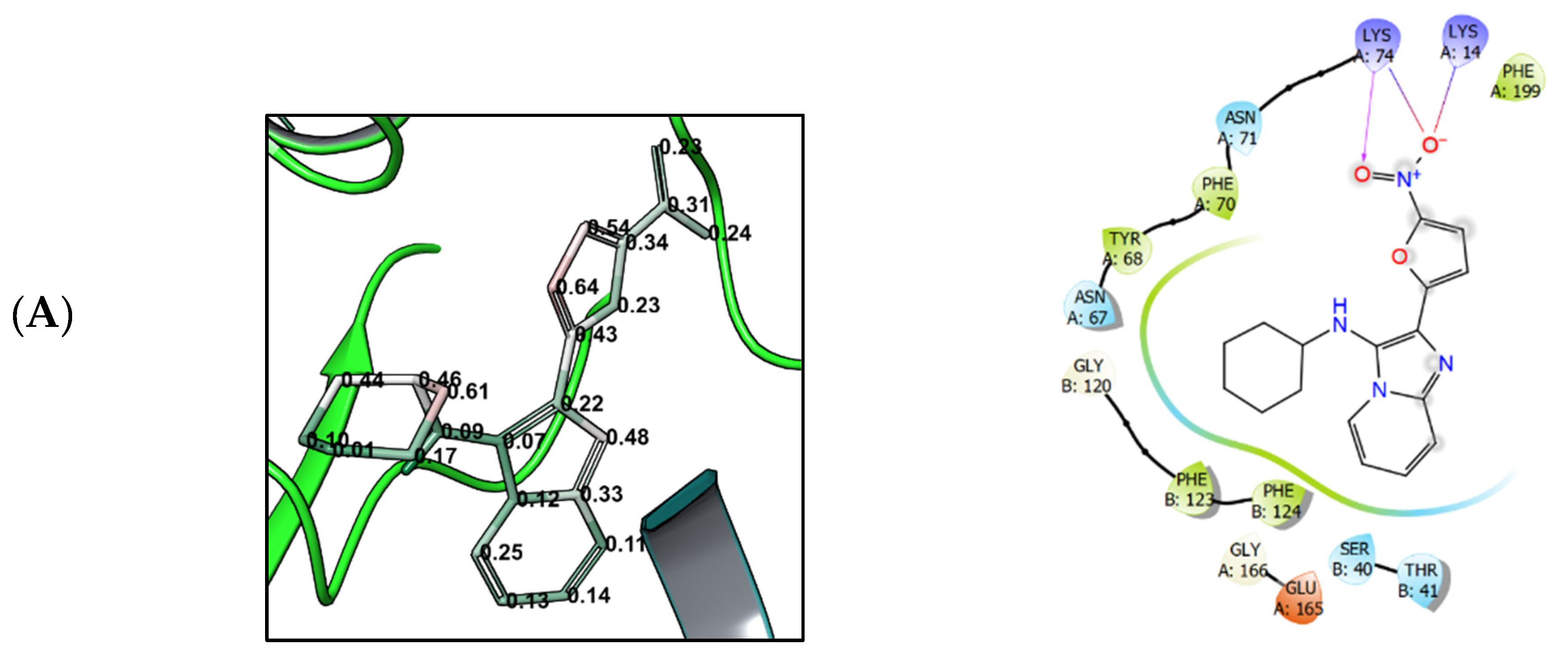
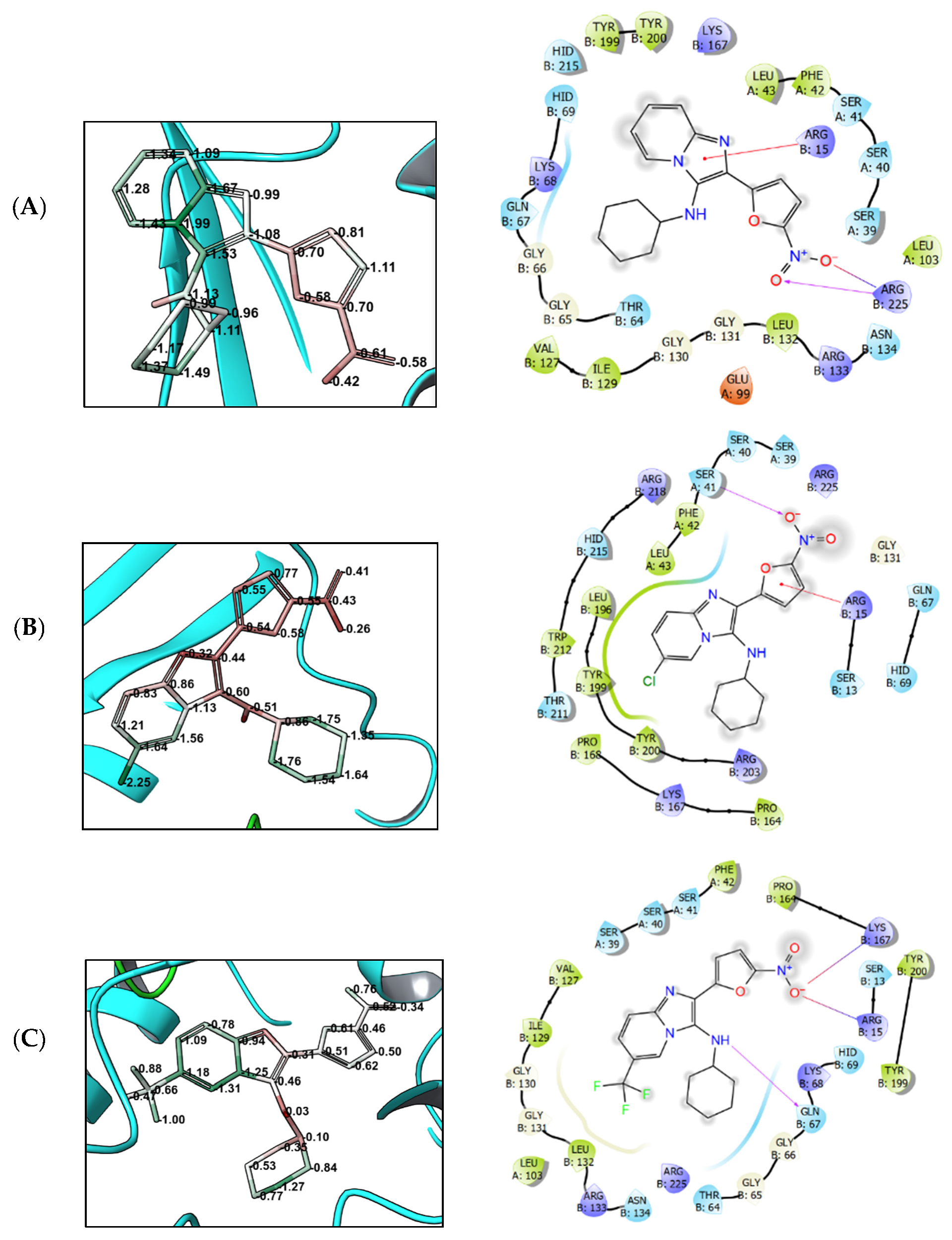
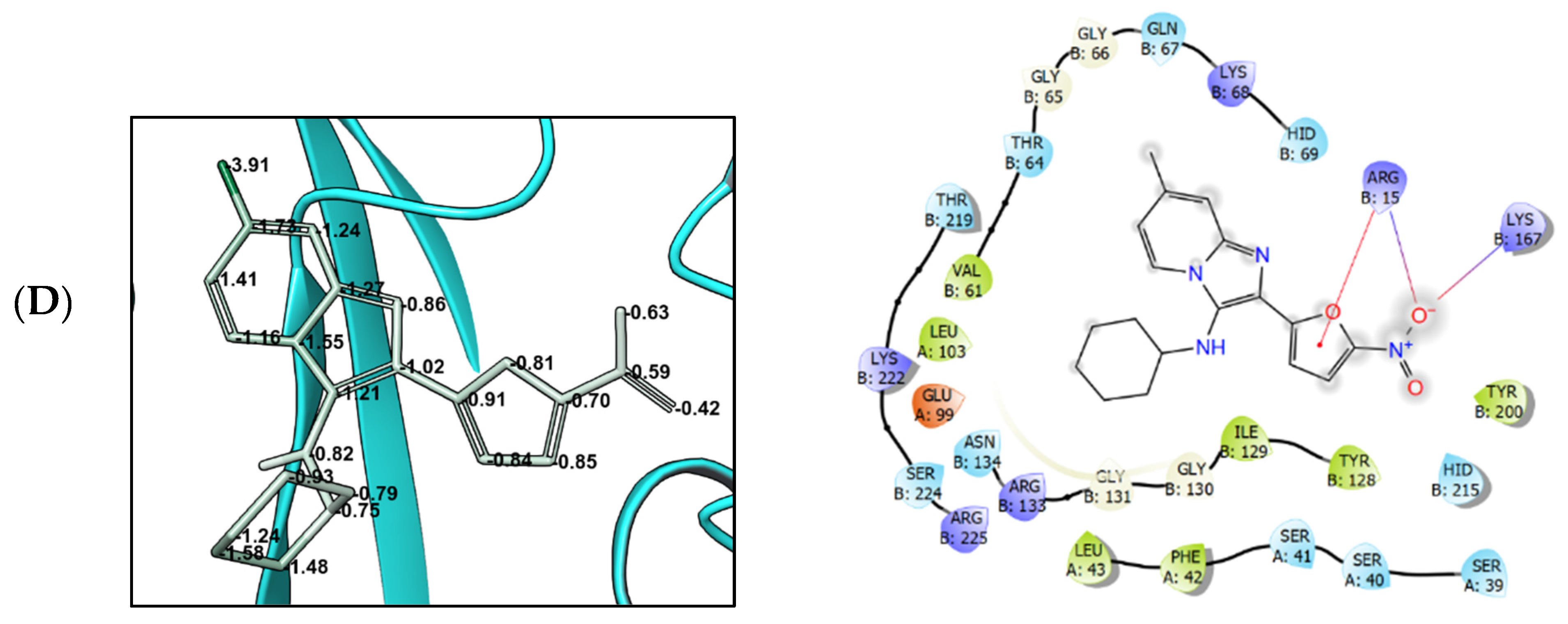
3.4. Molecular Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Swinney, D.C. Phenotypic vs. Target-Based Drug Discovery for First-in-Class Medicines. Clin. Pharmacol. Ther. 2013, 93, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Vincent, F.; Lee, J.A.; Eder, J.; Prunotto, M. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nat. Rev. Drug Discov. 2017, 16, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Langdon, S.R.; Brown, N.; Blagg, J. Scaffold Diversity of Exemplified Medicinal Chemistry Space. J. Chem. Inf. Model. 2011, 51, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.J.D.; Isidro-Llobet, A.; Spring, D.R. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat. Commun. 2010, 1, 80. [Google Scholar] [CrossRef]
- Ganem, B. Strategies for Innovation in Multicomponent Reaction Design. Acc. Chem. Res. 2009, 42, 463–472. [Google Scholar] [CrossRef]
- Blackburn, C.; Guan, B.; Fleming, P.; Shiosaki, K.; Tsai, S. Parallel synthesis of 3-aminoimidazo[1,2-a]pyridines and pyrazines by a new three-component condensation. Tetrahedron Lett. 1998, 39, 3635–3638. [Google Scholar] [CrossRef]
- Bienaymé, H.; Bouzid, K. A new heterocyclic multicomponent reaction for the combinatorial synthesis of fused 3-aminoimidazoles. Angew. Chem. Int. Ed. 1998, 37, 2234–2237. [Google Scholar] [CrossRef]
