Mu-Net a Light Architecture for Small Dataset Segmentation of Brain Organoid Bright-Field Images
Abstract
:1. Introduction
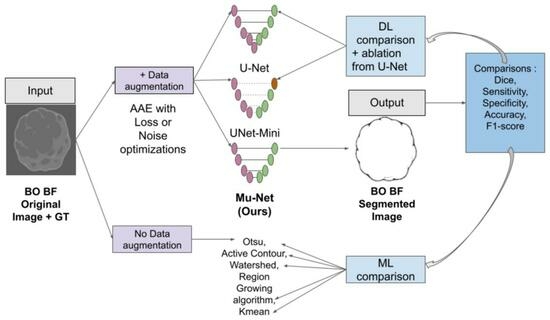
2. Materials and Methods
2.1. Resources
2.2. Segmentation Architectures
2.2.1. U-Net
2.2.2. UNet-Mini
2.2.3. Mu-Net
2.3. Training
2.4. Classical Segmentation Methods
2.5. Comparison of Segmentations
2.6. Visualisation
2.7. Ablation study
- Unet,
- “Layer”,
- “filter”,
- “kernel”,
- “Ewise”,
- “layer + filter”,
- “layer + kernel”,
- “layer + ewise”,
- “Filter + kernel”,
- “Filter + ewise”,
- “Kernel + ewise”,
- “layer + filter + kernel”,
- “layer + kernel + ewise”,
- “layer + filter + ewise”,
- “filter + kernel + ewise”,
- “layer + filter + kernel + ewise”,
- “layer + filter + kernel + stride” (Mu-Net),
- “layer + filter + kernel + ewise + stride + activation” (UNet-Mini).
3. Results
3.1. Qualitative
3.2. Quantitative
3.3. Computational Comparisons
3.4. Mu-Net and Machine Learning Comparisons
3.4.1. Qualitative Analysis
3.4.2. Quantitative Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BO | Brain organoid |
| AAE | Adversarial autoencoder |
| GAN | Generative adversarial network |
| ML | machine learning |
| DL | deep learning |
| GT | ground truth |
| BCE | Binary cross entropy |
| BCE + L1 | Binary cross entropy with L1 normalization |
| LS | Least square |
| Wass. | Wasserstein |
| P. Wass | Perceptual Wasserstein |
| Salt Pepp. | Salt and pepper noise |
References
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Kelava, I.; Lancaster, M.A. Dishing out mini-brains: Current progress and future prospects in brain organoid research. Dev. Biol. 2016, 420, 199–209. [Google Scholar] [CrossRef]
- Brémond Martin, C.; Simon Chane, C.; Clouchoux, C.; Histace, A. Recent Trends and Perspectives in Cerebral Organoids Imaging and Analysis. Front. Neurosci. 2021, 15, 629067. [Google Scholar] [CrossRef]
- Albanese, A.; Swaney, J.M.; Yun, D.H.; Evans, N.B.; Antonucci, J.M.; Velasco, S.; Sohn, C.H.; Arlotta, P.; Gehrke, L.; Chung, K. Multiscale 3D phenotyping of human cerebral organoids. Sci. Rep. 2020, 10, 21487. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Volume 18, pp. 234–241. [Google Scholar]
- Brémond Martin, C.; Simon Chane, C.; Clouchoux, C.; Histace, A. AAEGAN Loss Optimizations Supporting Data Augmentation on Cerebral Organoid Bright-Field Images. In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISAPPGRAPP), Virtual Event, 8–10 February 2021; p. 8. [Google Scholar]
- Siddique, N.; Sidike, P.; Elkin, C.; Devabhaktuni, V. U-Net and its variants for medical image segmentation: Theory and applications. IEEE Access 2021, 1118, 42. [Google Scholar] [CrossRef]
- van Rijthoven, M.; Balkenhol, M.; Siliņa, K.; van der Laak, J.; Ciompi, F. HookNet: Multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. arXiv 2020, arXiv:2006.12230. [Google Scholar] [CrossRef] [PubMed]
- El Jurdi, R.; Petitjean, C.; Honeine, P.; Abdallah, F. BB-UNet: U-Net With Bounding Box Prior. IEEE J. Sel. Top. Signal Process. 2020, 14, 1189–1198. [Google Scholar]
