Tuberculosis Severity Predictive Model Using Mtb Variants and Serum Biomarkers in a Colombian Cohort of APTB Patients
Abstract
:1. Introduction
2. Methods
2.1. Ethics Statement
2.2. Study Population
2.3. TB Score
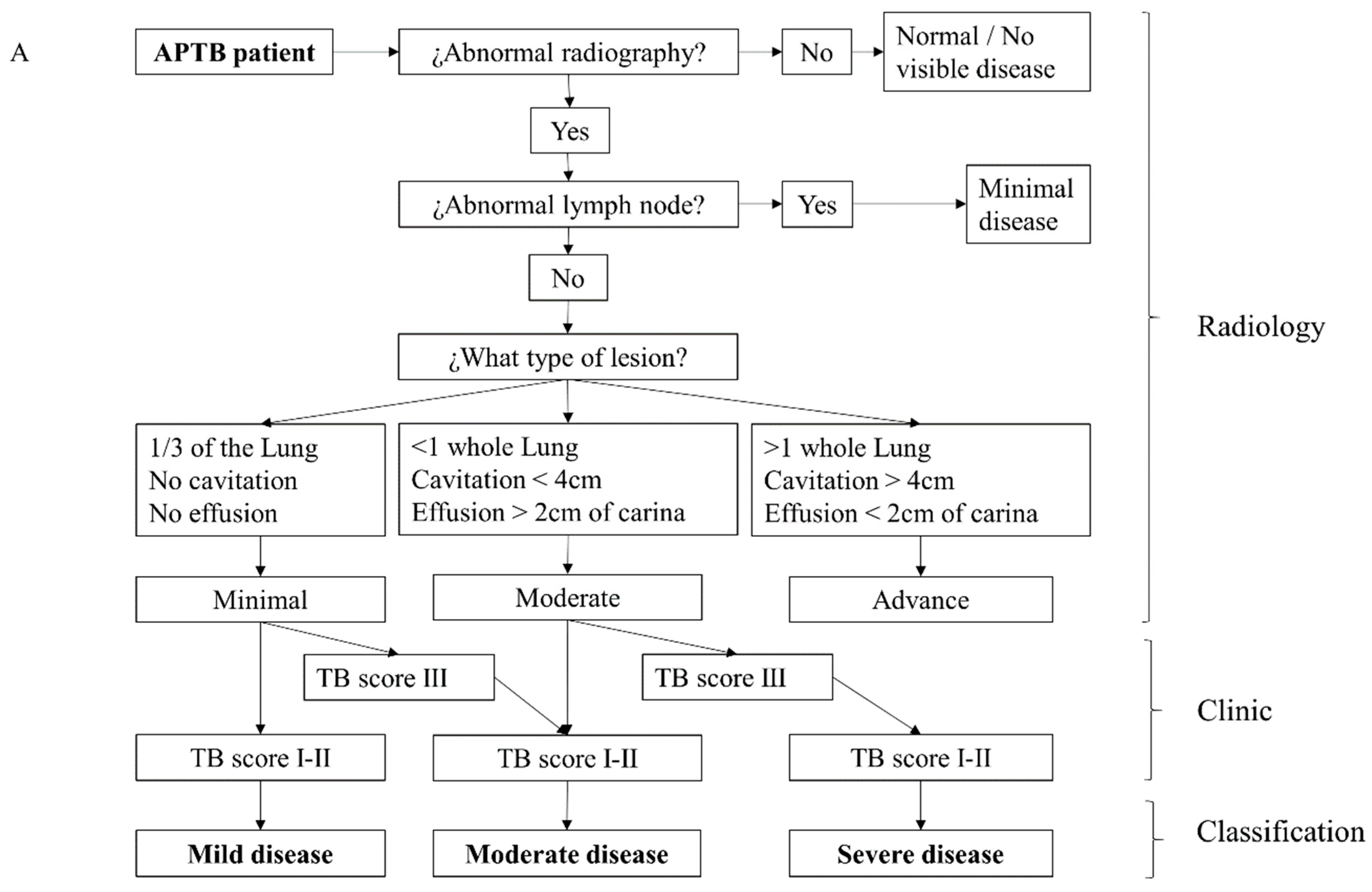
2.4. Clinical and Radiological Severity Classification
2.5. Recovery of Clinical Mtb Isolates
2.6. Whole-Genome Sequencing and Analysis
2.7. Cytokine Detection
2.8. Statistical Analysis
3. Results
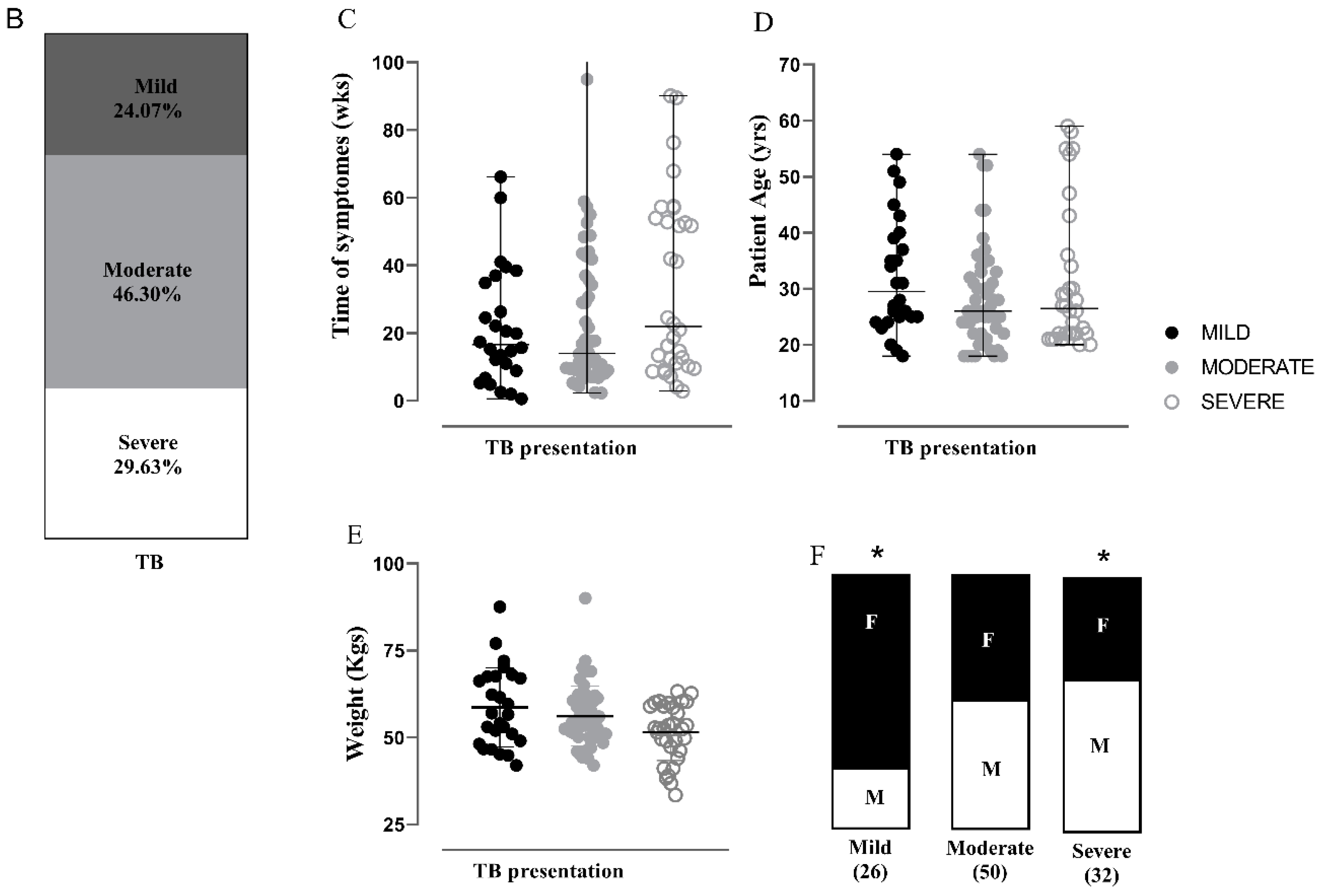
3.1. Characterization of the APTB Cohort in Antioquia, Colombia
3.2. Severity Classification for Individuals with APTB Using a Clinical-Radiological Algorithm at the Time of Diagnosis
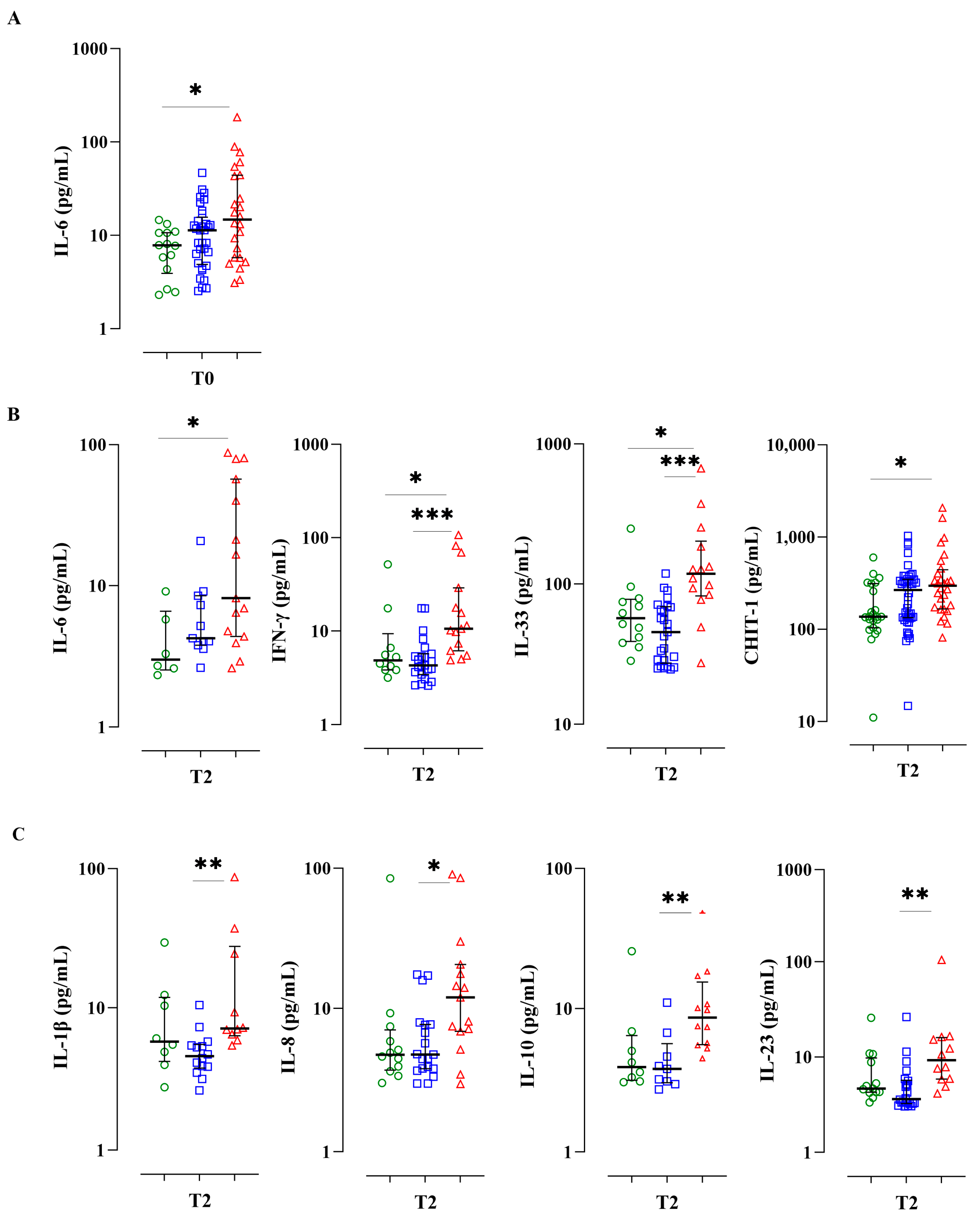
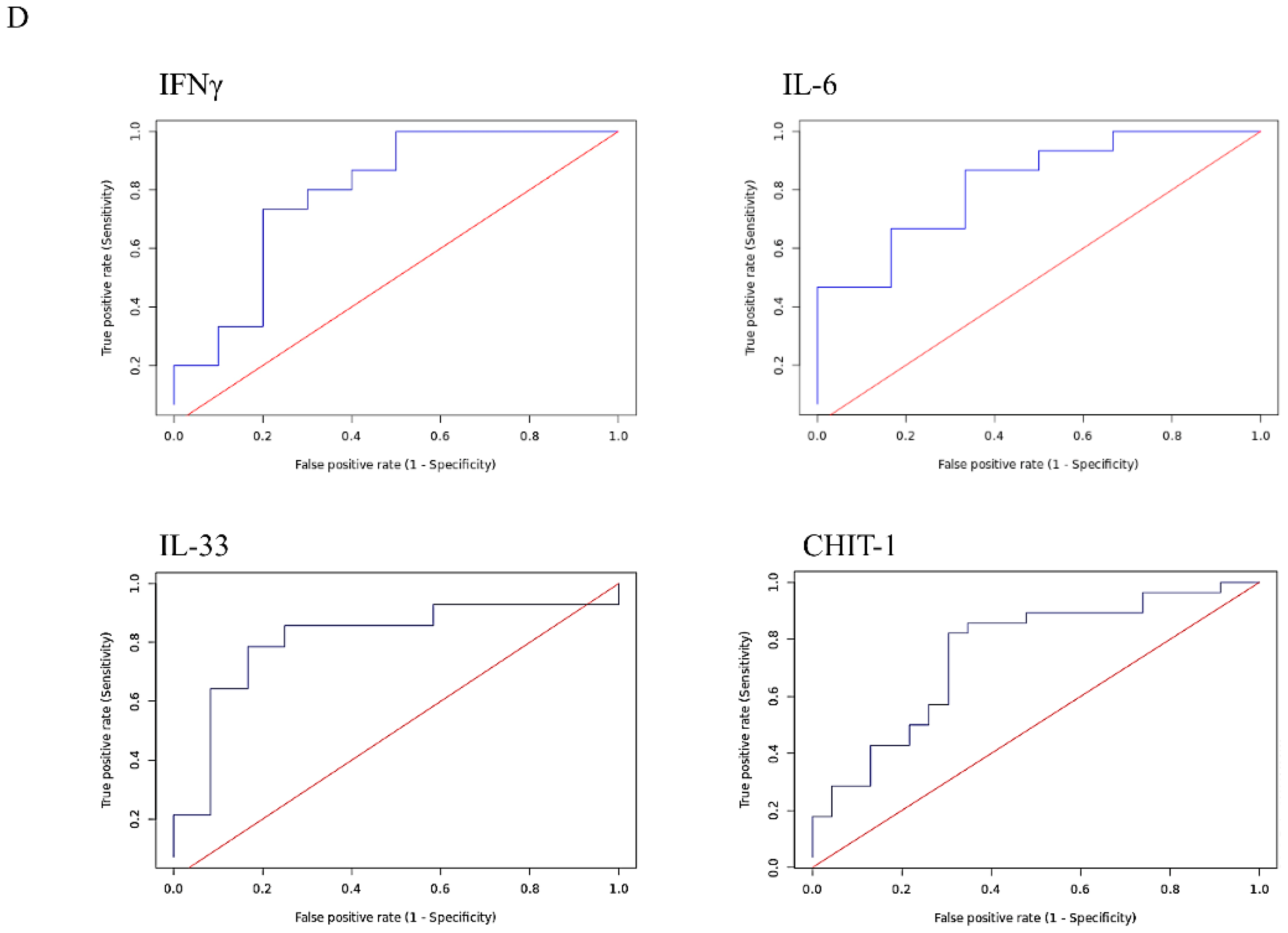
3.3. A Serum Bio-Profile Is Associated with Disease Severity in Individuals with APTB
3.4. Individuals with Severe APTB Exhibit Elevated Levels of Inflammatory Cytokines That Correlate with Regulatory Cytokines
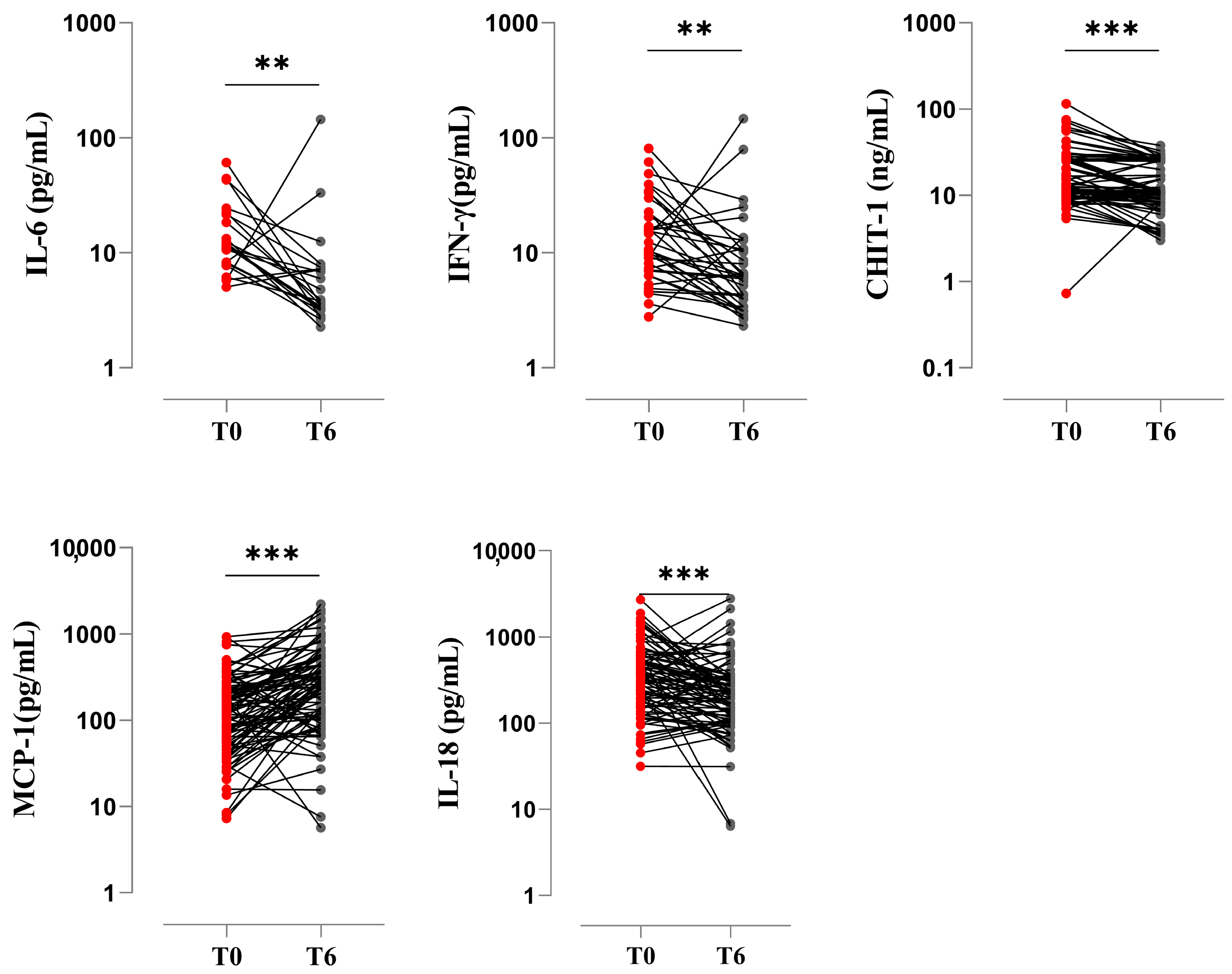
3.5. Pro-Inflammatory Biomarkers at the End of Treatment
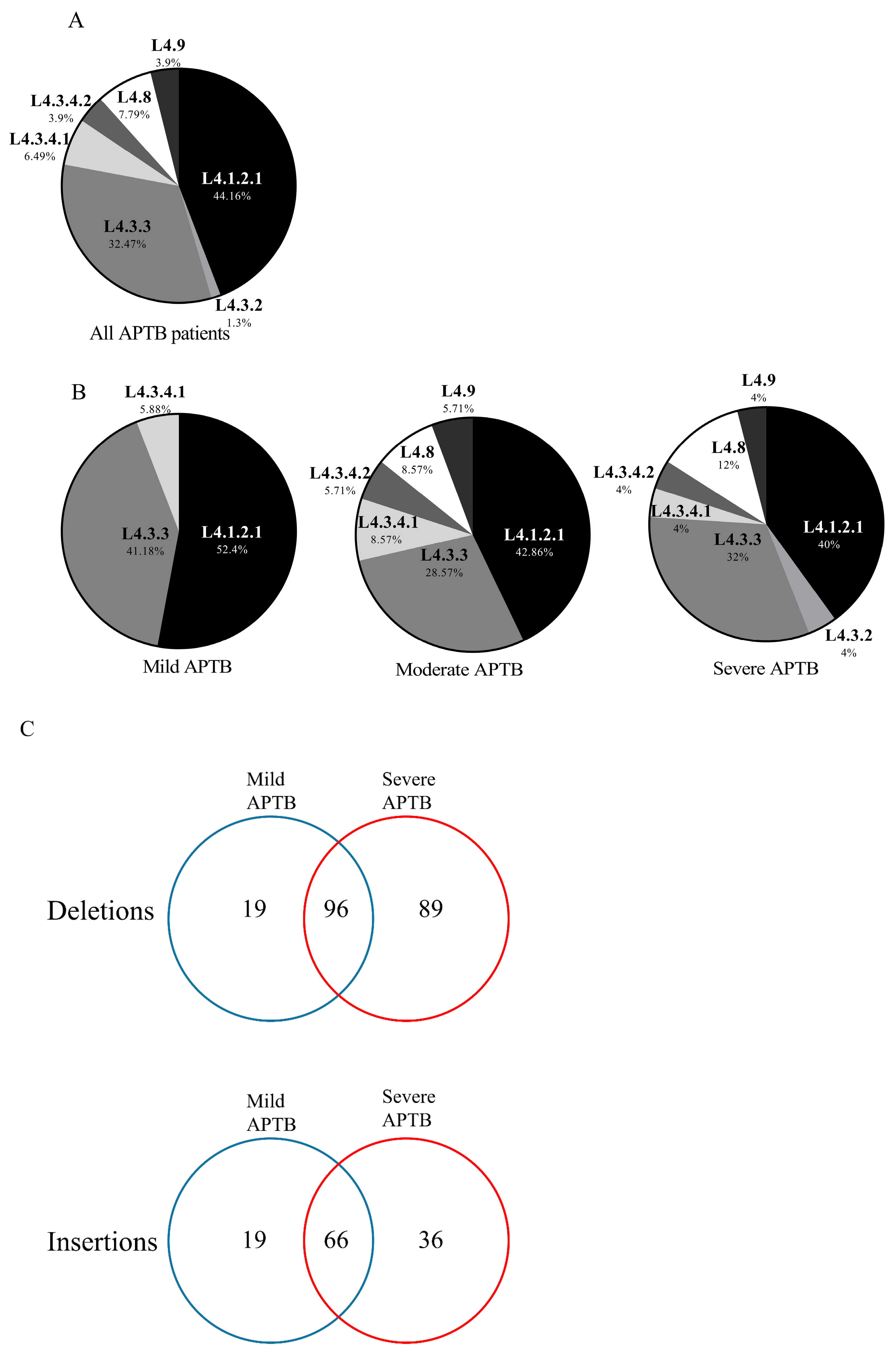
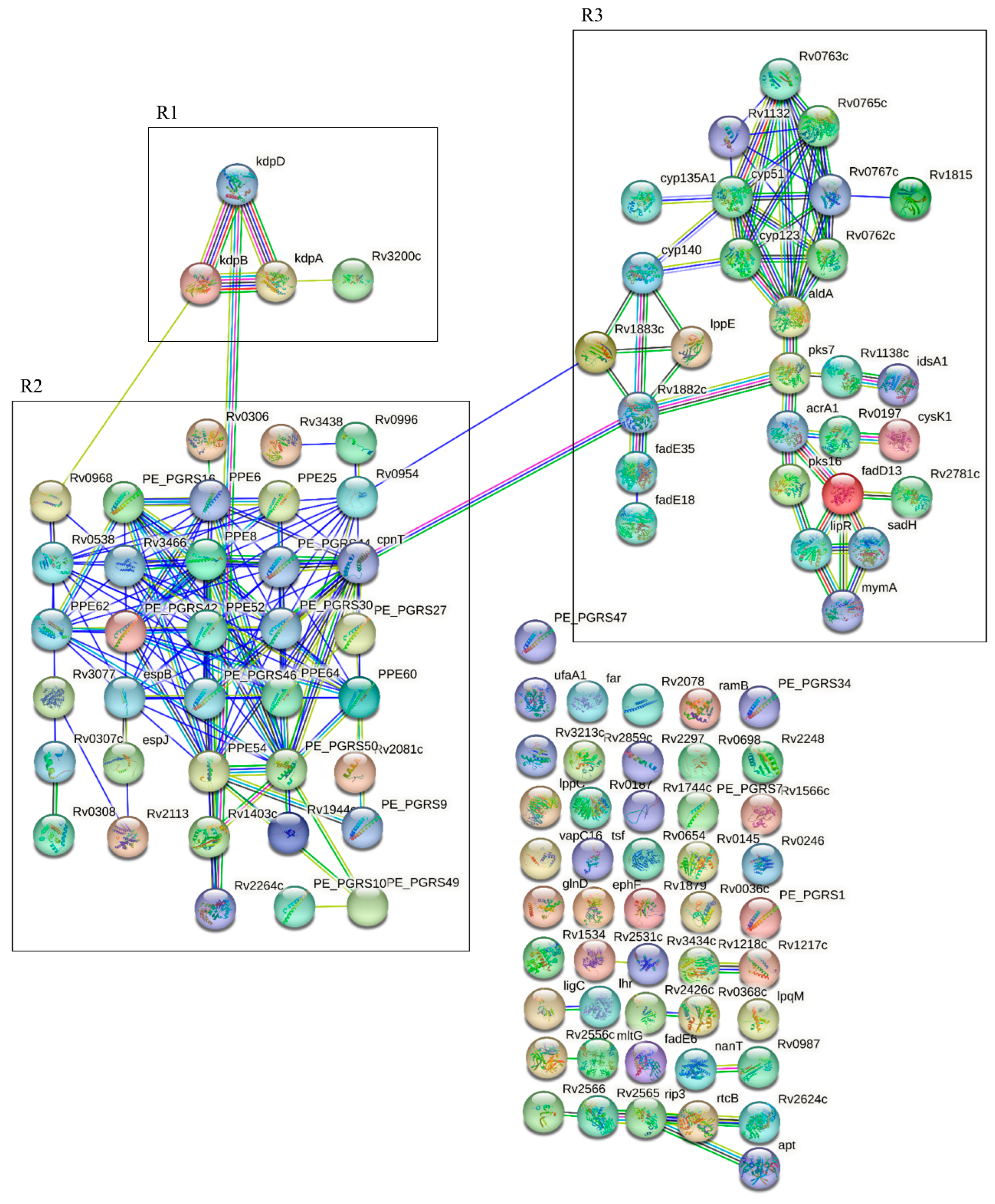
3.6. Genes Associated with Polymorphisms Present in Clinical Isolates of Mtb from Patients with Severe APTB
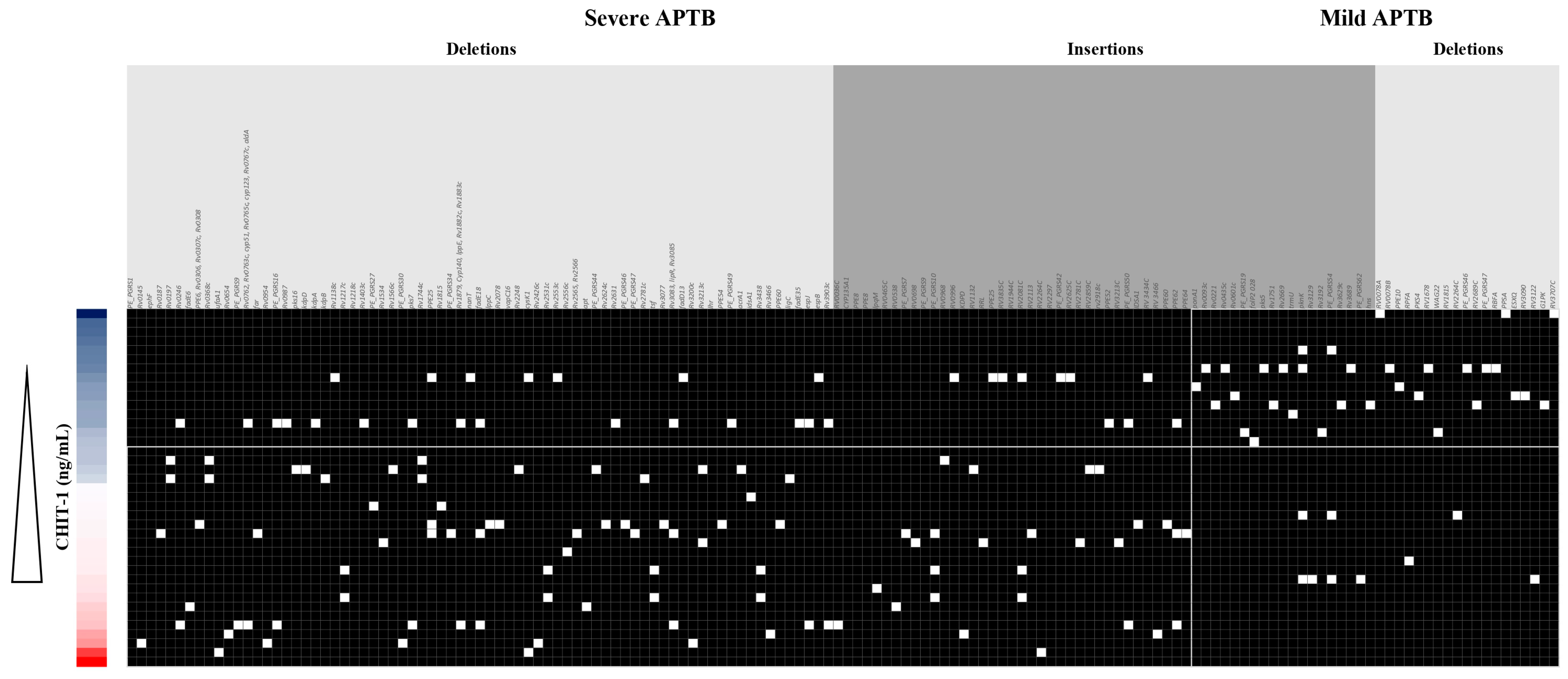
3.7. CHIT1 Serum Levels Are Associated with a Pattern of Mtb Indels According to Each APTB Patient’s Severity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Reid, M.J.A.; Arinaminpathy, N.; Bloom, A.; Bloom, B.R.; Boehme, C.; Chaisson, R.; Chin, D.P.; Churchyard, G.; Cox, H.; Ditiu, L.; et al. Building a tuberculosis-free world: The Lancet Commission on tuberculosis. Lancet 2019, 393, 1331–1384. [Google Scholar] [CrossRef] [PubMed]
- Riska, P.F.; Jacobs, W.R., Jr.; Alland, D. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuberc. Lung Dis. 2000, 4 (Suppl. 1), S4–S10. [Google Scholar] [PubMed]
- Russell, D.G. Who puts the tubercle in tuberculosis? Nat. Rev. Microbiol. 2007, 5, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Baena, A.; Porcelli, S.A. Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens 2009, 74, 189–204. [Google Scholar] [CrossRef]
- Ndong Sima, C.A.A.; Smith, D.; Petersen, D.C.; Schurz, H.; Uren, C.; Moller, M. The immunogenetics of tuberculosis (TB) susceptibility. Immunogenetics 2023, 75, 215–230. [Google Scholar] [CrossRef]
- WHO. Global Tuberculosis Report; Report No.: 978-92-4-006172-9; WHO: Geneva, Switzerland, 2023.
- Tiberi, S.; Torrico, M.M.; Rahman, A.; Krutikov, M.; Visca, D.; Silva, D.R.; Kunst, H.; Migliori, G.B. Managing severe tuberculosis and its sequelae: From intensive care to surgery and rehabilitation. J. Bras. Pneumol. 2019, 45, e20180324. [Google Scholar] [CrossRef]
- Sousa, J.; Ca, B.; Maceiras, A.R.; Simoes-Costa, L.; Fonseca, K.L.; Fernandes, A.I.; Ramos, A.; Carvalho, T.; Barros, L.; Magalhaes, C.; et al. Mycobacterium tuberculosis associated with severe tuberculosis evades cytosolic surveillance systems and modulates IL-1beta production. Nat. Commun. 2020, 11, 1949. [Google Scholar] [CrossRef]
