Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach
Abstract
1. Introduction
2. Methods
2.1. Identified Childhood Asthma Risk Genes
2.2. Gene Ontology Enrichment Analysis
2.3. KEGG Pathway Enrichment Analysis
2.4. Discovering Biomarker Gene of Childhood Asthma
2.5. Statistical Analysis
3. Results
3.1. Childhood Asthma Risk Genes Identification
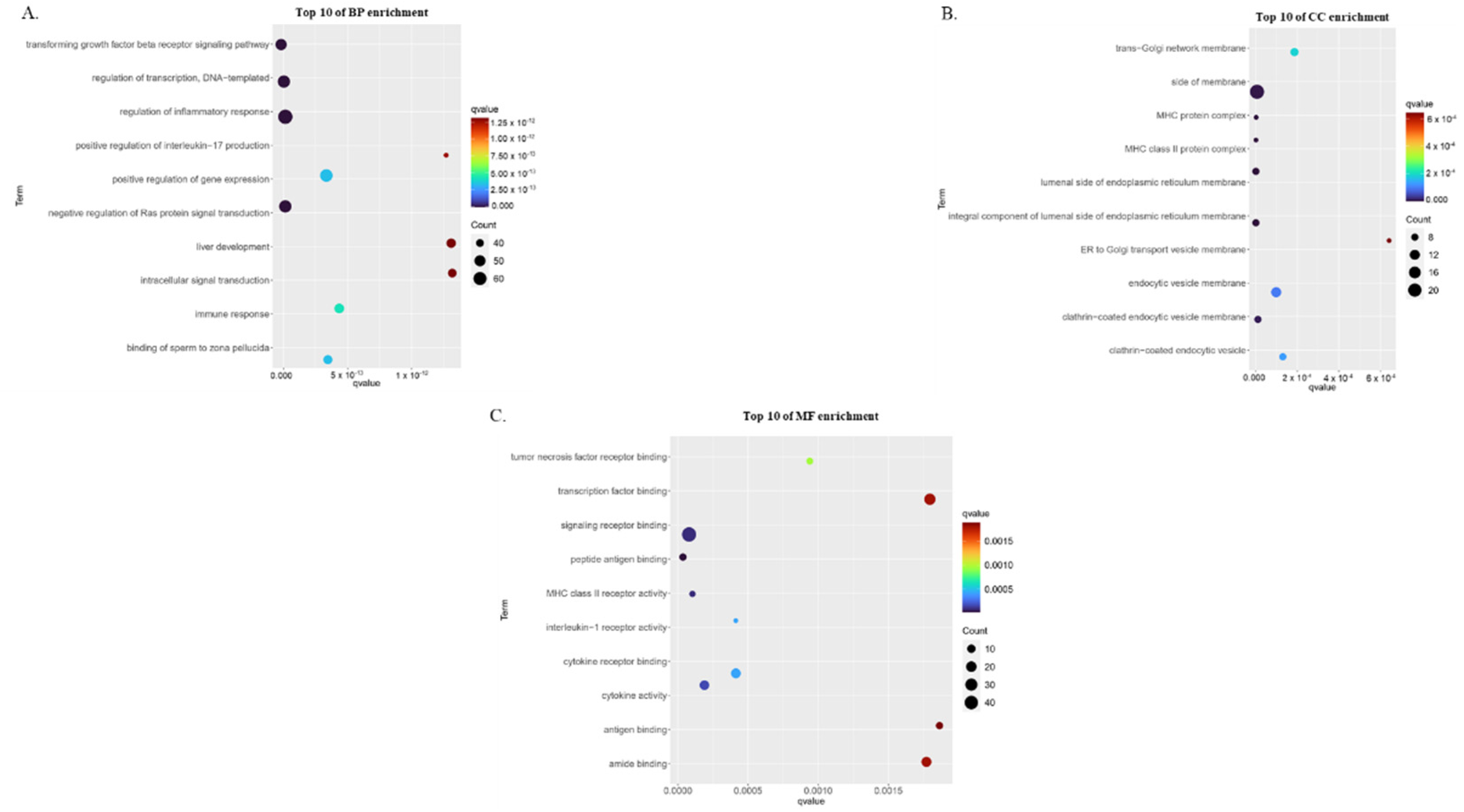
3.2. Gene Ontology Enrichment Analysis of Childhood Asthma Risk Genes
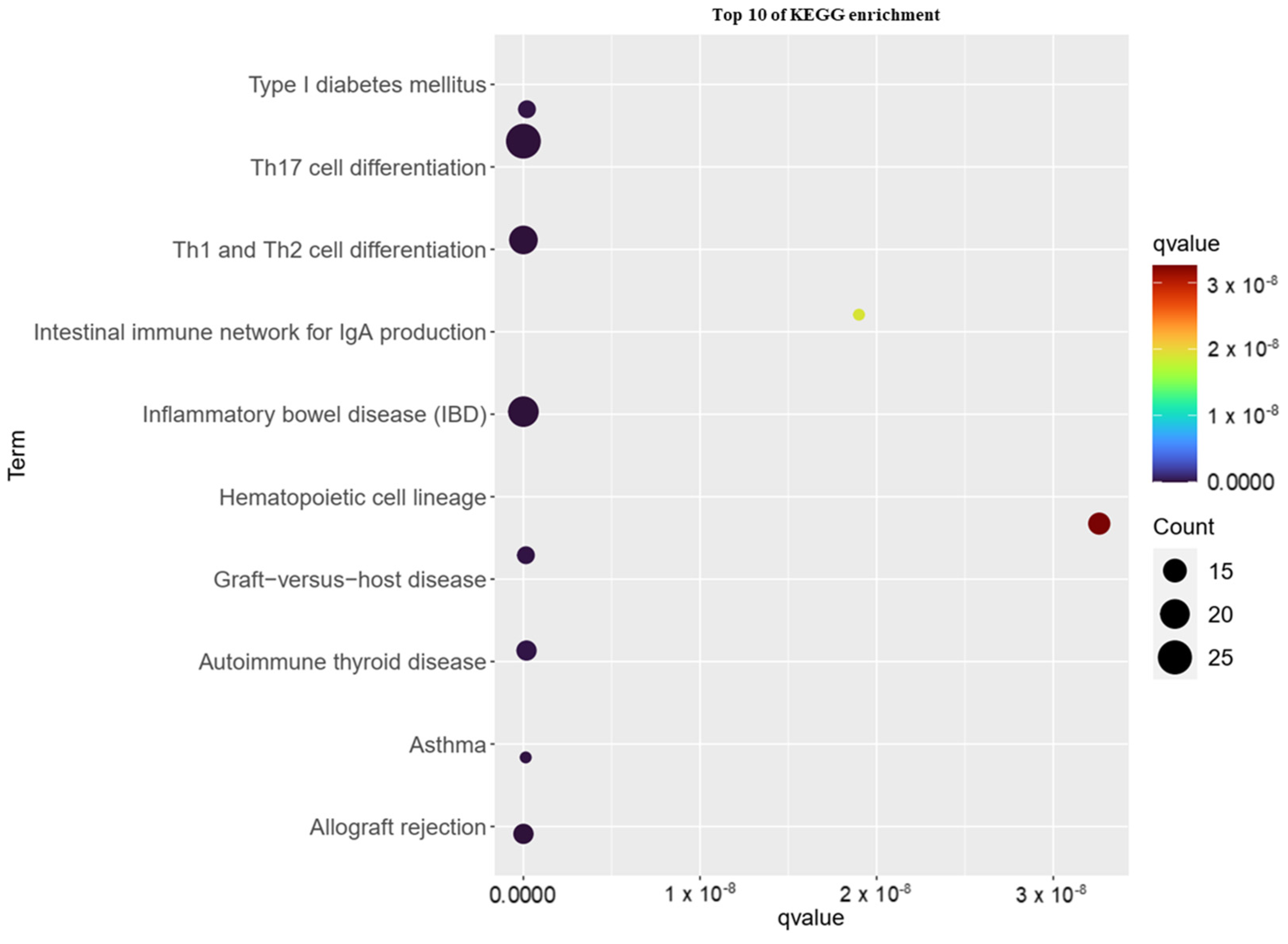
3.3. KEGG Pathway Analysis of Childhood Asthma Risk Genes
3.4. Identification of Potential Biomarkers of Childhood Asthma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Palmo, E.; Cantarelli, E.; Catelli, A.; Ricci, G.; Gallucci, M.; Miniaci, A.; Pession, A. The Predictive Role of Biomarkers and Genetics in Childhood Asthma Exacerbations. Int. J. Mol. Sci. 2021, 22, 4651. [Google Scholar] [CrossRef]