- Groebke, K.; Weber, L.; Mehlin, F. Synthesis of imidazo[1,2-a] annulated pyridines, pyrazines and pyrimidines by a novel threecomponent condensation. Synlett 1998, 6, 661–663. [Google Scholar] [CrossRef]
- Boltjes, A.; Dömling, A. The Groebke-Blackburn-Bienaymé Reaction. Eur. J. Org. Chem. 2019, 2019, 7007–7049. [Google Scholar] [CrossRef]
- Shaaban, S.; Abdel-Wahab, B.F. Groebke-Blackburn-Bienaymé multicomponent reaction: Emerging chemistry for drug discovery. Mol. Divers. 2016, 20, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Akritopoulou-Zanze, I.; Wakefield, B.D.; Gasiecki, A.; Kalvin, D.; Johnson, E.F.; Kovar, P.; Djuric, S.W. Scaffold oriented synthesis. Part 4: Design, synthesis and biological evaluation of novel 5-substituted indazoles as potent and selective kinase inhibitors employing heterocycle forming and multicomponent reactions. Bioorg. Med. Chem. Lett. 2011, 21, 1480–1483. [Google Scholar] [CrossRef] [PubMed]
- Odell, L.R.; Nilsson, M.T.; Gising, J.; Lagerlund, O.; Muthas, D.; Nordqvist, A.; Karlen, A.; Larhed, M. Functionalized 3-amino-imidazo[1,2-a]pyridines: A novel class of drug-like Mycobacterium Tuberculosis glutamine synthetase inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 4790–4793. [Google Scholar] [CrossRef] [PubMed]