- Abakay, O.; Abakay, A.; Sen, H.S.; Tanrikulu, A.C. The relationship between inflammatory marker levels and pulmonary tuberculosis severity. Inflammation 2015, 38, 691–696. [Google Scholar] [CrossRef]
- Al-Hajjaj, M.S.; Joharjy, I.A. Predictors of radiological sequelae of pulmonary tuberculosis. Acta Radiol. 2000, 41, 533–537. [Google Scholar] [CrossRef]
- Wiseman, C.A.; Gie, R.P.; Starke, J.R.; Schaaf, H.S.; Donald, P.R.; Cotton, M.F.; Hesseling, A.C. A proposed comprehensive classification of tuberculosis disease severity in children. Pediatr. Infect. Dis. J. 2012, 31, 347–352. [Google Scholar] [CrossRef]
- Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 202–213. [Google Scholar] [CrossRef] [PubMed]
- McHenry, M.L.; Bartlett, J.; Igo, R.P., Jr.; Wampande, E.M.; Benchek, P.; Mayanja-Kizza, H.; Fluegge, K.; Hall, N.B.; Gagneux, S.; Tishkoff, S.A.; et al. Interaction between host genes and Mycobacterium tuberculosis lineage can affect tuberculosis severity: Evidence for coevolution? PLoS Genet. 2020, 16, e1008728. [Google Scholar] [CrossRef]
- Bottai, D.; Frigui, W.; Sayes, F.; Di Luca, M.; Spadoni, D.; Pawlik, A.; Zoppo, M.; Orgeur, M.; Khanna, V.; Hardy, D.; et al. TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat. Commun. 2020, 11, 684. [Google Scholar] [CrossRef]
- Manca, C.; Tsenova, L.; Freeman, S.; Barczak, A.K.; Tovey, M.; Murray, P.J.; Barry, C.; Kaplan, G. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res. 2005, 25, 694–701. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, T.; Yin, J.; Jiang, H.; Takiff, H.E.; Gao, Q.; Liu, Q.; Li, W. The Global Success of Mycobacterium tuberculosis Modern Beijing Family Is Driven by a Few Recently Emerged Strains. Microbiol. Spectr. 2023, 25, e0333922. [Google Scholar] [CrossRef] [PubMed]
- Valway, S.E.; Sanchez, M.P.; Shinnick, T.F.; Orme, I.; Agerton, T.; Hoy, D.; Jones, J.S.; Westmoreland, H.; Onorato, I.M. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 1998, 338, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Connell, N.D.; Kreiswirth, B.N.; Whittam, T.S.; Musser, J.M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 1997, 94, 9869–9874. [Google Scholar] [CrossRef]
- Genestet, C.; Refregier, G.; Hodille, E.; Zein-Eddine, R.; Le Meur, A.; Hak, F.; Barbry, A.; Westeel, E.; Berland, J.L.; Engelmann, A.; et al. Mycobacterium tuberculosis genetic features associated with pulmonary tuberculosis severity. Int. J. Infect. Dis. 2022, 125, 74–83. [Google Scholar] [CrossRef]
- Aguilar, D.; Hanekom, M.; Mata, D.; Gey van Pittius, N.C.; van Helden, P.D.; Warren, R.M.; Hernandez-Pando, R. Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis 2010, 90, 319–325. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, A.; Wei, L.; Pang, Y.; Wu, B.; Luo, T.; Zhou, Y.; Zheng, H.X.; Jiang, Q.; Gan, M.; et al. China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat. Ecol. Evol. 2018, 2, 1982–1992. [Google Scholar] [CrossRef]
