Inflammation Regulation by Bacterial Molecular Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Care
2.2. Sample Administration to Mice
2.3. Isolation of Mononuclear Cells
2.4. Ethics Approvement and Voluntary Consent for Personal Data Processing and Publishing
2.5. In Vitro Studies
2.6. Quantitative RT-PCR
2.7. Statistical Analysis
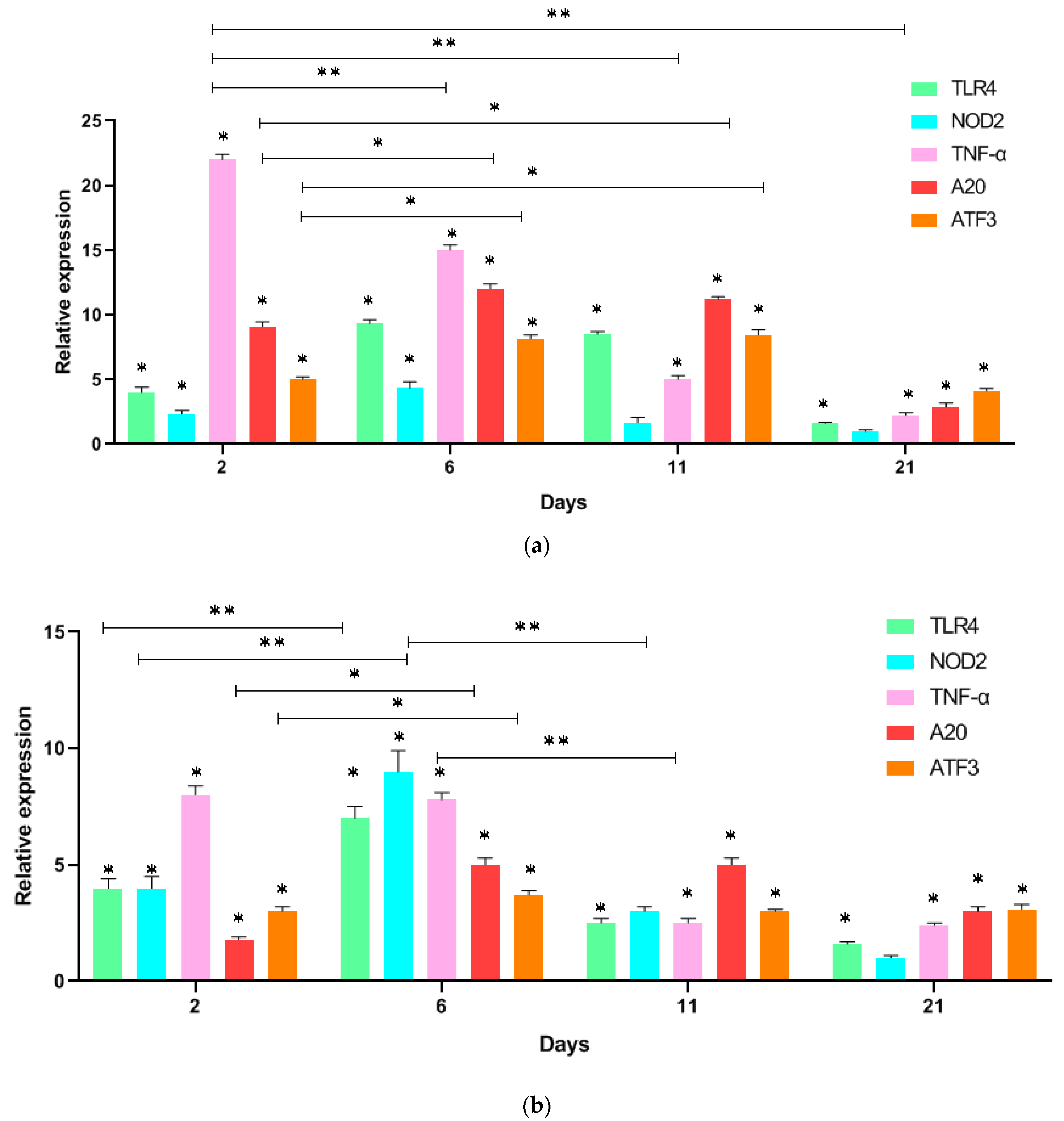
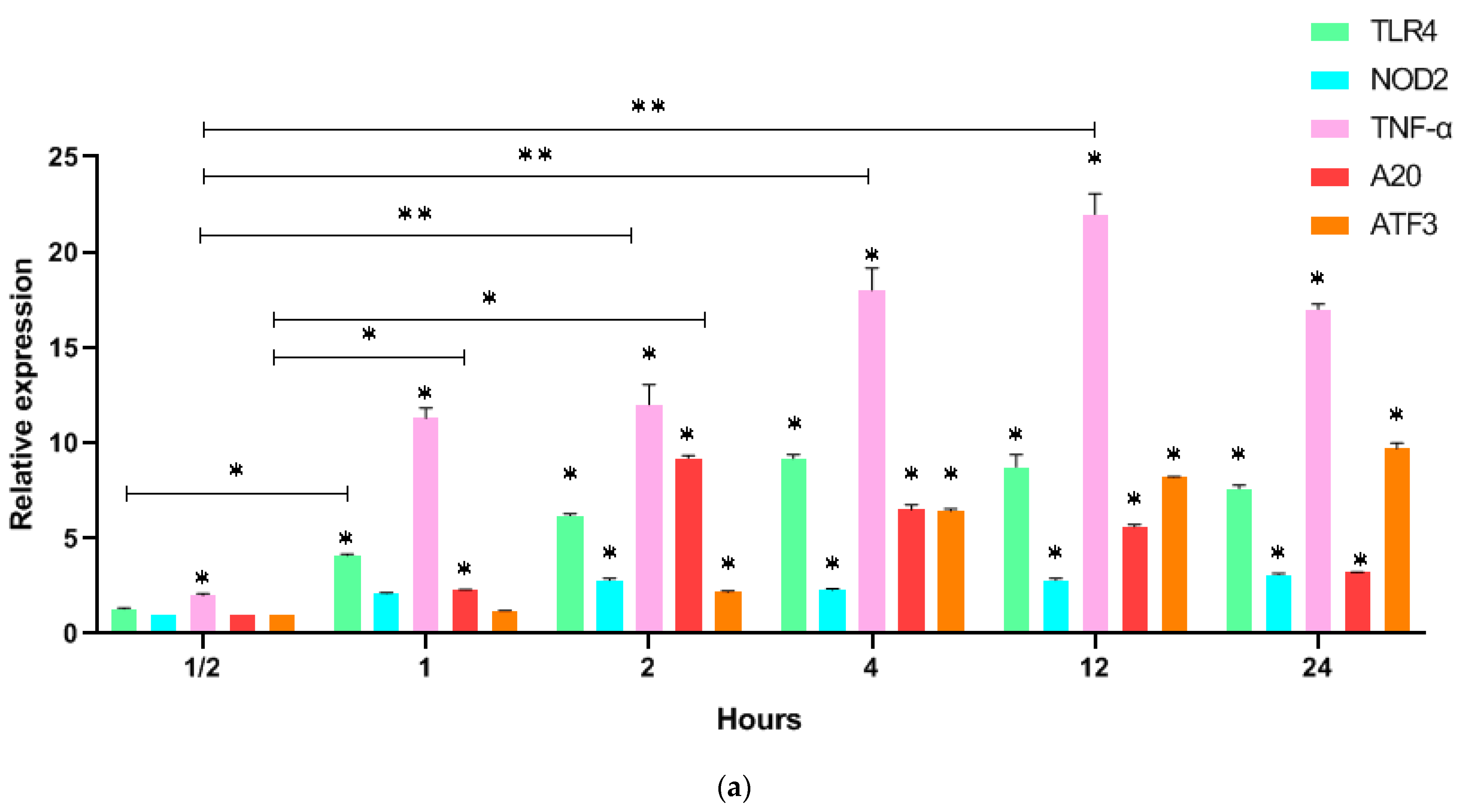
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Meunier, E.; Broz, P. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 2017, 38, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef]
- Netea, M.G. Training innate immunity: The changing concept of immunological memory in innate host defence. Eur. J. Clin. Investig. 2013, 43, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Larsen, S.B.; Gomez, N.C.; Alaverdyan, K.; Sendoel, A.; Yuan, S.; Polak, L.; Kulukian, A.; Chai, S.; Fuchs, E. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 2017, 550, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Ordovas-Montanes, J.; Dwyer, D.F.; Nyquist, S.K.; Buchheit, K.M.; Vukovic, M.; Deb, C.; Wadsworth, M.H., 2nd; Hughes, T.K.; Kazer, S.W.; Yoshimoto, E.; et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 2018, 560, 649–654. [Google Scholar] [CrossRef]
- Netea, M.G.; van Crevel, R. BCG-induced protection: Effects on innate immune memory. Semin. Immunol. 2014, 26, 512–517. [Google Scholar] [CrossRef]
- Xing, Z.; Afkhami, S.; Bavananthasivam, J.; Fritz, D.K.; D’Agostino, M.R.; Vaseghi-Shanjani, M.; Yao, Y.; Jeyanathan, M. Innate immune memory of tissue-resident macrophages and trained innate immunity: Re-vamping vaccine concept and strategies. J. Leukoc. Biol. 2020, 108, 825–834. [Google Scholar] [CrossRef]
- Medzhitov, R.; Schneider, D.S.; Soares, M.P. Disease tolerance as a defense strategy. Science 2012, 335, 936–941. [Google Scholar] [CrossRef]
- Shi, C.Y.; Yu, C.H.; Yu, W.Y.; Ying, H.Z. Gut-Lung Microbiota in Chronic Pulmonary Diseases: Evolution, Pathogenesis, and Therapeutics. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 9278441. [Google Scholar] [CrossRef]
- Gerretsen, V.I.V.; Schuijs, M.J. The role of LPS and CpG in the farm effect against allergies, and beyond. Allergol. Sel. 2022, 6, 104–110. [Google Scholar] [CrossRef]
- Feterowski, C.; Weighardt, H.; Emmanuilidis, K.; Hartung, T.; Holzmann, B. Immune protection against septic peritonitis in endotoxin-primed mice is related to reduced neutrophil apoptosis. Eur. J. Immunol. 2001, 31, 1268–1277. [Google Scholar] [CrossRef]
