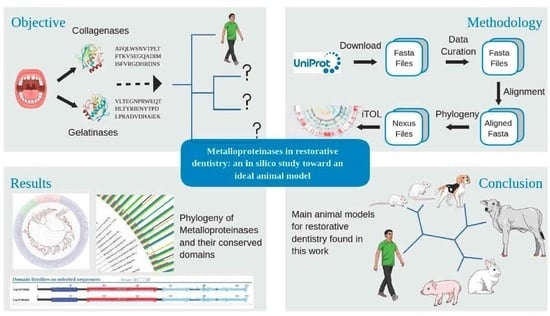
Metalloproteinases in Restorative Dentistry: An In Silico Study toward an Ideal Animal Model
Abstract
:1. Introduction
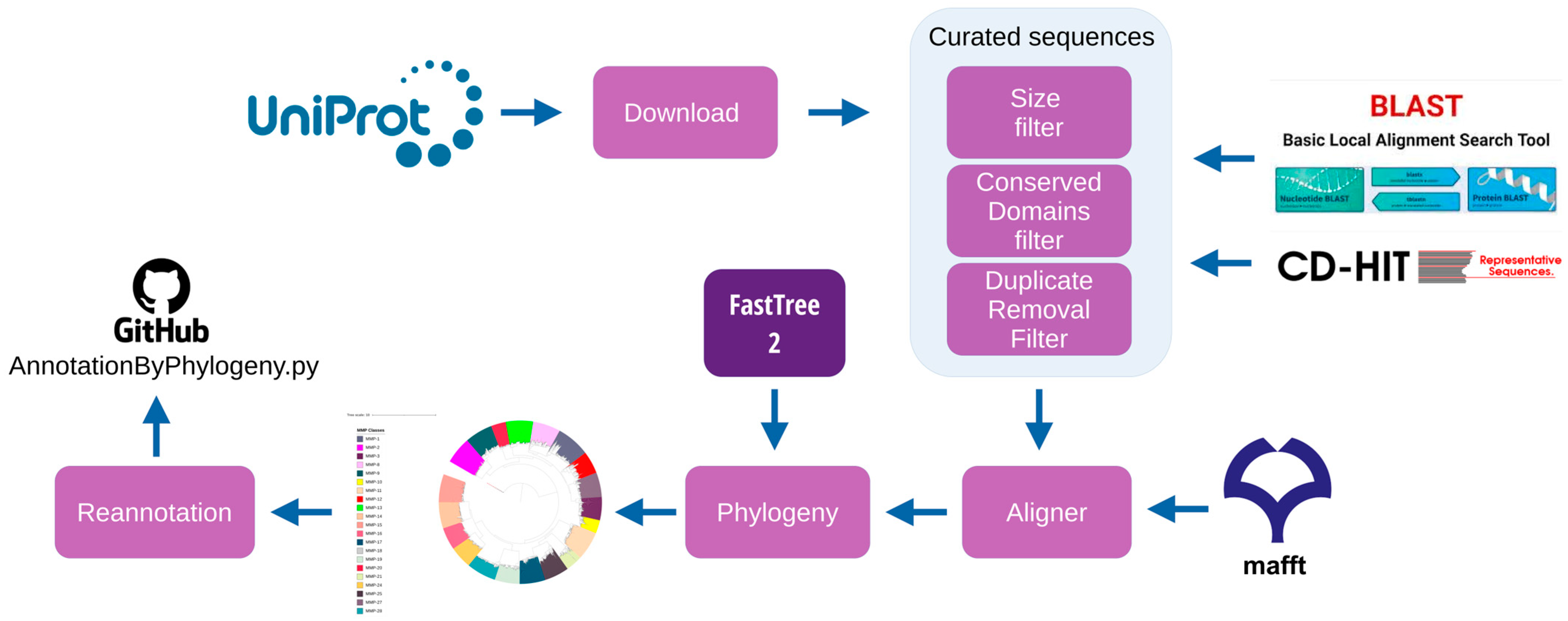
2. Materials and Methods
2.1. Retrieved Sequences
- (MMP OR collagenase OR matrix OR metalloproteinase OR metallopeptidase OR (interstitial AND collagenase) OR (neutrophil AND collagenase) OR stromelysin OR metalloelastase OR gelatinase OR matrilysin OR MT-MMP OR (Macrophage AND metalloelastase)) AND (taxonomy_id:40674)
- tr|A0A087RZB5|A0A087RZB5_9ARCH Matrix metalloproteinase-1 interstitial collagenase OS = Marine Group I thaumarchaeote SCGC AAA799-P11 OX = 1502295 GN = AAA799P11_00940 PE = 4 SV = 1
- MLQKKSDEFEQLYEKYDKLKMKVKELSQENQIYSHMCKKIETNSKDLKKSQSNLKKQLDQKLHSQLESEQEKLLLEKQLERSESSSKKSQKKYYVALVMAALSIAIISGAYSIMFAELAGQQYKIEVTPKPTGYTIQNLRGDTINTFLSWRLVPGDTLRVNIINSDNYDPEKIEVIKKTILSEKQLEIDNSLMHKGPKGTTSILYEGWLGALNDASKTDTNLFVPTNIEVIESNNGEGDITIELTNRKNADGFAGWTNSIADDSQNQILKSRITIFAVDSLSLAELETIVRHEMGHALGLAHSTDPEDLMYPTIQTNFPYISECDVDAIESLYDGQNTSEVICEI
2.2. Curated Sequences
2.2.1. Sequence Size Filter
2.2.2. Conserved Domains Filter
- matrixin (CDD:395334, CDD:239819, CDD:239806, CDD:239805, CDD:239804, CDD:239803, CDD:239796, CDD:238124 and CDD:214576).
- hemopexin (CDD:395000 and CDD:238046).
- pg-binding (CDD:396175).
- fibronectin (CDD:128373).
2.2.3. Duplicate Removal Filter
2.3. MSA Alignment and Phylogenetic Tree Construction
2.4. MMP Class (Re)annotation
2.5. Conserved Domain Tree
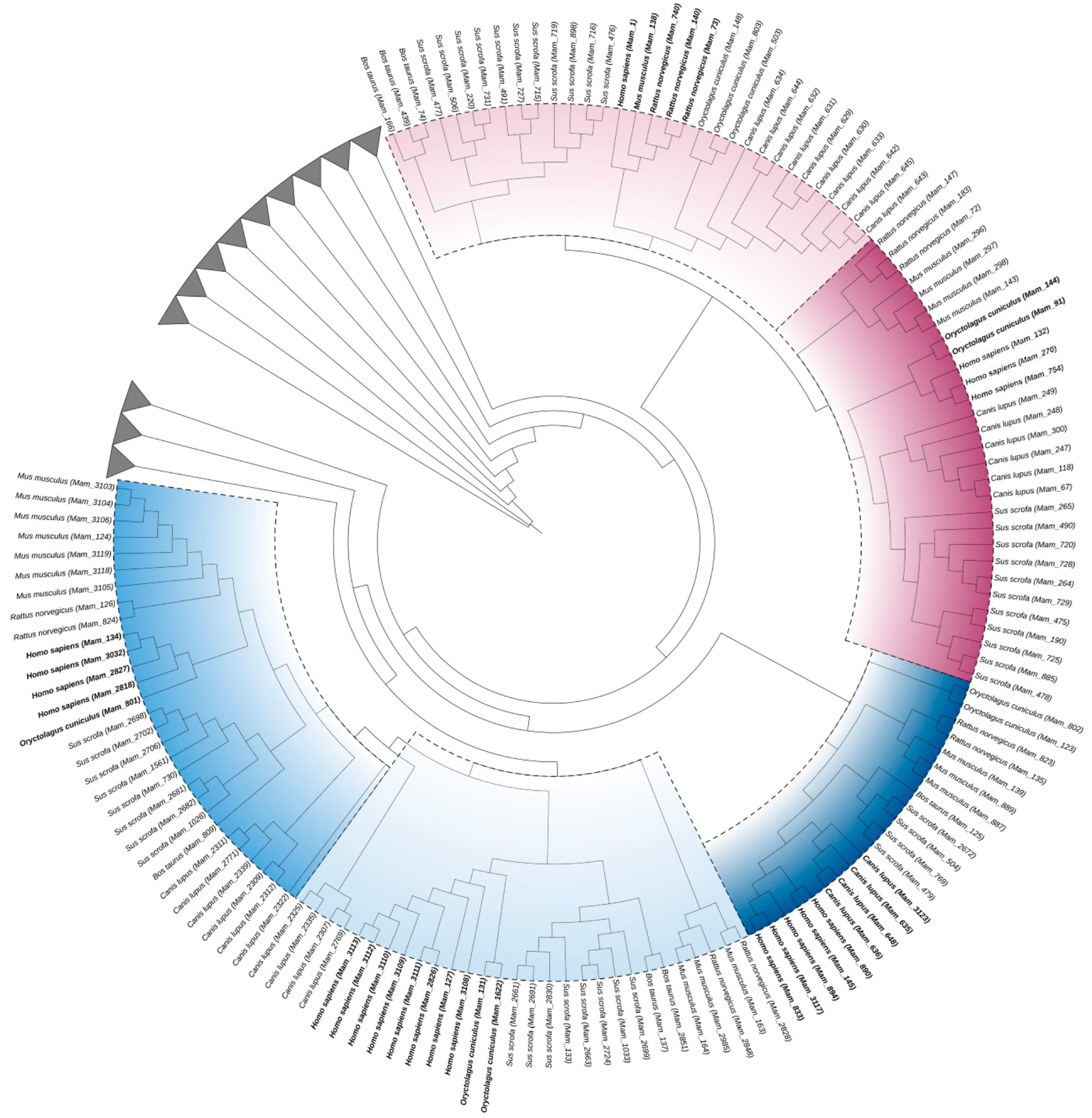
2.6. Model Organism Tree