- Kato-Maeda, M.; Shanley, C.A.; Ackart, D.; Jarlsberg, L.G.; Shang, S.; Obregon-Henao, A.; Harton, M.; Basaraba, R.J.; Henao-Tamayo, M.; Barrozo, J.C.; et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin. Vaccine Immunol. 2012, 19, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.C.; Gomes, L.L.; Amaral, E.P.; Andrade, M.R.; Almeida, F.M.; Rezende, A.L.; Lanes, V.R.; Carvalho, E.C.; Suffys, P.N.; Mokrousov, I.; et al. Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J. Clin. Microbiol. 2014, 52, 2615–2624. [Google Scholar] [CrossRef] [PubMed]
- Ramazanzadeh, R.; Sayhemiri, K. Prevalence of Beijing family in Mycobacterium tuberculosis in world population: Systematic Review and Meta-Analysis. Int. J. Mycobacteriol. 2014, 3, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Alix, E.; Godreuil, S.; Blanc-Potard, A.B. Identification of a Haarlem genotype-specific single nucleotide polymorphism in the mgtC virulence gene of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006, 44, 2093–2098. [Google Scholar] [CrossRef]
- Baena, A.; Cabarcas, F.; Alvarez-Eraso, K.L.F.; Isaza, J.P.; Alzate, J.F.; Barrera, L.F. Differential determinants of virulence in two Mycobacterium tuberculosis Colombian clinical isolates of the LAM09 family. Virulence 2019, 10, 695–710. [Google Scholar] [CrossRef]
- Romagnoli, A.; Petruccioli, E.; Palucci, I.; Camassa, S.; Carata, E.; Petrone, L.; Mariano, S.; Sali, M.; Dini, L.; Girardi, E.; et al. Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1beta-induced autophagy. Cell Death Dis. 2018, 9, 624. [Google Scholar] [CrossRef]
- Wang, C.; Peyron, P.; Mestre, O.; Kaplan, G.; van Soolingen, D.; Gao, Q.; Gicquel, B.; Neyrolles, O. Innate immune response to Mycobacterium tuberculosis Beijing and other genotypes. PLoS ONE 2010, 5, e13594. [Google Scholar] [CrossRef]
- Faksri, K.; Chaiprasert, A.; Pardieu, C.; Casali, N.; Palaga, T.; Prammananan, T.; Palittapongarnpim, P.; Prayoonwiwat, N.; Drobniewski, F. Heterogeneity of phenotypic characteristics of the modern and ancestral Beijing strains of Mycobacterium tuberculosis. Asian Pac. J. Allergy Immunol. 2014, 32, 124–132. [Google Scholar] [CrossRef]
- Van Laarhoven, A.; Mandemakers, J.J.; Kleinnijenhuis, J.; Enaimi, M.; Lachmandas, E.; Joosten, L.A.; Ottenhoff, T.H.; Netea, M.G.; van Soolingen, D.; van Crevel, R. Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect. Immun. 2013, 81, 3750–3756. [Google Scholar] [CrossRef]
- Duque, C.; Arroyo, L.; Ortega, H.; Montufar, F.; Ortiz, B.; Rojas, M.; Barrera, L.F. Different responses of human mononuclear phagocyte populations to Mycobacterium tuberculosis. Tuberculosis 2014, 94, 111–122. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kulkarni, S.; Rajan, R.; Sainis, K. Mycobacterium tuberculosis strains from ancient and modern lineages induce distinct patterns of immune responses. J. Infect. Dev. Ctries. 2018, 11, 904–911. [Google Scholar] [CrossRef]