- Al-Tel, T.H.; Al-Qawasmeh, R.A.; Zaarour, R. Design, synthesis and in vitro antimicrobial evaluation of novel Imidazo[1,2-a]pyridine and imidazo[2,1-b][1,3]benzothiazole motifs. Eur. J. Med. Chem. 2011, 46, 1874–1881. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, T.; Noushini, S.; Taban, S.; Mahdavi, M.; Khoshneviszadeh, M.; Saeedi, M.; Emami, S.; Eghtedari, M.; Sarrafi, Y.; Khoshneviszadeh, M.; et al. Synthesis and cytotoxic activity of novel poly-substituted imidazo[2,1-c][1,2,4]triazin-6-amines. Mol. Divers. 2015, 19, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Meirer, K.; Roedl, C.B.; Wisniewska, J.M.; George, S.; Haefner, A.-K.; Buscato, E.I.; Klingler, F.-M.; Hahn, S.; Berressem, D.; Wittmann, S.K.; et al. Synthesis and Structure—Activity Relationship Studies of Novel Dual Inhibitors of Soluble Epoxide Hydrolase and 5-Lipoxygenase. J. Med. Chem. 2013, 56, 1777–1781. [Google Scholar] [CrossRef]
- Shukla, N.M.; Salunke, D.B.; Yoo, E.; Mutz, C.A.; Balakrishna, R.; David, S.A. Antibacterial activities of Groebke-Blackburn-Bienaymé-derived imidazo[1,2-a]pyridin-3-amines. Bioorg. Med. Chem. 2012, 20, 5850–5863. [Google Scholar] [CrossRef]
- Kalinin, S.; Vedekhina, T.; Paramonova, P.; Krasavin, M. Antimicrobial activity of 5-membered nitroheteroaromatic compounds beyond nitrofurans and nitroimidazoles: Recent progress. Curr. Med. Chem. 2021, 28, 5926–5982. [Google Scholar] [CrossRef]