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robinson, N.B.; Krieger, K.; Khan, F.M.; Huffman, W.; Chang, M.; Naik, A.; Yongle, R.; Hameed, I.; Krieger, K.; Girardi, L.N.; et al. The current state of animal models in research: A review. Int. J. Surg. 2019, 72, 9–13. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Crim, M.J.; Franklin, C.L. A Brief History of Animal Modeling. Mo. Med. 2013, 110, 201–205. [Google Scholar]
- Committee on the Use of Laboratory Animals in; Commission on Life Sciences; National Research Council; Institute of Medicine. Use of Laboratory Animals in Biomedical and Behavioral Research [Internet]. In Use of Laboratory Animals in Biomedical and Behavioral Research; National Academies Press (US): Washington, DC, USA, 1988; 112p. Available online: https://www.ncbi.nlm.nih.gov/books/NBK218267/ (accessed on 14 February 2023).
- Pound, P.; Ritskes-Hoitinga, M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J. Transl. Med. 2018, 16, 304. [Google Scholar] [CrossRef]
- Perel, P.; Roberts, I.; Sena, E.; Wheble, P.; Briscoe, C.; Sandercock, P.; Macleod, M.; Mignini, L.E.; Jayaram, P.; Khan, K.S. Comparison of treatment effects between animal experiments and clinical trials: Systematic review. BMJ 2007, 334, 197. [Google Scholar] [CrossRef]
- Vieira, A.R. Genetics of Dental Caries: Controlled Animal Models. Monogr. Oral Sci. 2022, 30, 45–60. [Google Scholar]
- Blanc-Sylvestre, N.; Bouchard, P.; Chaussain, C.; Bardet, C. Pre-Clinical Models in Implant Dentistry: Past, Present, Future. Biomedicines 2021, 9, 1538. [Google Scholar] [CrossRef]
- Shaikh, M.; Sung, H.; Lopez, T.; Andra, R.; McKean, B.; Jesson, J.; Pascal, C.; Pascal, C.; Chavez, A.; Schwieterman, K.; et al. Effect of charcoal dentifrices on tooth whitening and enamel surface roughness. Am. J. Dent. 2021, 34, 295–299. [Google Scholar]
- Wang, S.; Liu, Y.; Fang, D.; Shi, S. The miniature pig: A useful large animal model for dental and orofacial research. Oral Dis. 2007, 13, 530–537. [Google Scholar] [CrossRef]