- Igbokwe, V.; Ruby, L.C.; Sultanli, A.; Belard, S. Post-tuberculosis sequelae in children and adolescents: A systematic review. Lancet Infect. Dis. 2023, 23, e138–e150. [Google Scholar] [CrossRef] [PubMed]
- Allwood, B.W.; Byrne, A.; Meghji, J.; Rachow, A.; van der Zalm, M.M.; Schoch, O.D. Post-Tuberculosis Lung Disease: Clinical Review of an Under-Recognised Global Challenge. Respiration 2021, 100, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Hulya, S.; Naz, I.; Karadeniz, G.; Erkan, S. Clinical effects of TB sequelae in patients with COPD. Int. J. Tuberc. Lung Dis. 2022, 26, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Sharma, L.; Dela Cruz, C.S. Chitotriosidase: A marker and modulator of lung disease. Eur. Respir. Rev. 2020, 29, 190143. [Google Scholar] [CrossRef]
- Bargagli, E.; Margollicci, M.; Nikiforakis, N.; Luddi, A.; Perrone, A.; Grosso, S.; Rottoli, P. Chitotriosidase activity in the serum of patients with sarcoidosis and pulmonary tuberculosis. Respiration 2007, 74, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Tasci, C.; Tapan, S.; Ozkaya, S.; Demirer, E.; Deniz, O.; Balkan, A.; Ozkan, M.; Inan, I.; Kurt, I.; Bilgic, H. Efficacy of serum chitotriosidase activity in early treatment of patients with active tuberculosis and a negative sputum smear. Ther. Clin. Risk Manag. 2012, 8, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Cakir, G.; Gumus, S.; Ucar, E.; Kaya, H.; Tozkoparan, E.; Akgul, E.O.; Karaman, B.; Deniz, O.; Kurt, I.; Ozkan, M.; et al. Serum chitotriosidase activity in pulmonary tuberculosis: Response to treatment and correlations with clinical parameters. Ann. Lab. Med. 2012, 32, 184–189. [Google Scholar] [CrossRef]
- Chen, M.; Deng, J.; Li, W.; Su, C.; Xia, Y.; Wang, M.; Li, X.; Abuaku, B.K.; Tan, H.; Wen, S.W. The relationship between chitotriosidase activity and tuberculosis. Epidemiol. Infect. 2015, 143, 3196–3202. [Google Scholar] [CrossRef]
- Wejse, C.; Gustafson, P.; Nielsen, J.; Gomes, V.F.; Aaby, P.; Andersen, P.L.; Sodemann, M. TBscore: Signs and symptoms from tuberculosis patients in a low-resource setting have predictive value and may be used to assess clinical course. Scand. J. Infect. Dis. 2008, 40, 111–120. [Google Scholar] [CrossRef]
- Berry, M.P.; Graham, C.M.; McNab, F.W.; Xu, Z.; Bloch, S.A.; Oni, T.; Wilkinson, K.A.; Banchereau, R.; Skinner, J.; Wilkinson, R.J.; et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010, 466, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Baena, A.; Cabarcas, F.; Ocampo, J.C.; Barrera, L.F.; Alzate, J.F. Large genomic deletions delineate Mycobacterium tuberculosis L4 sublineages in South American countries. PLoS ONE 2023, 18, e0285417. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Tedla, K.; Medhin, G.; Berhe, G.; Mulugeta, A.; Berhe, N. Factors associated with treatment initiation delay among new adult pulmonary tuberculosis patients in Tigray, Northern Ethiopia. PLoS ONE 2020, 15, e0235411. [Google Scholar] [CrossRef]