- Meshcheryakova, E.; Guryanova, S.; Makarov, E.; Alekseeva, L.; Andronova, T.; Ivanov, V. Prevention of experimental septic shock by pretreatment of mice with muramyl peptides. Int. Immunopharmacol. 2001, 1, 1857–1865. [Google Scholar] [CrossRef]
- Hedl, M.; Li, J.; Cho, J.H.; Abraham, C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc. Natl. Acad. Sci. USA 2007, 104, 19440–19445. [Google Scholar] [CrossRef]
- Bastos, P.A.D.; Wheeler, R.; Boneca, I.G. Uptake, Recognition and Responses to Peptidoglycan in the Mammalian Host. FEMS Microbiol. Rev. 2021, 45, fuaa044. [Google Scholar] [CrossRef]
- Cestero, J.J.; Castanheira, S.; Pucciarelli, M.G.; García-Del Portillo, F. A Novel Salmonella Periplasmic Protein Controlling Cell Wall Homeostasis and Virulence. Front. Microbiol. 2021, 12, 633701. [Google Scholar] [CrossRef]
- Irazoki, O.; Hernandez, S.B.; Cava, F. Peptidoglycan Muropeptides: Release, Perception, and Functions as Signaling Molecules. Front. Microbiol. 2019, 10, 500. [Google Scholar] [CrossRef]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Ma, J.; Sun, M.; Pan, Z.; Lu, C.; Yao, H. Diverse Toxic Effectors are Harbored by Vgrg Islands for Interbacterial Antagonism in Type VI Secretion System. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 1635–1643. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Khaitov, R.M. Strategies for Using Muramyl Peptides-Modulators of Innate Immunity of Bacterial Origin-in Medicine. Front. Immunol. 2021, 12, 607178. [Google Scholar] [CrossRef]
- Adam, A.; Lederer, E. Muramyl Peptides: Immunomodulators, Sleep Factors, and Vitamins. In Medical Research Reviews; Stevens, D.G., Ed.; Wiley and Sons: Hoboken, NJ, USA, 1984; Volume 4, pp. 111–152. [Google Scholar] [CrossRef]
- Guryanova, S.V. Regulation of Immune Homeostasis via Muramyl Peptides-Low Molecular Weight Bioregulators of Bacterial Origin. Microorganisms 2022, 10, 1526. [Google Scholar] [CrossRef] [PubMed]
- Fallah, A.; Sedighian, H.; Behzadi, E.; Havaei, S.A.; Kachuei, R.; Imani Fooladi, A.A. The role of serum circulating microbial toxins in severity and cytokine storm of COVID positive patients. Microb. Pathog. 2022, 16, 105888. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Susaki, E.A.; Nagaoka, I. Lipopolysaccharides and Cellular Senescence: Involvement in Atherosclerosis. Int. J. Mol. Sci. 2022, 23, 11148. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczak-Wiercioch, A.; Sałat, K. Lipopolysaccharide-Induced Model of Neuroinflammation: Mechanisms of Action, Research Application and Future Directions for Its Use. Molecules 2022, 27, 5481. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, H.; Hong, S.; Molagoda, I.M.N.; Jeong, J.W.; Jin, C.Y.; Kim, G.Y.; Choi, S.H.; Hong, S.H.; Choi, Y.H. Inhibition of Lipopolysaccharide-Induced Inflammatory and Oxidative Responses by Trans-cinnamaldehyde in C2C12 Myoblasts. Int. J. Med. Sci. 2021, 18, 2480–2492. [Google Scholar] [CrossRef]
- Liu, X.; Yin, S.; Chen, Y.; Wu, Y.; Zheng, W.; Dong, H.; Bai, Y.; Qin, Y.; Li, J.; Feng, S.; et al. LPS-induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF-κB, STAT3 or AP-1 activation. Mol. Med. Rep. 2018, 17, 5484–5491. [Google Scholar] [CrossRef]
- Torres-Espín, A.; Forero, J.; Fenrich, K.K.; Lucas-Osma, A.M.; Krajacic, A.; Schmidt, E.; Vavrek, R.; Raposo, P.; Bennett, D.J.; Popovich, P.G.; et al. Eliciting inflammation enables successful rehabilitative training in chronic spinal cord injury. Brain 2018, 141, 1946–1962. [Google Scholar] [CrossRef]
- Brooks, D.; Barr, L.C.; Wiscombe, S.; McAuley, D.F.; Simpson, A.J.; Rostron, A.J. Human lipopolysaccharide models provide mechanistic and therapeutic insights into systemic and pulmonary inflammation. Eur. Respir. J. 2020, 56, 1901298. [Google Scholar] [CrossRef]
- Buters, T.P.; Hameeteman, P.W.; Jansen, I.M.E.; van Hindevoort, F.C.; Ten Voorde, W.; Florencia, E.; Osse, M.; de Kam, M.L.; Grievink, H.W.; Schoonakker, M.; et al. Intradermal lipopolysaccharide challenge as an acute in vivo inflammatory model in healthy volunteers. Br. J. Clin. Pharmacol. 2022, 88, 680–690. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Gigani, O.B.; Gudima, G.O.; Kataeva, A.M.; Kolesnikova, N.V. Dual Effect of Low Molecular Weight Bioregulators of Bacterial Origin in Experimental Model of Asthma. Life 2022, 12, 192. [Google Scholar] [CrossRef]
- Kolesnikova, N.V.; Kozlov, I.G.; Guryanova, S.V.; Kokov, E.A.; Andronova, T.M. Clinical and immunological efficiency of muramyl dipeptide in the treatment of atopic diseases. Med. Immunol. 2016, 18, 15–20. (In Russian) [Google Scholar] [CrossRef]
- Manapova, E.R.; Fazylov VKh Guryanova, S.V. cytopenia and their correction in antiviral therapy of chronic hepatitis C in patients with genotype 1. Probl. Virol. 2017, 62, 174–178. [Google Scholar] [CrossRef]
- Guryanova, S.; Udzhukhu, V.; Kubylinsky, A. Pathogenetic Therapy of Psoriasis by Muramyl Peptide. Front. Immunol. 2019, 10, 1275. [Google Scholar] [CrossRef]