- Prasad, P.J.R.; Elle, O.J.; Lindseth, F.; Albregtsen, F.; Kumar, R.P. Modifying U-Net for small dataset: A simplified U-Net version for liver parenchyma segmentation. In Proceedings of the Medical Imaging 2021: Computer-Aided Diagnosis, Virtual Event, 15–20 February 2021; Mazurowski, M.A., Drukker, K., Eds.; International Society for Optics and Photonics (SPIE): Bellingham, WA, USA, 2021; Volume 11597, pp. 396–405. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop (DLMIA 2018) and 8th International Workshop (ML-CDS 2018), Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Gadosey, P.K.; Li, Y.; Agyekum, E.A.; Zhang, T.; Liu, Z.; Yamak, P.T.; Essaf, F. SD-UNet: Stripping down U-Net for Segmentation of Biomedical Images on Platforms with Low Computational Budgets. Diagnostics 2020, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Trebing, K.; Stanczyk, T.; Mehrkanoon, S. Smaat-UNet: Precipitation nowcasting using a small attention-UNet architecture. Pattern Recognit. Lett. 2021, 145, 178–186. [Google Scholar] [CrossRef]
- Wang, X.; Hu, Z.; Shi, S.; Hou, M.; Xu, L.; Zhang, X. A deep learning method for optimizing semantic segmentation accuracy of remote sensing images based on improved UNet. Sci. Rep. 2023, 13, 7600. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhao, S.X.; Yang, K.F.; Li, Y.J. A lightweight network guided with differential matched filtering for retinal vessel segmentation. Comput. Biol. Med. 2023, 160, 106924. [Google Scholar] [CrossRef] [PubMed]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Shuvo, M.B.; Ahommed, R.; Reza, S.; Hashem, M. CNL-UNet: A novel lightweight deep learning architecture for multimodal biomedical image segmentation with false output suppression. Biomed. Signal Process. Control. 2021, 70, 102959. [Google Scholar] [CrossRef]
- Xu, G.; Wu, X.; Zhang, X.; He, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. arXiv 2021, arXiv:2107.08623. [Google Scholar] [CrossRef]
- Yuan, D.; Xu, Z.; Tian, B.; Wang, H.; Zhan, Y.; Lukasiewicz, T. μ-Net: Medical image segmentation using efficient and effective deep supervision. Comput. Biol. Med. 2023, 160, 106963. [Google Scholar] [CrossRef]
- Jirik, M.; Gruber, I.; Moulisova, V.; Schindler, C.; Cervenkova, L.; Palek, R.; Rosendorf, J.; Arlt, J.; Bolek, L.; Dejmek, J.; et al. Semantic segmentation of intralobular and extralobular tissue from liver scaffold H&E images. Sensors 2020, 20, 7063. [Google Scholar]
- Brémond Martin, C.; Simon Chane, C.; Clouchoux, C.; Histace, A. AAEGAN Optimization by Purposeful Noise Injection for the Generation of Bright-Field Brain Organoid Images. In Proceedings of the Internal Conference On Image Proceccing Theory Tools and Application: Special Session Biological & Medical Image Analysis, Salzburg, Austria, 19–22 April 2022; p. 6. [Google Scholar]
- Gomez-Giro, G.; Arias-Fuenzalida, J.; Jarazo, J.; Zeuschner, D.; Ali, M.; Possemis, N.; Bolognin, S.; Halder, R.; Jäger, C.; Kuper, W.F.E.; et al. Synapse alterations precede neuronal damage and storage pathology in a human cerebral organoid model of CLN3-juvenile neuronal ceroid lipofuscinosis. Acta Neuropathol. Commun. 2019, 7, 222. [Google Scholar] [CrossRef]
