Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy
Abstract
:1. Introduction
2. Material and Methods
2.1. Research Objects and Treatment Procedures
2.2. Data Collection
2.3. SNP Selecting, DNA Extraction and Genotyping
2.4. Statistical Analysis
3. Results
3.1. Distribution of Characteristics in Lung Cancer Patients and Prognosis Analysis
3.2. Association between Polymorphisms and Prognosis in the Lung Cancer Patients
3.3. Stratification Analyses of Association between Polymorphisms and Prognosis in Lung Cancer Patients
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Roy-Chowdhuri, S. Molecular Pathology of Lung Cancer. Surg. Pathol. Clin. 2021, 14, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Vrana, D. Advances in the therapy of small cell lung cancer. Klin. Onkol. 2021, 34, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Hanna, N. Advances in systemic therapy for non-small cell lung cancer. BMJ 2021, 375, n2363. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C. Advances in Lung Cancer Treatment. Dtsch. Med. Wochenschr. 2021, 146, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, H.; Lee, B.; Lee, S.H.; Sun, J.M.; Park, W.Y.; Ahn, J.S.; Ahn, M.J.; Park, K. DNA Damage Response and Repair Pathway Alteration and Its Association with Tumor Mutation Burden and Platinum-Based Chemotherapy in SCLC. J. Thorac. Oncol. 2019, 14, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Huo, Z.; Li, C.; Wang, R.; Wang, R.; Liu, Y.; Chen, J.; Lu, Y.; Wen, Y.; Jiang, Y.; et al. Lung cancer risk in patients with multiple sclerosis: A Mendelian randomization analysis. Mult. Scler. Relat. Disord. 2021, 51, 102927. [Google Scholar] [CrossRef]
- Zou, T.; Yin, J.; Zheng, W.; Xiao, L.; Tan, L.; Chen, J.; Wang, Y.; Li, X.; Qian, C.; Cui, J.; et al. Rho GTPases: RAC1 polymorphisms affected platinum-based chemotherapy toxicity in lung cancer patients. Cancer Chemother. Pharmacol. 2016, 78, 249–258. [Google Scholar] [CrossRef]
- Yin, J.Y.; Zhang, J.T.; Zhang, W.; Zhou, H.H.; Liu, Z.Q. eIF3a: A new anticancer drug target in the eIF family. Cancer Lett. 2018, 412, 81–87. [Google Scholar] [CrossRef]
- He, J.; Wang, Z.; Wang, Y.; Zou, T.; Li, X.P.; Cao, L.; Chen, J. The Effects of WISP1 Polymorphisms on the Prognosis of Lung Cancer Patients with Platinum-Based Chemotherapy. Pharmacogenom. Pers. Med. 2021, 14, 1193–1203. [Google Scholar] [CrossRef]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Ashour, M.E.; Mosammaparast, N. Mechanisms of damage tolerance and repair during DNA replication. Nucleic Acids Res. 2021, 49, 3033–3047. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair 2019, 83, 102673. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T. DNA damage and regulation of protein homeostasis. DNA Repair 2021, 105, 103155. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.H.; Li, J.; Chen, P.; Jiang, H.G.; Wu, M.; Chen, Y.C. RNA interferences targeting the Fanconi anemia/BRCA pathway upstream genes reverse cisplatin resistance in drug-resistant lung cancer cells. J. Biomed. Sci. 2015, 22, 77. [Google Scholar] [CrossRef] [PubMed]
- Pilger, D.; Seymour, L.W.; Jackson, S.P. Interfaces between cellular responses to DNA damage and cancer immunotherapy. Genes Dev. 2021, 35, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Mao, Z.; Hine, C. Changes in DNA repair during aging. Nucleic Acids Res. 2007, 35, 7466–7474. [Google Scholar] [CrossRef]