- Karaman, G.E.; Emekli-Alturfan, E.; Akyüz, S. Zebrafish; an emerging model organism for studying toxicity and biocompatibility of dental materials. Cell. Mol. Biol. 2020, 66, 41–46. [Google Scholar] [CrossRef]
- Cavalcanti, A.L.; Lucena RN de Martins, V.M.; Granville-Garcia, A.F. Caracterização da pesquisa odontológica experimental em animais. RGO 2009, 57, 93–98. [Google Scholar]
- Franco, A.L.; Nogueira, M.N.M.; Sousa, N.G.K.; da Frota, M.F.; Fernandes, C.M.S.; Serra, M.d.C. Pesquisas em animais: Uma reflexão bioética. Acta Bioeth. 2014, 20, 247–253. [Google Scholar] [CrossRef]
- Darlu, P.; Tassy, P. La Reconstruction Phylogénétique: Concepts et Méthodes; Masson: Paris, France, 1993. [Google Scholar]
- Barton, N.H.; Briggs, D.E.; Eisen, J.A.; Goldstein, D.B.; Patel, N.H. Evolution; CSHL Press: Cold Spring Harbor, NY, USA, 2007; 836p. [Google Scholar]
- Mazzoni, A.; Breschi, L.; Carrilho, M.; Nascimento, F.D.; Orsini, G.; Ruggeri, A.; Gobbi, P.; Manzoli, L.; Tay, F.R.; Pashley, D.H.; et al. A review of the nature, role, and function of dentin non-collagenous proteins. Part II: Enzymes, serum proteins, and growth factors: A review of the nature, role, and function of dentin non-collagenous proteins. Endod. Top. 2009, 21, 19–40. [Google Scholar] [CrossRef]
- Checchi, V.; Maravic, T.; Bellini, P.; Generali, L.; Consolo, U.; Breschi, L.; Mazzoni, A. The Role of Matrix Metalloproteinases in Periodontal Disease. Int. J. Environ. Res. Public Health 2020, 17, 4923. [Google Scholar] [CrossRef]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Mazzoni, A.; Tjäderhane, L.; Checchi, V.; Di Lenarda, R.; Salo, T.; Tay, F.R.; Pashley, D.H.; Breschi, L. Role of dentin MMPs in caries progression and bond stability. J. Dent. Res. 2015, 94, 241–251. [Google Scholar] [CrossRef]