- Tedla, K.; Medhin, G.; Berhe, G.; Mulugeta, A.; Berhe, N. Delay in treatment initiation and its association with clinical severity and infectiousness among new adult pulmonary tuberculosis patients in Tigray, northern Ethiopia. BMC Infect. Dis. 2020, 20, 456. [Google Scholar] [CrossRef]
- Li, X.; Lv, X.; Lin, Y.; Zhen, J.; Ruan, C.; Duan, W.; Li, Y.; Xie, J. Role of two-component regulatory systems in intracellular survival of Mycobacterium tuberculosis. J. Cell. Biochem. 2019, 120, 12197–12207. [Google Scholar] [CrossRef]
- Cholo, M.C.; van Rensburg, E.J.; Osman, A.G.; Anderson, R. Expression of the Genes Encoding the Trk and Kdp Potassium Transport Systems of Mycobacterium tuberculosis during Growth In Vitro. Biomed. Res. Int. 2015, 2015, 608682. [Google Scholar] [CrossRef]
- Chatrath, S.; Gupta, V.K.; Dixit, A.; Garg, L.C. The Rv1651c-encoded PE-PGRS30 protein expressed in Mycobacterium smegmatis exhibits polar localization and modulates its growth profile. FEMS Microbiol. Lett. 2011, 322, 194–199. [Google Scholar] [CrossRef]
- Izquierdo Lafuente, B.; Ummels, R.; Kuijl, C.; Bitter, W.; Speer, A. Mycobacterium tuberculosis Toxin CpnT Is an ESX-5 Substrate and Requires Three Type VII Secretion Systems for Intracellular Secretion. mBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Saraav, I.; Singh, S.; Pandey, K.; Sharma, M.; Sharma, S.; Kothari, D.S. Effect of M. tuberculosis cellwall associated protein rrv3083 on toll like receptor expression in human macrophages. J. Clin. Immunol. 2015, 35, 308–309. [Google Scholar]
- Singh, A.; Gupta, R.; Vishwakarma, R.A.; Narayanan, P.R.; Paramasivan, C.N.; Ramanathan, V.D.; Tyagi, A.K. Correction for Singh et al., “Requirement of the mymA Operon for Appropriate Cell Wall Ultrastructure and Persistence of Mycobacterium tuberculosis in the Spleens of Guinea Pigs. J. Bacteriol. 2021, 203, e00094-21. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Jain, S.; Gupta, S.; Das, T.; Tyagi, A.K. mymA operon of Mycobacterium tuberculosis: Its regulation and importance in the cell envelope. FEMS Microbiol. Lett. 2003, 227, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, D.; Li, H.; Bi, L.; Deng, J.; Zhu, G.; Zhang, J.; Li, C.; Li, M.; Fang, Y.; et al. FattyacylCoA synthetase FadD13 regulates proinflammatory cyto-kine secretion dependent on the NF-kB signalling pathway bybinding to eEF1A1. Cell. Microbiol. 2019, 21, e13090. [Google Scholar] [CrossRef]
- Saraav, I.; Singh, S.; Pandey, K.; Sharma, M.; Sharma, S. Mycobacterium tuberculosis MymA is a TLR2 agonist that activate macrophages and a TH1 response. Tuberculosis 2017, 106, 16–24. [Google Scholar] [CrossRef]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. Te sigma factors of Mycobacterium tuberculosis: Regulation of the regulators. FEBS J. 2010, 277, 605–626. [Google Scholar] [CrossRef] [PubMed]