- Muniesa-Vargas, A.; Theil, A.F.; Ribeiro-Silva, C.; Vermeulen, W.; Lans, H. XPG: A multitasking genome caretaker. Cell. Mol. Life Sci. 2022, 79, 166. [Google Scholar] [CrossRef]
- Kiyohara, C.; Yoshimasu, K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: A meta-analysis. Int. J. Med. Sci. 2007, 4, 59–71. [Google Scholar] [CrossRef]
- Lan, X.; Li, Y.; Wu, Y.; Li, X.; Xu, L. The Association of ERCC1 and ERCC5 Polymorphisms with Lung Cancer Risk in Han Chinese. J. Cancer 2022, 13, 517–526. [Google Scholar] [CrossRef]
- Liu, D.; Wu, J.; Shi, G.Y.; Zhou, H.F.; Yu, Y. Role of XRCC1 and ERCC5 polymorphisms on clinical outcomes in advanced non-small cell lung cancer. Genet. Mol. Res. 2014, 13, 3100–3107. [Google Scholar] [CrossRef]
- Pradhan, S.; Sarma, H.; Mattaparthi, V.S.K. Investigation of the probable homo-dimer model of the Xeroderma pigmentosum complementation group A (XPA) protein to represent the DNA-binding core. J. Biomol. Struct. Dyn. 2019, 37, 3322–3336. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yu, C.; Yu, K. Association of human XPA rs1800975 polymorphism and cancer susceptibility: An integrative analysis of 71 case-control studies. Cancer Cell Int. 2020, 20, 164. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Hou, M.; Guo, S.; Hua, F.; Zheng, D.; Xu, F.; Jiang, Y.; Li, L.; Qiao, Y.; Fan, Y.; et al. Polymorphisms in DNA repair genes of XRCC1, XPA, XPC, XPD and associations with lung cancer risk in Chinese people. Thorac. Cancer 2014, 5, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, S.; Hong, X.; Li, X.; Zhao, X.; Huai, C.; Chen, H.; Gao, Z.; Qian, J.; Wang, J.; et al. Single nucleotide polymorphisms of nucleotide excision repair pathway are significantly associated with outcomes of platinum-based chemotherapy in lung cancer. Sci. Rep. 2017, 7, 11785. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, H.; Xu, F.; Kong, J.; Yu, H.; Qian, B. DNA Repair Gene Polymorphisms in Relation to Non-Small Cell Lung Cancer Survival. Cell. Physiol. Biochem. 2015, 36, 1419–1429. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, L.; Wang, Y.; Yin, J.; Li, X.; Wang, Z.; Li, H.; Zou, T.; Qian, C.; Li, C.; et al. Effect of transporter and DNA repair gene polymorphisms to lung cancer chemotherapy toxicity. Tumor Biol. 2016, 37, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Piljic Burazer, M.; Mladinov, S.; Matana, A.; Kuret, S.; Bezic, J.; Glavina Durdov, M. Low ERCC1 expression is a good predictive marker in lung adenocarcinoma patients receiving chemotherapy based on platinum in all TNM stages—A single-center study. Diagn. Pathol. 2019, 14, 105. [Google Scholar] [CrossRef]
- Grenda, A.; Blach, J.; Szczyrek, M.; Krawczyk, P.; Nicos, M.; Kuznar Kaminska, B.; Jakimiec, M.; Balicka, G.; Chmielewska, I.; Batura-Gabryel, H.; et al. Promoter polymorphisms of TOP2A and ERCC1 genes as predictive factors for chemotherapy in non-small cell lung cancer patients. Cancer Med. 2020, 9, 605–614. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Tang, L.L.; Long, J.T.; Zhu, J.; Hua, R.X.; Li, J. XPG gene polymorphisms and cancer susceptibility: Evidence from 47 studies. Oncotarget 2017, 8, 37263–37277. [Google Scholar] [CrossRef]