- Castillo, J.R.; Peters, S.P.; Busse, W.W. Asthma Exacerbations: Pathogenesis, Prevention, and Treatment. J. Allergy Clin. Immunol. Pract. 2017, 5, 918–927. [Google Scholar] [CrossRef]
- Singh, M. The Burden of Asthma in Children: An Asian Perspective. Paediatr. Respir. Rev. 2005, 6, 14–19. [Google Scholar] [CrossRef]
- Serebrisky, D.; Wiznia, A. Pediatric Asthma: A Global Epidemic. Ann. Glob. Health 2019, 85, 1–6. [Google Scholar] [CrossRef]
- Beasley, R.; Semprini, A.; Mitchell, E.A. Risk Factors for Asthma: Is Prevention Possible? Lancet 2015, 386, 1075–1085. [Google Scholar] [CrossRef]
- Bønnelykke, K.; Ober, C. Leveraging Gene-Environment Interactions and Endotypes for Asthma Gene Discovery. J. Allergy Clin. Immunol. 2016, 137, 667–679. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Kere, M.; Melén, E. Gene-environment Interactions in Childhood Asthma Revisited; Expanding the Interaction Concept. Pediatric Allergy Immunol. 2022, 33. [Google Scholar] [CrossRef]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef]
- Moore, W.C.; Meyers, D.A.; Wenzel, S.E.; Teague, W.G.; Li, H.; Li, X.; D’Agostino, R.; Castro, M.; Curran-Everett, D.; Fitzpatrick, A.M.; et al. Identification of Asthma Phenotypes Using Cluster Analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010, 181, 315–323. [Google Scholar] [CrossRef]
- Wenzel, S.E. Severe Adult Asthmas: Integrating Clinical Features, Biology, and Therapeutics to Improve Outcomes. Am. J. Respir. Crit. Care Med. 2021, 203, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ovando, S.; Simpson, J.L.; Barker, D.; Baines, K.J.; Wark, P.A.B. Transcriptomics of Biopsies Identifies Novel Genes and Pathways Linked to Neutrophilic Inflammation in Severe Asthma. Clin. Exp. Allergy 2021, 51, 1279–1294. [Google Scholar] [CrossRef]
- Global Initiative for Asthma Global Strategy for Asthma Management and Prevention (2019 Update). Available online: www.ginasthma.org (accessed on 25 April 2022).