- Zorzi, R.R.; Jorge, S.D.; Palace-Berl, F.; Pascualoto, K.F.M.; de Sá Bortolozo, L.; de Castro Siqueira, A.M.; Tavares, L.C. Exploring 5-nitrofuran derivatives against nosocomial pathogens: Synthesis, antimicrobial activity and chemometric analysis. Bioorg. Med. Chem. 2014, 22, 2844–2854. [Google Scholar] [CrossRef]
- Murugasu-Oei, B.; Dick, T. Bactericidal activity of nitrofurans against growing and dormant Mycobacterium bovis BCG. J. Antimicrob. Chemother. 2000, 46, 917–919. [Google Scholar] [CrossRef] [Green Version]
- Verbitskiy, E.V.; Baskakova, S.A.; Rusinov, G.L.; Charushin, V.N. New approach to 5-arylamino-4-(5-aryloxyfuran-2-yl)pyrimidines: Synthesis and antibacterial activity. Russ. Chem. Bull. 2021, 70, 937–942. [Google Scholar] [CrossRef]
- Krasavin, M.; Lukin, A.; Vedekhina, T.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Zabolotnykh, N.; Rogacheva, E.; Kraeva, L.; Yablonsky, P. Conjugation of a 5-nitrofuran-2-oyl moiety to aminoalkylimidazoles produces non-toxic nitrofurans that are efficacious in vitro and in vivo against multidrug-resistant Mycobacterium tuberculosis. Eur. J. Med. Chem. 2018, 157, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Krasavin, M.; Shetnev, A.; Panova, V.; Ivanovskyi, S.; Kalinin, S.; Vinogradova, T.; Sharoyko, V.; Yablonsky, P. Hetaryl- and heteroarylvinyl-substituted nitrofurans identified as non-cytotoxic selective antitubercular agents. Mendeleev Commun. 2022, 32, 452–453. [Google Scholar] [CrossRef]