- Lui, G.; Wong, C.K.; Ip, M.; Chu, Y.J.; Yung, I.M.; Cheung, C.S.; Zheng, L.; Lam, J.S.; Wong, K.T.; Sin, W.W.; et al. HMGB1/RAGE Signaling and Pro-Inflammatory Cytokine Responses in Non-HIV Adults with Active Pulmonary Tuberculosis. PLoS ONE 2016, 11, e0159132. [Google Scholar] [CrossRef] [PubMed]
- Wallis, R.S.; Wang, C.; Meyer, D.; Thomas, N. Month 2 culture status and treatment duration as predictors of tuberculosis relapse risk in a meta-regression model. PLoS ONE 2013, 8, e71116. [Google Scholar] [CrossRef]
- Shuaib, Y.A.; Khalil, E.A.G.; Schaible, U.E.; Wieler, L.H.; Bakheit, M.A.M.; Mohamed-Noor, S.E.; Abdalla, M.A.; Homolka, S.; Andres, S.; Hillemann, D.; et al. Smear Microscopy for Diagnosis of Pulmonary Tuberculosis in Eastern Sudan. Tuberc. Res. Treat. 2018, 2018, 8038137. [Google Scholar] [CrossRef]
- Nagabhushanam, V.; Solache, A.; Ting, L.M.; Escaron, C.J.; Zhang, J.Y.; Ernst, J.D. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 2003, 171, 4750–4757. [Google Scholar] [CrossRef]
- Park, S.J.; Nakagawa, T.; Kitamura, H.; Atsumi, T.; Kamon, H.; Sawa, S.; Kamimura, D.; Ueda, N.; Iwakura, Y.; Ishihara, K.; et al. IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J. Immunol. 2004, 173, 3844–3854. [Google Scholar] [CrossRef]
- Tsao, T.C.; Huang, C.C.; Chiou, W.K.; Yang, P.Y.; Hsieh, M.J.; Tsao, K.C. Levels of interferon-gamma and interleukin-2 receptor-alpha for bronchoalveolar lavage fluid and serum were correlated with clinical grade and treatment of pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 2002, 6, 720–727. [Google Scholar]
- Schmitz, J.; Owyang, A.; Oldham, E.; Song, Y.; Murphy, E.; McClanahan, T.K.; Zurawski, G.; Moshrefi, M.; Qin, J.; Li, X.; et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 2005, 23, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Milovanovic, M.; Volarevic, V.; Radosavljevic, G.; Jovanovic, I.; Pejnovic, N.; Arsenijevic, N.; Lukic, M.L. IL-33/ST2 axis in inflammation and immunopathology. Immunol. Res. 2012, 52, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Griesenauer, B.; Paczesny, S. The ST2/IL-33 Axis in Immune Cells during Inflammatory Diseases. Front. Immunol. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Herzog, E.L.; Ahangari, F.; Zhou, Y.; Gulati, M.; Lee, C.M.; Peng, X.; Feghali-Bostwick, C.; Jimenez, S.A.; Varga, J.; et al. Chitinase 1 is a biomarker for and therapeutic target in scleroderma-associated interstitial lung disease that augments TGF-beta1 signaling. J. Immunol. 2012, 189, 2635–2644. [Google Scholar] [CrossRef] [PubMed]