- Boguszewska-Czubara, A.; Budzynska, B.; Skalicka-Wozniak, K.; Kurzepa, J. Perspectives and New Aspects of Metalloproteinases’ Inhibitors in the Therapy of CNS Disorders: From Chemistry to Medicine. Curr. Med. Chem. 2019, 26, 3208–3224. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Robinson, M.R.; Dibas, M.; Stamer, W.D. Matrix Metalloproteinases and Glaucoma Treatment. J. Ocul. Pharmacol. Ther. 2020, 36, 208–228. [Google Scholar] [CrossRef]
- Goldberg, M.; Smith, A.J. Cells and Extracellular Matrices of Dentin and Pulp: A Biological Basis for Repair and Tissue Engineering. Crit. Rev. Oral Biol. Med. 2004, 15, 13–27. [Google Scholar] [CrossRef]
- Sulkala, M.; Tervahartiala, T.; Sorsa, T.; Larmas, M.; Salo, T.; Tjäderhane, L. Matrix metalloproteinase-8 (MMP-8) is the major collagenase in human dentin. Arch. Oral Biol. 2007, 52, 121–127. [Google Scholar] [CrossRef]
- Mazzoni, A.; Mannello, F.; Tay, F.R.; Tonti, G.A.M.; Papa, S.; Mazzotti, G.; Di Lenarda, R.; Pashley, D.H.; Breschi, L. Zymographic Analysis and Characterization of MMP-2 and -9 Forms in Human Sound Dentin. J. Dent. Res. 2007, 86, 436–440. [Google Scholar] [CrossRef]
- Breschi, L.; Maravic, T.; Cunha, S.R.; Comba, A.; Cadenaro, M.; Tjäderhane, L.; Pashley, D.H.; Tay, F.R.; Mazzoni, A. Dentin bonding systems: From dentin collagen structure to bond preservation and clinical applications. Dent. Mater. 2018, 34, 78–96. [Google Scholar] [CrossRef]
- Uniprot Consortium. UniProtKB/Swiss-Prot Protein Knowledgebase Release 2022_05 Statistics [Internet]. 2023. Available online: https://web.expasy.org/docs/relnotes/relstat.html (accessed on 14 February 2023).