- Gilroy, K.D. Overcoming Shot noise Limitations with Bright Field Mode. Vis. Res. 2019, 1, 74242. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Boyat, A.K.; Joshi, B.K. A Review Paper: Noise Models in Digital Image Processing. Signal Image Process. Int. J. 2015, 6, 63–75. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man, Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Adams, R.; Bischof, L. Seeded region growing. IEEE Trans. Pattern Anal. Mach. Intell. 1994, 16, 641–647. [Google Scholar] [CrossRef]
- Vora, P.; Oza, B. A survey on k-mean clustering and particle swarm optimization. Int. J. Sci. Mod. Eng. 2013, 1, 24–26. [Google Scholar]
- Menet, S.; Saint-Marc, P.; Medioni, G. Active contour models: Overview, implementation and applications. In Proceedings of the 1990 IEEE International Conference on Systems, Man, and Cybernetics, Los Angeles, CA, USA, 4–7 November 1990; pp. 194–199. [Google Scholar]
- Meziou, L.; Histace, A.; Precioso, F.; Matuszewski, B.; Carreiras, F. 3D Confocal Microscopy data analysis using level-set segmentation with alpha-divergence similarity measure. In Proceedings of the International Conference on Computer Vision Theory and Applications, Rome, Italy, 24–26 February 2012; pp. 861–864. [Google Scholar]
- Sharma, A.K.; Nandal, A.; Dhaka, A.; Koundal, D.; Bogatinoska, D.C.; Alyami, H. Enhanced watershed segmentation algorithm-based modified ResNet50 model for brain tumor detection. BioMed Res. Int. 2022, 2022, 7348344. [Google Scholar] [CrossRef]
- Malhotra, P.; Gupta, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. Deep neural networks for medical image segmentation. J. Healthc. Eng. 2022, 2022, 9580991. [Google Scholar] [CrossRef]
- El Jurdi, R.; Petitjean, C.; Honeine, P.; Cheplygina, V.; Abdallah, F. High-level prior-based loss functions for medical image segmentation: A survey. Comput. Vis. Image Underst. 2021, 210, 103248. [Google Scholar] [CrossRef]
- Hoang, H.H.; Trinh, H.H. Improvement for Convolutional Neural Networks in Image Classification Using Long Skip Connection. Appl. Sci. 2021, 11, 2092. [Google Scholar] [CrossRef]
- Qi, G.J.; Luo, J. Small data challenges in big data era: A survey of recent progress on unsupervised and semi-supervised methods. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 2168–2187. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Walia, E.; Babyn, P. Generative Adversarial Network in Medical Imaging: A Review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Hao, X.; Yin, L.; Li, X.; Zhang, L.; Yang, R. A Multi-Objective Semantic Segmentation Algorithm Based on Improved U-Net Networks. Remote. Sens. 2023, 15, 1838. [Google Scholar] [CrossRef]
- Arano-Martinez, J.A.; Martínez-González, C.L.; Salazar, M.I.; Torres-Torres, C. A framework for biosensors assisted by multiphoton effects and machine learning. Biosensors 2022, 12, 710. [Google Scholar] [CrossRef] [PubMed]



| Dataset | Effectives | Sample |
|---|---|---|
| (1) Original | 40 |  |
| (2) Transformed | 40 |  |