- Letuve, S.; Kozhich, A.; Humbles, A.; Brewah, Y.; Dombret, M.C.; Grandsaigne, M.; Adle, H.; Kolbeck, R.; Aubier, M.; Coyle, A.J.; et al. Lung chitinolytic activity and chitotriosidase are elevated in chronic obstructive pulmonary disease and contribute to lung inflammation. Am. J. Pathol. 2010, 176, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Correale, J.; Fiol, M. Chitinase effects on immune cell response in neuromyelitis optica and multiple sclerosis. Mult. Scler. 2011, 17, 521–531. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, T.; Homer, R.J.; Kim, Y.K.; Chen, N.Y.; Cohn, L.; Hamid, Q.; Elias, J.A. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 2004, 304, 1678–1682. [Google Scholar] [CrossRef]
- Van Dyken, S.J.; Liang, H.E.; Naikawadi, R.P.; Woodruff, P.G.; Wolters, P.J.; Erle, D.J.; Locksley, R.M. Spontaneous Chitin Accumulation in Airways and Age-Related Fibrotic Lung Disease. Cell 2017, 169, 497–509.e13. [Google Scholar] [CrossRef]
- Kiran, D.; Podell, B.K.; Chambers, M.; Basaraba, R.J. Host-directed therapy targeting the Mycobacterium tuberculosis granuloma: A review. Semin. Immunopathol. 2016, 38, 167–183. [Google Scholar] [CrossRef]
- Young, C.; Walzl, G.; Du Plessis, N. Therapeutic host-directed strategies to improve outcome in tuberculosis. Mucosal. Immunol. 2020, 13, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Djoba Siawaya, J.F.; Beyers, N.; van Helden, P.; Walzl, G. Differential cytokine secretion and early treatment response in patients with pulmonary tuberculosis. Clin. Exp. Immunol. 2009, 156, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Verbon, A.; Juffermans, N.; Van Deventer, S.J.; Speelman, P.; Van Deutekom, H.; Van Der Poll, T. Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin. Exp. Immunol. 1999, 115, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Kinjo, Y.; Kawakami, K.; Uezu, K.; Yara, S.; Miyagi, K.; Koguchi, Y.; Hoshino, T.; Okamoto, M.; Kawase, Y.; Yokota, K.; et al. Contribution of IL-18 to Th1 response and host defense against infection by Mycobacterium tuberculosis: A comparative study with IL-12p40. J. Immunol. 2002, 169, 323–329. [Google Scholar] [CrossRef]
- Parish, T.; Smith, D.A.; Kendall, S.; Casali, N.; Bancroft, G.J.; Stoker, N.G. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 2003, 71, 1134–1140. [Google Scholar] [CrossRef]
- Singh, A.; Gupta, R.; Vishwakarma, R.A.; Narayanan, P.R.; Paramasivan, C.N.; Ramanathan, V.D.; Tyagi, A.K. Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J. Bacteriol. 2005, 187, 4173–4186. [Google Scholar] [CrossRef]
- Iantomasi, R.; Sali, M.; Cascioferro, A.; Palucci, I.; Zumbo, A.; Soldini, S.; Rocca, S.; Greco, E.; Maulucci, G.; De Spirito, M.; et al. PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cell. Microbiol. 2012, 14, 356–367. [Google Scholar] [CrossRef]
- Mi, Y.; Bao, L.; Gu, D.; Luo, T.; Sun, C.; Yang, G. Mycobacterium tuberculosis PPE25 and PPE26 proteins expressed in Mycobacterium smegmatis modulate cytokine secretion in mouse macrophages and enhance mycobacterial survival. Res. Microbiol. 2017, 168, 234–243. [Google Scholar] [CrossRef]
- Mi, Y.J.; Liang, L.J.; Ci, C.H.; Wang, Y.Y. The Role of PPE25 in Mycobacterial Infections of Polymorphonuclear Neutrophils. Sichuan Da Xue Xue Bao Yi Xue Ban 2018, 49, 871–875. [Google Scholar]