- Robinson, D.; Humbert, M.; Buhl, R.; Cruz, A.A.; Inoue, H.; Korom, S.; Hanania, N.A.; Nair, P. Revisiting Type 2-High and Type 2-Low Airway Inflammation in Asthma: Current Knowledge and Therapeutic Implications. Clin. Exp. Allergy 2017, 47, 161–175. [Google Scholar] [CrossRef]
- Sánchez-García, S.; Habernau Mena, A.; Quirce, S. Biomarkers in Inflammometry Pediatric Asthma: Utility in Daily Clinical Practice. Eur. Clin. Respir. J. 2017, 4, 1356160. [Google Scholar] [CrossRef] [PubMed]
- Licari, A.; Castagnoli, R.; Brambilla, I.; Marseglia, A.; Tosca, M.A.; Marseglia, G.L.; Ciprandi, G. Asthma Endotyping and Biomarkers in Childhood Asthma. Pediatr. Allergy Immunol. Pulmonol. 2018, 31, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhong, Z.; Zhang, W.; Su, G.; Yang, P. Integrated Analysis of Key Pathways and Drug Targets Associated With Vogt-Koyanagi-Harada Disease. Front. Immunol 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Xia, Z.; Zhang, M.; Han, Q.; Hu, D.; Qi, S.; Xing, D.; Chen, Y.; Zhao, X. Integrated Bioinformatics-Based Identification of Potential Diagnostic Biomarkers Associated with Diabetic Foot Ulcer Development. J. Diabetes Res. 2021, 2021, 1–7. [Google Scholar] [CrossRef]
- Zhao, C.; Quan, X.; He, J.; Zhao, R.; Zhang, Y.; Li, X.; Sun, S.; Ma, R.; Zhang, Q. Identification of Significant Gene Biomarkers of Low Back Pain Caused by Changes in the Osmotic Pressure of Nucleus Pulposus Cells. Sci. Rep. 2020, 10, 1–19. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Hu, L.; Shen, X.; Liu, C. Identification of Potential Genetic Biomarkers and Target Genes of Peri-Implantitis Using Bioinformatics Tools. Biomed. Res. Int. 2021, 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Buniello, A.; Macarthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Blake, J.A.; Christie, K.R.; Dolan, M.E.; Drabkin, H.J.; Hill, D.P.; Ni, L.; Sitnikov, D.; Burgess, S.; Buza, T.; Gresham, C.; et al. Gene Ontology Consortium: Going Forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive Gene and Pathway Analysis of Cervical Cancer Progression. Oncol. Lett. 2020, 19, 3316–3332. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. V An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. CytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst Biol 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Wang, J.; Liao, Y.; Jaehnig, E.; Shi, Z.; Sheng, Q. Package ‘WebGestaltR’. Available online: https://cran.r-project.org/web/packages/WebGestaltR/index.html (accessed on 8 August 2022).
- Wickham, H. Ggplot2; Springer Science+Business Media: New York, NY, USA, 2009. [Google Scholar]
- Ober, C. Asthma Genetics in the Post-GWAS Era. In Proceedings of the Annals of the American Thoracic Society, New York, NY, USA, 1 March 2016; Volume 13, pp. S85–S90. [Google Scholar] [CrossRef]
- Beck, T.; Hastings, R.K.; Gollapudi, S.; Free, R.C.; Brookes, A.J. GWAS Central: A Comprehensive Resource for the Comparison and Interrogation of Genome-Wide Association Studies. Eur. J. Hum. Genet. 2014, 22, 949–952. [Google Scholar] [CrossRef]
- Macarthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; Mcmahon, A.; Milano, A.; Morales, J.; et al. The New NHGRI-EBI Catalog of Published Genome-Wide Association Studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, 896–901. [Google Scholar] [CrossRef]