| (3) Loss optimization AAE | 240 |  |
| (4) Noise injection AAE | 160 |  |
| Update Name | Unet | UNet-Mini | Mu-Net |
|---|---|---|---|
| Layer Number | 5 | 4 | 4 |
| Filter | [64, 128, 256, 512, 1024] | [16, 32, 64, 64] | [16, 32, 64, 64] |
| Stride | default (1) | default (1) | 1 or 2 |
| Activation | Sigmoid | Softmax | Sigmoid |
| Skip connections | Concatenation | Addition | Concatenation |
| Name | Filter | Parameters |
|---|---|---|
| Conv | 16 | [k = 3, s = 1, a = relu] |
| Conv | 16 | [k = 3, s = 2, a = relu] |
| BatchNorm | ||
| Conv | 32 | [k = 3, s = 1, a = relu] |
| Conv | 32 | [k = 3, s = 2, a = relu] |
| BatchNorm | ||
| Conv | 64 | [k = 3, s = 1, a = relu] |
| Conv | 64 | [k = 3, s = 2, a = relu] |
| BatchNorm | ||
| Conv | 64 | [k = 3, s = 1, a = relu] |
| Conv | 64 | [k = 3, s = 2, a = relu] |
| BatchNorm | ||
| Dropout | [d = 0.5] | |
| Maxpooling | [p = 2.2] | |
| Deconv | 64 | [k = 3, s = 1, a = relu] |
| Deconv | 64 | [k = 3, s = 2, a = relu] |
| Upsampling | 64 | [k = 2, a = relu, sz = 2.2] |
| Concatenation | ||
| Deconv | 64 | [k = 3, a = relu] |
| Deconv | 64 | [k = 3, a = relu] |
| Upsampling | 32 | [k = 2, a = relu, sz = 2.2] |
| Concatenation | ||
| Deconv | 32 | [k = 3, a = relu] |
| Deconv | 32 | [k = 3, a = relu] |
| Upsampling | 16 | [k = 2, a = relu, sz = 2.2] |
| Concatenation | ||
| Deconv | 16 | [k = 3, a = relu] |
| Deconv | 16 | [k = 3, a = relu] |
| Conv2D | 2 | [k = 3, a = relu] |
| Conv2D | 1 | [k = 1, a = sigmoid] |
| GT | Classic | Gaussian | Salt & Pepp. | Speckle | Shot | |
| U-Net |  |  |  |  |  |  |
| Unet-Mini |  |  |  |  |  |  |
| Mu-Net |  |  |  |  |  |  |
| Legend |  |  |  |  |
| GT | Classic | BCE | BCE + L1 | LS | Poisson | Wass. | P.Wass. | |
| U-Net |  |  |  |  |  |  |  |  |
| Unet-Mini |  |  |  |  |  |  |  |  |
| Mu-Net |  |  |  |  |  |  |  |  |
| Classic | BCE | BCE + L1 | LS | Poisson | Wass. | P. Wass. | |
|---|---|---|---|---|---|---|---|
| U-Net | 0.68 | 0.87 | 0.87 | 0.86 | 0.87 | 0.88 | 0.90 |
| Layer | 0.83 | 0.89 | 0.88 | 0.87 | 0.90 | 0.88 | 0.91 |
| Filter | 0.85 | 0.56 | 0.84 | 0.82 | 0.83 | 0.85 | 0.83 |
| Kernel | 0.85 | 0.17 | 0.84 | 0.85 | 0.68 | 0.85 | 0.64 |
| Ewise | 0.51 | 0.69 | 0.72 | 0.80 | 0.66 | 0.68 | 0.73 |
| Layer + Filter | 0.84 | 0.87 | 0.87 | 0.85 | 0.88 | 0.87 | 0.88 |
| Layer + Kernel | 0.34 | 0.35 | 0.68 | 0.68 | 0.35 | 0.50 | 0.70 |
| Layer + Ewise | 0.33 | 0.21 | 0.28 | 0.28 | 0.25 | 0.32 | 0.37 |
| Filter + Kernel | 0.71 | 0.78 | 0.72 | 0.59 | 0.70 | 0.71 | 0.74 |
| Filter + Ewise | 0.67 | 0.39 | 0.77 | 0.76 | 0.59 | 0.31 | 0.65 |
| Kernel + Ewise | 0.17 | 0.65 | 0.55 | 0.59 | 0.64 | 0.65 | 0.56 |
| Layer + Filter + Kernel | 0.83 | 0.91 | 0.88 | 0.86 | 0.87 | 0.88 | 0.87 |
| Layer + Kernel + Ewise | 0.35 | 0.33 | 0.16 | 0.16 | 0.21 | 0.31 | 0.37 |
| Layer + Filter + Ewise | 0.42 | 0.28 | 0.19 | 0.36 | 0.13 | 0.29 | 0.41 |
| Filter + Kernel + Ewise | 0.50 | 0.34 | 0.29 | 0.29 | 0.45 | 0.37 | 0.21 |
| Layer + Filter + Kernel + Ewise (Mu-Net) | 0.85 | 0.88 | 0.87 | 0.89 | 0.87 | 0.89 | 0.88 |