- Poux, S.; Arighi, C.N.; Magrane, M.; Bateman, A.; Wei, C.H.; Lu, Z.; Boutet, E.; Bye-A-Jee, H.; Famiglietti, M.L.; Roechert, B.; et al. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 2017, 33, 3454–3460. [Google Scholar] [CrossRef]
- Chen, Q.; Britto, R.; Erill, I.; Jeffery, C.J.; Liberzon, A.; Magrane, M.; Onami, J.-I.; Robinson-Rechavi, M.; Sponarova, J.; Zobel, J.; et al. Quality Matters: Biocuration Experts on the Impact of Duplication and Other Data Quality Issues in Biological Databases. Genom. Proteom. Bioinform. 2020, 18, 91–103. [Google Scholar] [CrossRef]
- Kapoor, C.; Vaidya, S.; Wadhwan, V.; Hitesh Kaur, G.; Pathak, A. Seesaw of matrix metalloproteinases (MMPs). J. Cancer Res. Ther. 2016, 12, 28. [Google Scholar] [CrossRef]
- Farris, J.S. Outgroups and Parsimony. Syst. Biol. 1982, 31, 328–334. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Mieliauskaitė, D.; Kontenis, V.; Šiaurys, A. Lessons from Animal Models in Sjögren’s Syndrome. Int. J. Mol. Sci. 2023, 24, 12995. [Google Scholar] [CrossRef]
- de Carvalho, M.F.F.; Leijôto-Lannes, A.C.N.; Rodrigues MCN de Nogueira, L.C.; Ferraz, N.K.L.; Moreira, A.N.; Yamauti, M.; Zina, L.G.; de Magalhães, C.S. Viability of Bovine Teeth as a Substrate in Bond Strength Tests: A Systematic Review and Meta-analysis. J. Adhes. Dent. 2018, 20, 471–479. [Google Scholar]
- Huang, H.; Okamoto, M.; Watanabe, M.; Matsumoto, S.; Moriyama, K.; Komichi, S.; Ali, M.; Matayoshi, S.; Nomura, R.; Nakano, K.; et al. Development of Rat Caries-Induced Pulpitis Model for Vital Pulp Therapy. J. Dent. Res. 2023, 102, 574–582. [Google Scholar] [CrossRef]
- Ancuta, D.L.; Alexandru, D.M.; Crivineanu, M.; Coman, C. Induction of Periodontitis Using Bacterial Strains Isolated from the Human Oral Microbiome in an Experimental Rat Model. Biomedicines 2023, 11, 2098. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Bode, W.; Gomis-Rüth, F.X.; Stöckler, W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the “metzincins”. FEBS Lett. 1993, 331, 134–140. [Google Scholar] [CrossRef]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef]
- Subbaraj, G.K.; Elangovan, H.; Chandramouli, P.; Yasam, S.K.; Chandrasekaran, K.; Kulanthaivel, L.; Pandi, S.; Subramanian, S. Antiangiogenic Potential of Troxerutin and Chitosan Loaded Troxerutin on Chorioallantoic Membrane Model. Biomed. Res. Int. 2023, 2023, 5956154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, C.; He, B.; Zhang, X.; Hao, H.; Hou, Y.; Li, A.; Wang, Y.; Wang, Y. Macrophage Migration Inhibitory Factor Promotes Expression of Matrix Metalloproteinases 1 and 3 in Spinal Cord Astrocytes following Gecko Tail Amputation. J. Integr. Neurosci. 2023, 22, 29. [Google Scholar] [CrossRef]
- Gimenes, S.N.C.; Sachett, J.A.G.; Colombini, M.; Freitas-de-Sousa, L.A.; Ibiapina, H.N.S.; Costa, A.G.; Santana, M.F.; Park, J.J.; Sherman, N.E.; Ferreira, L.C.L.; et al. Observation of Bothrops atrox Snake Envenoming Blister Formation from Five Patients: Pathophysiological Insights. Toxins 2021, 13, 800. [Google Scholar] [CrossRef]