- Irham, L.M.; Adikusuma, W.; Perwitasari, D.A.; Dania, H.; Maliza, R.; Faridah, I.N.; Santri, I.N.; Phiri, Y.V.A.; Cheung, R. The Use of Genomic Variants to Drive Drug Repurposing for Chronic Hepatitis B. Biochem. Biophys. Rep. 2022, 31, 101307. [Google Scholar] [CrossRef]
- Adikusuma, W.; Irham, L.M.; Chou, W.H.; Wong, H.S.C.; Mugiyanto, E.; Ting, J.; Perwitasari, D.A.; Chang, W.P.; Chang, W.C. Drug Repurposing for Atopic Dermatitis by Integration of Gene Networking and Genomic Information. Front. Immunol 2021, 12, 724277. [Google Scholar] [CrossRef]
- Adikusuma, W.; Chou, W.H.; Lin, M.R.; Ting, J.; Irham, L.M.; Perwitasari, D.A.; Chang, W.P.; Chang, W.C. Identification of Druggable Genes for Asthma by Integrated Genomic Network Analysis. Biomedicines 2022, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, A.; De Ferrari, L.; Folli, C.; Mauri, P.; Riccio, A.M.; Canonica, G.W. Biomarkers and Severe Asthma: A Critical Appraisal. Clin. Mol. Allergy 2015, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hachim, M.Y.; Elemam, N.M.; Ramakrishnan, R.K.; Salameh, L.; Olivenstein, R.; Hachim, I.Y.; Venkatachalam, T.; Mahboub, B.; Al Heialy, S.; Hamid, Q.; et al. Derangement of Cell Cycle Markers in Peripheral Blood Mononuclear Cells of Asthmatic Patients as a Reliable Biomarker for Asthma Control. Sci. Rep. 2021, 11, 1–24. [Google Scholar] [CrossRef]
- Peters, M.C.; Mcgrath, K.W.; Hawkins, G.A.; Ph, D.; Hastie, T.; Ph, D.; Levy, B.D.; Israel, E.; Phillips, B.R.; Mauger, T.; et al. Plasma IL6 Levels, Metabolic Dysfunction, and Asthma Severity: A Cross-Sectional Analysis of Two Cohorts. Lancet Respir. Med. 2017, 4, 574–584. [Google Scholar] [CrossRef]
- Jackson, D.J.; Bacharier, L.B.; Calatroni, A.; Gill, M.A.; Hu, J.; Liu, A.H.; Wheatley, L.M.; Gern, J.E.; Gruchalla, R.S.; Hershey, G.K.K.; et al. Serum IL-6: A Biomarker in Childhood Asthma? J. Allergy Clin. Immunol. 2021, 145, 1701–1704. [Google Scholar] [CrossRef]
- Neveu, W.A.; Allard, J.L.; Raymond, D.M.; Bourassa, L.M.; Burns, S.M.; Bunn, J.Y.; Irvin, C.G.; Kaminsky, D.A.; Rincon, M. Elevation of IL-6 in the Allergic Asthmatic Airway Is Independent of Inflammation but Associates with Loss of Central Airway Function. Respir. Res. 2010, 11, 1–10. [Google Scholar] [CrossRef]
- Jevnikar, Z.; Östling, J.; Ax, E.; Calvén, J.; Thörn, K.; Israelsson, E.; Öberg, L.; Singhania, A.; Lau, L.C.K.; Wilson, S.J.; et al. Epithelial IL-6 Trans-Signaling Defines a New Asthma Phenotype with Increased Airway Inflammation. J. Allergy Clin. Immunol. 2019, 143, 577–590. [Google Scholar] [CrossRef]
- Permaul, P.; Mas, M.C.P.; Ma, C.R.P.; Carlos, J.; Mph, C.; Ly, N.P.; Mph, S.K.R.; Ross, K.; Fitzpatrick, A.; Israel, E.; et al. The Association of Plasma IL-6 with Measures of Asthma Morbidity in a Moderate-Severe Pediatric Cohort Aged 6–18 Years. J. Allergy Clin. Immunol. Pract. 2021, 9, 2916–2919.e2. [Google Scholar] [CrossRef]
- Massey, O.; Suphioglu, C. Recent Advances in the Inhibition of the IL-4 Cytokine Pathway for the Treatment of Allergen-Induced Asthma. Int. J. Mole 2021, 22, 13655. [Google Scholar] [CrossRef]
- Wang, R.; Jin, H.; Shang, S.; Liu, X.; Chen, S.; Jin, Z. Associations of IL-2 and IL-4 Expression and Polymorphisms With the Risks of Mycoplasma Pneumoniae Infection and Asthma in Children. Arch. Bronconeumol 2015, 51, 571–578. [Google Scholar] [CrossRef]