- Verbitskiy, E.V.; Baskakova, S.A.; Belyaev, D.V.; Vakhrusheva, D.V.; Eremeeva, N.I.; Rusinov, G.L.; Charushin, V.N. Renaissance of 4-(5-nitrofuran-2-yl)-5-arylamino substituted pyrimidines: Microwave-assisted synthesis and antitubercular activity. Mendeleev Commun. 2021, 31, 210–212. [Google Scholar] [CrossRef]
- Schneider, P.; Schneider, G. Privileged Structures Revisited. Angew. Chem. Int. Ed. 2017, 56, 7971–7974. [Google Scholar] [CrossRef]
- Huttner, A.; Verhaegh, E.M.; Harbarth, S.; Muller, A.E.; Theuretzbacher, U.; Mouton, J.W. Nitrofurantoin revisited: A systematic review and meta-analysis of controlled trials. J. Antimicrob. Chemother. 2015, 70, 2456–2464. [Google Scholar] [CrossRef]
- Männistö, P.; Karttunen, P. Pharmacokinetics of furagin, a new nitrofurantoin congener, on human volunteers. Int. J. Clin. Pharmacol. Biopharm. 1979, 17, 264–270. [Google Scholar] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- EUCAST SOP 9.2; Procedure for Establishing Zone Diameter Breakpoints and Quality Control Criteria. EUCAST: Växjö, Sweden, 2020. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/2020/EUCAST_SOP_9.2_Disk_diffusion_breakpoints_and_QC_ranges_final.pdf (accessed on 28 July 2022).