- Feng, G.; Wei, L.; Che, H.; Shen, Y.; Mi, K.; Bian, H.; Yang, H.; Wu, J.; Mu, L. Cathelicidin-NV from Nanorana ventripunctata effectively protects HaCaT cells, ameliorating ultraviolet B-induced skin photoaging. Peptides 2022, 150, 170712. [Google Scholar] [CrossRef] [PubMed]
- Bindhani, B.; Maity, S.; Chakrabarti, I.; Saha, S.K. Roles of matrix metalloproteinases in development, immunology, and ovulation in fruit Fly (Drosophila). Arch. Insect. Biochem. Physiol. 2022, 109, e21849. [Google Scholar] [CrossRef] [PubMed]
- Dolmatov, I.Y.; Nizhnichenko, V.A.; Dolmatova, L.S. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions. Cells 2021, 10, 2331. [Google Scholar] [CrossRef]
- Chang, A.B.; Lin, R.; Keith Studley, W.; Tran, C.V.; Saier, M.H. Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 2004, 21, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zobel, J.; Verspoor, K. Duplicates, redundancies and inconsistencies in the primary nucleotide databases: A descriptive study. Database 2017, 2017, baw163. [Google Scholar] [CrossRef]
- Rembeza, E.; Engqvist, M.K.M. Experimental and computational investigation of enzyme functional annotations uncovers misannotation in the EC 1.1.3.15 enzyme class. PLOS Comput. Biol. 2021, 17, e1009446. [Google Scholar] [CrossRef]
- Vialle, R.A.; Tamuri, A.U.; Goldman, N. Alignment Modulates Ancestral Sequence Reconstruction Accuracy. Mol. Biol. Evol. 2018, 35, 1783–1797. [Google Scholar] [CrossRef]
- Varón-González, C.; Whelan, S.; Klingenberg, C.P. Estimating Phylogenies from Shape and Similar Multidimensional Data: Why It Is Not Reliable. Syst. Biol. 2020, 69, 863–883. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Linder, C.R.; Warnow, T. RAxML and FastTree: Comparing Two Methods for Large-Scale Maximum Likelihood Phylogeny Estimation. PLoS ONE 2011, 6, e27731. [Google Scholar] [CrossRef]
- Massova, I.; Kotra, L.P.; Fridman, R.; Mobashery, S. Matrix metalloproteinases: Structures, evolution, and diversification. FASEB J. 1998, 12, 1075–1095. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.; Abu-Elmagd, M.; Grocott, T.; Yates, C.; Gavrilovic, J.; Wheeler, G.N. Matrix metalloproteinase genes inXenopus development. Dev. Dyn. 2004, 231, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Molière, S.; Jaulin, A.; Tomasetto, C.L.; Dali-Youcef, N. Roles of Matrix Metalloproteinases and Their Natural Inhibitors in Metabolism: Insights into Health and Disease. Int. J. Mol. Sci. 2023, 24, 10649. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.E.; Lee, D.H.; Yamakoshi, Y.; Klingenhoff, A.; Reichenberger, E.; Wright, J.T.; Simmer, J.P.; Komisarof, J.A.; Cantley, L.C.; Bartlett, J.D. MMP-20 is Predominately a Tooth-specific Enzyme with a Deep Catalytic Pocket that Hydrolyzes Type V Collagen. Biochemistry 2006, 45, 3863–3874. [Google Scholar] [CrossRef]