- Saini, N.K.; Baena, A.; Ng, T.W.; Venkataswamy, M.M.; Kennedy, S.C.; Kunnath-Velayudhan, S.; Carreno, L.J.; Xu, J.; Chan, J.; Larsen, M.H.; et al. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat. Microbiol. 2016, 1, 16133. [Google Scholar] [CrossRef]


| Variable | Descriptor | Cases |
|---|---|---|
| Sociodemographic Factors | ||
| Age | Median (IQR) | 26.5 (22–34.7) |
| Sex | Male n (%) | 50 (46.2%) |
| Female | 58 (53.7%) | |
| Clinical-Biological Factors | ||
| Weight (kg) | Median (IQR) | 53.9 (49.5–60.4) |
| Height (m) | 1.64 (1.5–1.7) | |
| BMI (kg/m2) | 19.7 (17.8–22.05) | |
| BMI Classification (WHO) | Underweight BMI < 18.5, n (%) | 39 (36.1%) |
| Normal BMI 18.5–24.9 | 57 (52.7%) | |
| Overweight BMI 25–29.9 | 12 (11.1%) | |
| Previous APTB episode | No n (%) | 107 (99.07%) |
| Yes | 1 (0.9%) | |
| Contact with APTB patient | No n (%) | 94 (87.03%) |
| Yes | 14 (12.9%) | |
| Positive radiological findings | No n (%) | 2 (1.85%) |
| Yes | 106 (98.14%) | |
| Extent of parenchymal involvement | ||
| <1/3 of one lung n (%) | 33 (31.1%) | |
| <1 complete lung | 47 (44.3%) | |
| 2 lungs | 26 (24.5%) | |
| Lung cavitation | ||
| Yes, n (%) | 38 (35.18%) | |
| No, n (%) | 70 (64.81%) | |
| Pleural effusion | ||
| Yes, n (%) | 2 (1.85%) | |
| No, n (%) | 106 (98.14%) | |
| Total diagnostic delay in weeks | Median (IQR) | 17.4 (9.14–41.6) |
| Bacilloscopy | Total samples | 99 |
| 1 to 10 AFB in 100 fields n (%) | 11 (11.1%) | |
| 10 to 99 AFB in 100 fields (1+) | 36 (36.36%) | |
| 1 to 10 AFB per field (2+) | 27 (27.27%) | |
| 10 AFB per field (3+) | 25 (25.25%) | |
| TB score-GICIG | SC I 1–4 n (%) | 33 (30.5%) |
| SC II 5–8 | 51 (47.2%) | |
| SC III >= 9 | 24 (22.2%) | |
| Past Medical History | Alcohol consumption n (%) | 10 (9.2%) |
| Smoking | 9 (8.3%) | |
| Respiratory | 3 (2.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocampo, J.C.; Alzate, J.F.; Barrera, L.F.; Baena, A. Tuberculosis Severity Predictive Model Using Mtb Variants and Serum Biomarkers in a Colombian Cohort of APTB Patients. Biomedicines 2023, 11, 3110. https://doi.org/10.3390/biomedicines11123110
Ocampo JC, Alzate JF, Barrera LF, Baena A. Tuberculosis Severity Predictive Model Using Mtb Variants and Serum Biomarkers in a Colombian Cohort of APTB Patients. Biomedicines. 2023; 11(12):3110. https://doi.org/10.3390/biomedicines11123110
Chicago/Turabian StyleOcampo, Juan C., Juan F. Alzate, Luis F. Barrera, and Andres Baena. 2023. "Tuberculosis Severity Predictive Model Using Mtb Variants and Serum Biomarkers in a Colombian Cohort of APTB Patients" Biomedicines 11, no. 12: 3110. https://doi.org/10.3390/biomedicines11123110
APA StyleOcampo, J. C., Alzate, J. F., Barrera, L. F., & Baena, A. (2023). Tuberculosis Severity Predictive Model Using Mtb Variants and Serum Biomarkers in a Colombian Cohort of APTB Patients. Biomedicines, 11(12), 3110. https://doi.org/10.3390/biomedicines11123110