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Chuprun, S.; Dar’in, D.; Rogacheva, E.; Kraeva, L.; Levin, O.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Bakulina, O.; Krasavin, M. Mutually isomeric 2- and 4-(3-nitro-1,2,4-triazol-1-yl)pyrimidines inspired by an antimycobacterial screening hit: Synthesis and biological activity against the ESKAPE panel of pathogens. Antibiotics 2020, 9, 666. [Google Scholar] [CrossRef]
- Stevens, M.; Howe, C.; Ray, A.-M.; Washburn, A.; Chitre, S.; Sivinski, J.; Park, Y.; Hoang, Q.Q.; Chapman, E.; Johnson, S.M. Analogs of nitrofuran antibiotics are potent GroEL/ES inhibitor pro-drugs. Bioorg. Med. Chem. 2020, 28, 115710. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.; Kaplan, E.; Laurieri, N.; Lowe, E.; Sim, E. Activation of nitrofurazone by azoreductases: Multiple activities in one enzyme. Sci. Rep. 2011, 1, 63. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols and influence on virtual screening enrichments. J. Comp. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Suenaga, A.; Okimoto, N.; Hirano, Y.; Fukui, K. An efficient computational method for calculating ligand binding affinities. PLoS ONE 2012, 7, e42846. [Google Scholar] [CrossRef]
- Shao, T.; Gong, Z.; Su, T.; Hao, W.; Che, C. A practical and efficient approach to imidazo[1,2-a]pyridine-fused isoquinolines through the post-GBB transformation strategy. Beilstein J. Org. Chem. 2017, 13, 817–824. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Donaldson, P.M.; Pallett, A.P.; Carroll, M.P. Ciprofloxacin in general practice. Brit. Med. J. 1994, 308, 1437. [Google Scholar] [CrossRef] [PubMed]
- Krasavin, M.; Lukin, A.; Vedekhina, T.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Zabolotnykh, N.; Rogacheva, E.; Kraeva, L.; Sharoyko, V.; et al. Attachment of a 5-nitrofuroyl moiety to spirocyclic piperidines produces non-toxic nitrofurans that are efficacious in vitro against multidrug-resistant Mycobacterium tuberculosis. Eur. J. Med. Chem. 2019, 166, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Available online: https://www.molinspiration.com/ (accessed on 7 August 2022).



| Compound | E1 | S | K | A | P | E2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | |
| 4a | 17 | 0.25 | 17 | 0.06 | 7 | 0.25 | 0 | NT | 0 | NT | 9 | 0.25 |
| 4b | 0 | NT | 0 | NT | 0 | NT | 0 | NT | 0 | NT | 0 | NT |
| 4c | 0 | NT | 0 | NT | 0 | NT | 0 | NT | 0 | NT | 0 | NT |
| 4d | 0 | NT | 10 | 0.43 | 6 | 1.75 | 0 | NT | 0 | NT | 0 | NT |
| 4e | 14 | 0.45 | 15 | 0.23 | 7 | 0.45 | 0 | NT | 0 | NT | 0 | NT |
| 4f | 11 | 0.78 | 11 | 1.55 | 0 | NT | 0 | NT | 0 | NT | 13 | 1.55 |
| 4g | 10 | 1 | 17 | 0.5 | 8 | 0.5 | 0 | NT | 0 | NT | 13 | 0.5 |
| 4h | 0 | NT | 9 | 0.43 | 7 | 0.43 | 0 | NT | 0 | NT | 0 | NT |
| 4i | 0 | NT | 16 | 0.39 | 6 | 0.78 | 0 | NT | 0 | NT | 9 | 1.55 |
| 4j | 12 | 2.1 | 19 | 0.53 | 6 | 2.1 | 0 | NT | 0 | NT | 13 | 1.05 |