- Ahokas, K.; Lohi, J.; Lohi, H.; Elomaa, O.; Karjalainen-Lindsberg, M.L.; Kere, J.; Saarialho-Kere, U. Matrix metalloproteinase-21, the human orthologue for XMMP, is expressed during fetal development and in cancer. Gene 2002, 301, 31–41. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Tang, H.; Finn, R.D.; Thomas, P.D. TreeGrafter: Phylogenetic tree-based annotation of proteins with Gene Ontology terms and other annotations. Bioinformatics 2019, 35, 518–520. [Google Scholar] [CrossRef]
- Wolfoviz-Zilberman, A.; Houri-Haddad, Y.; Beyth, N. A Novel Dental Caries Model Replacing, Refining, and Reducing Animal Sacrifice. Appl. Sci. 2021, 11, 7141. [Google Scholar] [CrossRef]


| Filter | Description |
|---|---|
| Sequence size | Sequences with a length within 50% of the average length. |
| Conserved domains | Sequences with at least 3 domains characteristic of MMPs. |
| Duplicate removal | Removal of sequences with 100% identity. |
| Statistics/Tasks | Amount Sequences | Smaller Sequence Length | Bigger Sequence Length | Mean | Standard Deviation |
|---|---|---|---|---|---|
| Downloaded sequences | 176,077 | 8 | 14,507 | 655 | 653 |
| Sequences above sequence size threshold | 82,539 | 328 | 984 | 575 | 179 |
| Sequences after conserved domains filter | 3647 | 330 | 980 | 541 | 88 |
| Sequences after duplicated sequences | 3178 | 330 | 980 | 540 | 90 |
| Class Name | MMP | Amount Sequences | Class Annotation | Reannotation |
|---|---|---|---|---|
| Collagenases | MMP-1 | 218 | 44 (20.2%) | 3 (1.4%) |
| MMP-8 | 178 | 19 (10.7%) | 2 (1.1%) | |
| MMP-13 | 176 | 1 (0.6%) | - | |
| MMP-18 | 1 | - | - | |
| Gelatinases | MMP-2 | 174 | - | - |
| MMP-9 | 180 | 2 (1.1%) | 2 (1.1%) | |
| Transmembrane | MMP-14 | 171 | 2 (1.2%) | - |
| MMP-15 | 182 | 22 (12.1%) | 2 (1.1%) | |
| MMP-16 | 145 | 6 (4.1%) | - | |
| MMP-17 | 165 | 17 (10.3%) | - | |
| MMP-24 | 144 | 14 (9.7%) | 1 (0.7%) | |
| MMP-25 | 160 | 11 (6.9%) | - | |
| Stromelysins | MMP-3 | 150 | 14 (9.3%) | 2 (1.3%) |
| MMP-10 | 85 | 7 (8.2%) | 10 (11.8%) | |
| MMP-11 | 187 | 13 (7.0%) | - | |
| Other | MMP-12 | 145 | 14 (9.7%) | 1 (0.7%) |
| MMP-19 | 162 | 11 (6.8%) | 1 (0.6%) | |
| MMP-20 | 96 | 5 (5.2%) | - | |
| MMP-21 | 108 | 6 (5.6%) | - | |
| MMP-27 | 160 | 8 (5.0%) | 3 (1.9%) | |
| MMP-28 | 191 | 19 (9.9%) | - | |
| Total | 3178 | 235 (7.4%) | 27 (0.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, S.G.; Kotowski, N.; Sampaio-Filho, H.R.; Aguiar, F.H.B.; Dávila, A.M.R.; Jardim, R. Metalloproteinases in Restorative Dentistry: An In Silico Study toward an Ideal Animal Model. Biomedicines 2023, 11, 3042. https://doi.org/10.3390/biomedicines11113042
de Oliveira SG, Kotowski N, Sampaio-Filho HR, Aguiar FHB, Dávila AMR, Jardim R. Metalloproteinases in Restorative Dentistry: An In Silico Study toward an Ideal Animal Model. Biomedicines. 2023; 11(11):3042. https://doi.org/10.3390/biomedicines11113042
Chicago/Turabian Stylede Oliveira, Simone Gomes, Nelson Kotowski, Helio Rodrigues Sampaio-Filho, Flávio Henrique Baggio Aguiar, Alberto Martín Rivera Dávila, and Rodrigo Jardim. 2023. "Metalloproteinases in Restorative Dentistry: An In Silico Study toward an Ideal Animal Model" Biomedicines 11, no. 11: 3042. https://doi.org/10.3390/biomedicines11113042