| Layer + Filter + Kernel + Stride | 0.84 | 0.83 | 0.85 | 0.78 | 0.84 | 0.82 | 0.81 |
| Layer + Filter + Kernel + Stride + Activation + Ewise (Mini-UNet) | 0.86 | 0.84 | 0.87 | 0.88 | 0.85 | 0.88 | 0.82 |
| Classic | Gaussian | Salt & Pepp. | Speckle | Shot | |
|---|---|---|---|---|---|
| U-Net | 0.68 | 0.83 | 0.82 | 0.74 | 0.74 |
| Layer | 0.83 | 0.91 | 0.83 | 0.83 | 0.85 |
| Filter | 0.85 | 0.83 | 0.52 | 0.73 | 0.74 |
| Kernel | 0.85 | 0.64 | 0.72 | 0.58 | 0.75 |
| Ewise | 0.51 | 0.73 | 0.22 | 0.18 | 0.38 |
| Layer + Filter | 0.84 | 0.88 | 0.83 | 0.69 | 0.80 |
| Layer + Kernel | 0.34 | 0.70 | 0.77 | 0.49 | 0.40 |
| Layer + Ewise | 0.33 | 0.20 | 0.54 | 0.20 | 0.32 |
| Filter + Kernel | 0.71 | 0.74 | 0.59 | 0.55 | 0.62 |
| Filter + Ewise | 0.67 | 0.65 | 0.45 | 0.10 | 0.21 |
| Kernel + Ewise | 0.17 | 0.56 | 0.45 | 0.11 | 0.26 |
| Layer + Filter + Kernel | 0.83 | 0.87 | 0.81 | 0.77 | 0.74 |
| Layer + Kernel + Ewise | 0.35 | 0.42 | 0.37 | 0.26 | 0.32 |
| Layer + Filter + Ewise | 0.42 | 0.18 | 0.14 | 0.08 | 0.14 |
| Filter + Kernel + Ewise | 0.50 | 0.21 | 0.29 | 0.26 | 0.16 |
| Layer + Filter + Kernel + Ewise (Mu-Net) | 0.85 | 0.88 | 0.80 | 0.81 | 0.83 |
| Layer + Filter + Kernel + Stride | 0.84 | 0.81 | 0.59 | 0.75 | 0.81 |
| Layer + Filter + Kernel + Stride + Activation + Ewise (Mini-UNet) | 0.86 | 0.82 | 0.74 | 0.48 | 0.77 |
| Architecture | Time 1 Launch (s) | Time LOO (s) | Memory GPU (GB) | Memory Model (MB) | Model Param. | Flops (MFLOP) | Flops/ Param. |
|---|---|---|---|---|---|---|---|
| U-Net [5] | 990 | 39,613 | 35 | 118 | 17 M | 1092 | 9 |
| UNet-Mini [20] | 157 | 6282 | 7.35 | 1.31 | 128 k | 40 | 316 |
| Mu-Net (ours) | 151 | 6044 | 7.83 | 2.60 | 680 k | 45 | 58 |
| Threshold | Active | Kmean | Region | Watershed | Mu-Net | |
|---|---|---|---|---|---|---|
| Dice | <0.1 | 0.62 | 0.89 | <0.1 | 0.61 | 0.88 |
| F1-score | 0.40 | 0.66 | 0.72 | 0.20 | 0.61 | 0.77 |
| Sensitivity | 0.39 | 0.69 | 0.77 | 0.19 | 0.53 | 0.90 |
| Specificity | 0.96 | 0.84 | 0.89 | 0.86 | 0.94 | 0.86 |
| Accuracy | 0.83 | 0.90 | 0.90 | 0.7 | 0.88 | 0.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brémond Martin, C.; Simon Chane, C.; Clouchoux, C.; Histace, A. Mu-Net a Light Architecture for Small Dataset Segmentation of Brain Organoid Bright-Field Images. Biomedicines 2023, 11, 2687. https://doi.org/10.3390/biomedicines11102687
Brémond Martin C, Simon Chane C, Clouchoux C, Histace A. Mu-Net a Light Architecture for Small Dataset Segmentation of Brain Organoid Bright-Field Images. Biomedicines. 2023; 11(10):2687. https://doi.org/10.3390/biomedicines11102687
Chicago/Turabian StyleBrémond Martin, Clara, Camille Simon Chane, Cédric Clouchoux, and Aymeric Histace. 2023. "Mu-Net a Light Architecture for Small Dataset Segmentation of Brain Organoid Bright-Field Images" Biomedicines 11, no. 10: 2687. https://doi.org/10.3390/biomedicines11102687
APA StyleBrémond Martin, C., Simon Chane, C., Clouchoux, C., & Histace, A. (2023). Mu-Net a Light Architecture for Small Dataset Segmentation of Brain Organoid Bright-Field Images. Biomedicines, 11(10), 2687. https://doi.org/10.3390/biomedicines11102687