- Davis, K.; Nakamura, S.; Weiser, J.N. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. J. Clin. Investig. 2011, 121, 3666–3676. [Google Scholar] [CrossRef]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 is a General Sensor of Peptidoglycan Through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef]
- Brosbøl-Ravnborg, A.; Hvas, C.L.; Agnholt, J.; Dahlerup, J.F.; Vind, I.; Till, A.; Rosenstiel, P.; Höllsberg, P. Toll-Like Receptor-Induced Granulocyte-Macrophage Colony-Stimulating Factor Secretion is Impaired in Crohn’s Disease by Nucleotide Oligomerization Domain 2-Dependent and -Independent Pathways. Clin. Exp. Immunol. 2009, 155, 487–495. [Google Scholar] [CrossRef]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef]
- Wicherska-Pawłowska, K.; Wróbel, T.; Rybka, J. Toll-Like Receptors (TLRs), NOD-Like Receptors (NLRs), and RIG-I-Like Receptors (RLRs) in Innate Immunity. TLRs, NLRs, and RLRs Ligands as Immunotherapeutic Agents for Hematopoietic Diseases. Int. J. Mol. Sci. 2021, 22, 13397. [Google Scholar] [CrossRef]
- Sizyakina, L.P.; Andreeva, I.I.; Petruchik, S.V. Optimization of Therapy of Patient with Genetic Defect Antibody Production. RUDN J. Med. 2019, 3, 405–411. [Google Scholar] [CrossRef]
- Rosenstiel, P.; Fantini, M.; Seegert, D.; Schreiberdoi, S. TNF-α and IFN-γ Regulate the Expression of the NOD2 (CARD15) Gene in Human Intestinal Epithelial Cells. Gastroenterology 2003, 124, 1001–1009. [Google Scholar] [CrossRef]
- Takahashi, Y.; Isuzugawa, K.; Murase, Y.; Imai, M.; Yamamoto, S.; Iizuka, M.; Akira, S.; Bahr, G.M.; Momotani, E.; Hori, M.; et al. Up-regulation of NOD1 and NOD2 through TLR4 and TNF-alpha in LPS-treated murine macrophages. J. Vet. Med. Sci. 2006, 68, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Tewari, R.; Choudhury, S.R.; Ghosh, S.; Mehta, V.S.; Sen, E. Involvement of TNFα-induced TLR4–NF-κB and TLR4–HIF-1α feed-forward loops in the regulation of inflammatory responses in glioma. J. Mol. Med. 2012, 90, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, X.; Fu, D.; Gu, Y.; Fan, R.; Yi, H.; He, X.; Wang, C.; Ouyang, B.; Zhao, P.; et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe 2022, 30, 1139–1150.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, Y.; Bao, P.; Wu, J.; Zeng, G.; Hu, X. β1,4-galactosyltransferase-I protects chondrocytes against TNF-induced apoptosis by blocking the TLR4 signaling pathway. Am. J. Transl. Res. 2019, 11, 4358–4366. [Google Scholar]