| 4k | 0 | NT | 18 | 0.66 | 9 | 0.83 | 0 | NT | 0 | NT | 15 | 0.66 |
| 4l | 15 | 0.93 | 9 | 0.93 | 5 | 1.85 | 0 | NT | 0 | NT | 0 | NT |
| 4m | 7 | 0.61 | 11 | 0.61 | 9 | 2.45 | 0 | NT | 0 | NT | 0 | NT |
| Furazidine | 13 | 32 | 23 | 8 | 14 | 32 | 0 | NT | 0 | NT | 21 | 2 |
| Nitrofurantoin | 15 | 8 | 21 | 4 | 12 | 64 | 0 | NT | 0 | NT | 24 | 4 |
| Ciprofloxacin | NT | 1.25 | NT | 1.25 | NT | 0.6 | NT | 2.5 | NT | 0.6 | NT | 0.3 |
| Compound | MIC (µg/mL) | Compound | MIC (µg/mL) |
|---|---|---|---|
| 4a | >100 | 4h | 100 |
| 4b | >100 | 4i | 6.2 |
| 4c | >100 | 4j | 50 |
| 4d | >100 | 4k | 50 |
| 4e | 50 | 4l | 100 |
| 4f | >100 | 4m | 100 |
| 4g | 100 | Isoniazid | 0.10 |
| Compound | Total System Energy (kcal/mol) | MM-GBSA ΔG (kcal/mol) | ||
|---|---|---|---|---|
| 1YKI | 7NB9 | 1YKI | 7NB9 | |
| native-ligand | −17,665.90 | −19,712.29 | −75.40 | −47.54 |
| 4a | −17,677.63 | −19,737.57 | −62.04 | −51.57 |
| 4b | −15,897.41 | −17,646.36 | −11.97 | −27.15 |
| 4c | −15,842.04 | −17,638.79 | −15.63 | −34.85 |
| 4d | −17,641.98 | −19,842.16 | −58.44 | −53.58 |
| Compound | MW | cLogP | HBA | HBD | NRotB |
|---|---|---|---|---|---|
| 4a | 326.4 | 4.08 | 7 | 1 | 4 |
| 4b | 360.8 | 4.71 | 7 | 1 | 4 |
| 4c | 394.4 | 4.93 | 7 | 1 | 5 |
| 4d | 340.4 | 4.49 | 7 | 1 | 4 |
| 4e | 314.4 | 3.20 | 7 | 1 | 4 |
| 4f | 406.4 | 4.97 | 8 | 1 | 7 |
| 4g | 327.3 | 3.16 | 8 | 1 | 4 |
| 4h | 355.4 | 3.82 | 8 | 1 | 4 |
| 4i | 332.4 | 3.82 | 7 | 1 | 4 |
| 4j | 334.4 | 3.54 | 7 | 1 | 4 |
| 4k | 392.4 | 3.77 | 9 | 1 | 7 |
| 4l | 392.4 | 3.76 | 9 | 1 | 8 |
| 4m | 321.4 | 2.48 | 8 | 1 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapegin, A.; Rogacheva, E.; Kraeva, L.; Gureev, M.; Dogonadze, M.; Vinogradova, T.; Yablonsky, P.; Balalaie, S.; Baykov, S.V.; Krasavin, M. Novel 5-Nitrofuran-Tagged Imidazo-Fused Azines and Azoles Amenable by the Groebke–Blackburn–Bienaymé Multicomponent Reaction: Activity Profile against ESKAPE Pathogens and Mycobacteria. Biomedicines 2022, 10, 2203. https://doi.org/10.3390/biomedicines10092203
Sapegin A, Rogacheva E, Kraeva L, Gureev M, Dogonadze M, Vinogradova T, Yablonsky P, Balalaie S, Baykov SV, Krasavin M. Novel 5-Nitrofuran-Tagged Imidazo-Fused Azines and Azoles Amenable by the Groebke–Blackburn–Bienaymé Multicomponent Reaction: Activity Profile against ESKAPE Pathogens and Mycobacteria. Biomedicines. 2022; 10(9):2203. https://doi.org/10.3390/biomedicines10092203
Chicago/Turabian StyleSapegin, Alexander, Elizaveta Rogacheva, Lyudmila Kraeva, Maxim Gureev, Marine Dogonadze, Tatiana Vinogradova, Petr Yablonsky, Saeed Balalaie, Sergey V. Baykov, and Mikhail Krasavin. 2022. "Novel 5-Nitrofuran-Tagged Imidazo-Fused Azines and Azoles Amenable by the Groebke–Blackburn–Bienaymé Multicomponent Reaction: Activity Profile against ESKAPE Pathogens and Mycobacteria" Biomedicines 10, no. 9: 2203. https://doi.org/10.3390/biomedicines10092203