- Takayama, G.; Arima, K.; Kanaji, T.; Toda, S.; Tanaka, H.; Shoji, S.; McKenzie, A.N.J.; Nagai, H.; Hotokebuchi, T.; Izuhara, K. Periostin: A Novel Component of Subepithelial Fibrosis of Bronchial Asthma Downstream of IL-4 and IL-13 Signals. J. Allergy Clin. Immunol. 2006, 118, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Munitz, A.; Brandt, E.B.; Mingler, M.; Finkelman, F.D.; Rothenberg, M.E. Distinct Roles for IL-13 and IL-4 via IL-13 Receptor Alpha1 and the Type II IL-4 Receptor in Asthma Pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 7240–7245. [Google Scholar] [CrossRef]
- Truyen, E.; Coteur, L.; Dilissen, E.; Overbergh, L.; Dupont, L.J.; Ceuppens, J.L.; Bullens, D.M.A. Evaluation of Airway Inflammation by Quantitative Th1/Th2 Cytokine MRNA Measurement in Sputum of Asthma Patients. Thorax 2006, 61, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.S.; Lowe, A.J.; Samarasinghe, P.; Lodge, C.J.; Huang, Y.; Abramson, M.J.; Dharmage, S.C.; Jaffe, A. Exhaled Breath Condensate in Pediatric Asthma: Promising New Advance or Pouring Cold Water on a Lot of Hot Air? A Systematic Review. Pediatr. Pulmonol. 2013, 48, 419–442. [Google Scholar] [CrossRef] [PubMed]
- Robroeks, C.M.H.H.T.; van de Kant, K.D.G.; Jöbsis, Q.; Hendriks, H.J.E.; van Gent, R.; Wouters, E.F.M.; Damoiseaux, J.G.M.C.; Bast, A.; Wodzig, W.K.W.H.; Dompeling, E. Exhaled Nitric Oxide and Biomarkers in Exhaled Breath Condensate Indicate the Presence, Severity and Control of Childhood Asthma. Clin. Exp. Allergy 2007, 37, 1303–1311. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.K.; Kharitonov, S.A.; Wilson, N.M.; Bush, A.; Barnes, P.J. Increased Interleukin-4 and Decreased Interferon-Gamma in Exhaled Breath Condensate of Children with Asthma. Am. J. Respir. Crit. Care Med. 2002, 165, 1290–1293. [Google Scholar] [CrossRef] [PubMed]


| Rank | Gene ID | Gene Name | Score |
|---|---|---|---|
| 1 | IL6 | Interleukin 6 | 22,370,717,281 |
| 2 | IL4 | Interleukin 4 | 22,370,701,130 |
| 3 | IL2 | Interleukin 2 | 22,370,622,650 |
| 4 | IL13 | Interleukin 13 | 22,369,670,594 |
| 5 | PTPRC | Protein Tyrosine Phosphatase Receptor Type C | 22,363,673,434 |
| 6 | IL5 | Interleukin 5 | 22,340,430,000 |
| 7 | IL33 | Interleukin 33 | 21,784,097,718 |
| 8 | TBX21 | T-Box Transcription Factor 21 | 21,767,214,138 |
| 9 | IL2RA | Interleukin 2 Receptor Subunit Alpha | 21,701,358,536 |
| 10 | STAT6 | Signal Transducer and Activator of Transcription 6 | 21,233,178,024 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santri, I.N.; Irham, L.M.; Djalilah, G.N.; Perwitasari, D.A.; Wardani, Y.; Phiri, Y.V.A.; Adikusuma, W. Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach. Biomedicines 2022, 10, 2311. https://doi.org/10.3390/biomedicines10092311
Santri IN, Irham LM, Djalilah GN, Perwitasari DA, Wardani Y, Phiri YVA, Adikusuma W. Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach. Biomedicines. 2022; 10(9):2311. https://doi.org/10.3390/biomedicines10092311
Chicago/Turabian StyleSantri, Ichtiarini Nurullita, Lalu Muhammad Irham, Gina Noor Djalilah, Dyah Aryani Perwitasari, Yuniar Wardani, Yohane Vincent Abero Phiri, and Wirawan Adikusuma. 2022. "Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach" Biomedicines 10, no. 9: 2311. https://doi.org/10.3390/biomedicines10092311
APA StyleSantri, I. N., Irham, L. M., Djalilah, G. N., Perwitasari, D. A., Wardani, Y., Phiri, Y. V. A., & Adikusuma, W. (2022). Identification of Hub Genes and Potential Biomarkers for Childhood Asthma by Utilizing an Established Bioinformatic Analysis Approach. Biomedicines, 10(9), 2311. https://doi.org/10.3390/biomedicines10092311