- Dagil, Y.A.; Arbatsky, N.P.; Alkhazova, B.I.; L’vov, V.L.; Mazurov, D.V.; Pashenkov, M.V. The Dual NOD1/NOD2 Agonism of Muropeptides Containing a Meso-Diaminopimelic Acid Residue. PLoS ONE 2016, 11, e0160784. [Google Scholar] [CrossRef]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M. Tumour necrosis factor signalling in health and disease. F1000Research 2019, 8, 111. [Google Scholar] [CrossRef]
- Sethi, J.K.; Hotamisligil, G.S. Metabolic Messengers: Tumour necrosis factor. Nat. Metab. 2021, 3, 1302–1312. [Google Scholar] [CrossRef]
- Chu, H.M.; Tan, Y.; Kobierski, L.A.; Balsam, L.B.; Comb, M.J. Activating transcription factor-3 stimulates 3’,5’-cyclic adenosine monophosphate-dependent gene expression. Mol. Endocrinol. 1994, 8, 59–68. [Google Scholar] [CrossRef]
- Thompson, M.R.; Xu, D.; Williams, B.R. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. 2009, 87, 1053–1060. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Sun, X.; Liu, J.; Fu, Y.; Liu, B.; Qiu, J.; Lian, J.; Zhou, J. ATF3 in atherosclerosis: A controversial transcription factor. J. Mol. Med. 2022, 100, 1557–1568. [Google Scholar] [CrossRef]
- Kwon, J.-W.; Kwon, H.-K.; Shin, H.-J.; Choi, Y.-M.; Anwar, M.A.; Choi, S. Activating transcription factor 3 represses inflammatory responses by binding to the p65 subunit of NF-κB. Sci. Rep. 2015, 5, 14470. [Google Scholar] [CrossRef]
- Shembade, N.; Harhaj, E. Regulation of NF-κB signaling by the A20 deubiquitinase. Cell Mol. Immunol. 2012, 9, 123–130. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- McKeown-Longo, P.J.; Higgins, P.J. Integration of Canonical and Noncanonical Pathways in TLR4 Signaling: Complex Regulation of the Wound Repair Program. Adv. Wound Care 2017, 6, 320–329. [Google Scholar] [CrossRef]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef]
- The sbv IMPROVER project team (in alphabetical order); Boué, S.; Fields, B.; Hoeng, J.; Park, J.; Peitsch, M.C.; Schlage, W.K.; Talikka, M.; Binenbaum, I.; Bondarenko, V.; et al. Enhancement of COPD biological networks using a web-based collaboration interface. F1000Research 2015, 4, 32. [Google Scholar] [CrossRef]
- Namasivayam, A.A.; Morales, A.F.; Lacave, M.F.; Tallam, A.; Simovic, B.; Alfaro, D.G.; Bobbili, D.R.; Martin, F.; Androsova, G.; Shvydchenko, I.; et al. Community-Reviewed Biological Network Models for Toxicology and Drug Discovery Applications. Gene Regul. Syst. Biol. 2016, 10, 51–66. [Google Scholar] [CrossRef]
- Günthner, R.; Kumar, V.R.; Lorenz, G.; Anders, H.J.; Lech, M. Pattern-recognition receptor signaling regulator mRNA expression in humans and mice, and in transient inflammation or progressive fibrosis. Int J Mol Sci. 2013, 14, 18124–18147. [Google Scholar] [CrossRef]
- Primer-BLAST. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 5 January 2023).
- King, A.E.; Horne, A.W.; Hombach-Klonisch, S.; Mason, J.I.; Critchley, H.O. Differential expression and regulation of nuclear oligomerization domain proteins NOD1 and NOD2 in human endometrium: A potential role in innate immune protection and menstruation. Mol Hum Reprod. 2009, 15, 311–319. [Google Scholar] [CrossRef]
- Wan, J.; Shan, Y.; Fan, Y.; Fan, C.; Chen, S.; Sun, J.; Zhu, L.; Qin, L.; Yu, M.; Lin, Z. NF-κB inhibition attenuates LPS-induced TLR4 activation in monocyte cells. Mol. Med. Rep. 2016, 14, 4505–4510. [Google Scholar] [CrossRef]
- Hassan-Nejhad, M.; Bagheri, M.; Khadem-Vatani, K.; Seyed Mohammad Zad, M.H.; Abdi Rad, I.; Rahimi, B.; Rostamzadeh, A.; Rahimlou, A. Tumor Necrosis Factor-alpha Gene Expression in PBMCs of Iranian Azeri Turkish Patients with Premature Coronary Artery Disease (Age.50 Years). Maedica 2018, 13, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Yoon, H.M.; Kwon, O.; Lee, W.J. The Effect of Pueraria Lobata/Rehmannia Glutinosa and Exercise on Fatty Acid Transporters Expression in Ovariectomized Rats Skeletal Muscles. J. Exerc. Nutrition Biochem. 2016, 20, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.origene.com/catalog/gene-expression/qpcr-primer-pairs/mp200254/atf3-mouse-qpcr-primer-pair-nm_007498 (accessed on 5 January 2023).
- Bowcutt, R.; Bramhall, M.; Logunova, L.; Wilson, J.; Booth, C.; Carding, S.R.; Grencis, R.; Cruickshank, S. A role for the pattern recognition receptor Nod2 in promoting recruitment of CD103+ dendritic cells to the colon in response to Trichuris muris infection. Mucosal. Immunol. 2014, 7, 1094–1105. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guryanova, S.V.; Kataeva, A. Inflammation Regulation by Bacterial Molecular Patterns. Biomedicines 2023, 11, 183. https://doi.org/10.3390/biomedicines11010183
Guryanova SV, Kataeva A. Inflammation Regulation by Bacterial Molecular Patterns. Biomedicines. 2023; 11(1):183. https://doi.org/10.3390/biomedicines11010183
Chicago/Turabian StyleGuryanova, Svetlana V., and Anastasiya Kataeva. 2023. "Inflammation Regulation by Bacterial Molecular Patterns" Biomedicines 11, no. 1: 183. https://doi.org/10.3390/biomedicines11010183
APA StyleGuryanova, S. V., & Kataeva, A. (2023). Inflammation Regulation by Bacterial Molecular Patterns. Biomedicines, 11(1), 183. https://doi.org/10.3390/biomedicines11010183