- Liutkeviciene, R.; Vilkeviciute, A.; Morkunaite, G.; Glebauskiene, B.; Kriauciuniene, L. SIRT1 (rs3740051) role in pituitary adenoma development. BMC Med. Genet. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, J.Y.; Li, X.P.; Chen, J.; Qian, C.Y.; Zheng, Y.; Fu, Y.L.; Chen, Z.Y.; Zhou, H.H.; Liu, Z.Q. The association of transporter genes polymorphisms and lung cancer chemotherapy response. PLoS ONE 2014, 9, e91967. [Google Scholar] [CrossRef]
- Cibeira, M.T.; de Larrea, C.F.; Navarro, A.; Diaz, T.; Fuster, D.; Tovar, N.; Rosinol, L.; Monzo, M.; Blade, J. Impact on response and survival of DNA repair single nucleotide polymorphisms in relapsed or refractory multiple myeloma patients treated with thalidomide. Leuk. Res. 2011, 35, 1178–1183. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Liu, J.Y.; She, L.; Yin, J.Y.; Li, X.; Li, X.P.; Zhou, H.H.; Chen, J.; Liu, Z.Q. The Association Between Heat-Shock Protein Polymorphisms and Prognosis in Lung Cancer Patients Treated With Platinum-Based Chemotherapy. Front. Pharmacol. 2020, 11, 1029. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.P.; Zhou, Y.; Chen, Y.; Li, N.N.; Wu, X.T. XRCC3 Thr241Met polymorphism and gastric cancer susceptibility: A meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Li, G.; Wu, L.; Huang, M. Perspectives of ERCC1 in early-stage and advanced cervical cancer: From experiments to clinical applications. Front. Immunol. 2022, 13, 1065379. [Google Scholar] [CrossRef] [PubMed]
- Rasekhian, M.; Roohvand, F.; Habtemariam, S.; Marzbany, M.; Kazemimanesh, M. The Role of 3’UTR of RNA Viruses on mRNA Stability and Translation Enhancement. Mini-Rev. Med. Chem. 2021, 21, 2389–2398. [Google Scholar] [CrossRef]
- Xu, E.; Zhang, J.; Zhang, M.; Jiang, Y.; Cho, S.J.; Chen, X. RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol. Cancer Res. 2014, 12, 359–369. [Google Scholar] [CrossRef]
- Mayr, C. What Are 3′ UTRs Doing? Cold Spring Harb. Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef]
- Berkovits, B.D.; Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367. [Google Scholar] [CrossRef]
- Gu, S.; Rong, H.; Zhang, G.; Kang, L.; Yang, M.; Guan, H. Functional SNP in 3′-UTR MicroRNA-Binding Site of ZNF350 Confers Risk for Age-Related Cataract. Hum. Mutat. 2016, 37, 1223–1230. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Su, C.; Zhao, X.; Tang, L.; Zhou, C. The association between polymorphisms in the DNA nucleotide excision repair genes and RRM1 gene and lung cancer risk. Thorac. Cancer 2012, 3, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Bhat, G.R.; Sethi, I.; Bhat, A.; Verma, S.; Bakshi, D.; Sharma, B.; Nazir, M.; Dar, K.A.; Abrol, D.; Shah, R.; et al. Genetic evaluation of the variants using MassARRAY in non-small cell lung cancer among North Indians. Sci. Rep. 2021, 11, 11291. [Google Scholar] [CrossRef]
- Minina, V.I.; Bakanova, M.L.; Soboleva, O.A.; Ryzhkova, A.V.; Titov, R.A.; Savchenko, Y.A.; Sinitsky, M.Y.; Voronina, E.N.; Titov, V.A.; Glushkov, A.N. Polymorphisms in DNA repair genes in lung cancer patients living in a coal-mining region. Eur. J. Cancer Prev. 2019, 28, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ramirez, C.; Canadas-Garre, M.; Molina, M.A.; Robles, A.I.; Faus-Dader, M.J.; Calleja-Hernandez, M.A. Contribution of genetic factors to platinum-based chemotherapy sensitivity and prognosis of non-small cell lung cancer. Mutat. Res. Rev. Mutat. Res. 2017, 771, 32–58. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Duan, Z.; Li, P.; Xu, Q.; Yuan, Y. Role of ERCC5 promoter polymorphisms in response to platinum-based chemotherapy in patients with advanced non-small-cell lung cancer. Anticancer Drugs 2013, 24, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, G.; Li, X.; Ren, S.; Li, A.; Xu, J.; Zhang, J.; Zhou, C. Association between single nucleotide polymorphisms (SNPs) and toxicity of advanced non-small-cell lung cancer patients treated with chemotherapy. PLoS ONE 2012, 7, e48350. [Google Scholar] [CrossRef]
- Landi, L.; Cappuzzo, F. HER2 and lung cancer. Expert Rev. Anticancer. Ther. 2013, 13, 1219–1228. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Sharma, B.; Kanwar, S.S. Phosphatidylserine: A cancer cell targeting biomarker. Semin. Cancer Biol. 2018, 52, 17–25. [Google Scholar] [CrossRef]
- Diao, W.Y.; Ding, C.L.; Yuan, B.Y.; Li, Z.; Sun, N.; Huang, J.B. Clinical Characteristics and Prognosis of HER2 Gene Phenotype in Patients with Non-Small Cell Lung Cancer. Int. J. Gen. Med. 2021, 14, 9153–9161. [Google Scholar] [CrossRef] [PubMed]
- Baraibar, I.; Mezquita, L.; Gil-Bazo, I.; Planchard, D. Novel drugs targeting EGFR and HER2 exon 20 mutations in metastatic NSCLC. Crit. Rev. Oncol. Hematol. 2020, 148, 102906. [Google Scholar] [CrossRef] [PubMed]
- Abdalkhalek, E.S.; Wakeel, L.M.E.; Nagy, A.A.; Sabri, N.A. Variants of ERCC5 and the outcome of platinum-based regimens in non-small cell lung cancer: A prospective cohort study. Med. Oncol. 2022, 39, 152. [Google Scholar] [CrossRef] [PubMed]
- Zienolddiny, S.; Campa, D.; Lind, H.; Ryberg, D.; Skaug, V.; Stangeland, L.; Phillips, D.H.; Canzian, F.; Haugen, A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 2006, 27, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Han, X.; Liu, C.; Li, X.; Lu, S.; Jiang, Q.; Yao, J. Long Non-Coding RNA PNKY Modulates the Development of Choroidal Neovascularization. Front. Cell Dev. Biol. 2022, 10, 836031. [Google Scholar] [CrossRef]
- Liu, T.; Yang, Q.; Wei, W.; Wang, K.; Wang, E. Toll/IL-1 receptor-containing proteins STIR-1, STIR-2 and STIR-3 synergistically assist Yersinia ruckeri SC09 immune escape. Fish Shellfish Immunol. 2020, 103, 357–365. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, Z.; Li, X.; Wei, S.; Yuan, K. Gene expression and prognosis of x-ray repair cross-complementing family members in non-small cell lung cancer. Bioengineered 2021, 12, 6210–6228. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Yang, Y.; Yang, H.; Guo, Q.; Guo, J.L.; Liu, H.S.; Zhang, J.; Li, D. The roles of risk model based on the 3-XRCC genes in lung adenocarcinoma progression. Transl. Cancer Res. 2021, 10, 4413–4431. [Google Scholar] [CrossRef]
- Saviozzi, S.; Ceppi, P.; Novello, S.; Ghio, P.; Lo Iacono, M.; Borasio, P.; Cambieri, A.; Volante, M.; Papotti, M.; Calogero, R.A.; et al. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009, 69, 3390–3396. [Google Scholar] [CrossRef]

| Patient Characteristics | N (%) |
|---|---|
| Total no. of patients | 345 |
| Histology | |
| Adenocarcinoma | 112 (32.5) |
| Squamous cell | 111 (32.2) |
| Small cell | 99 (28.7) |
| Age | |
| ≤56 | 172 (49.9) |
| >56 | 173 (50.1) |
| Clinical stage | |
| I/II/LD | 41 (11.9) |
| III/IV/ED | 298 (86.4) |
| Smoking status | |
| Non-smoker | 122 (35.4) |
| Smoker | 223 (64.6) |
| Gender | |
| Male | 285 (82.6) |
| Female | 60 (17.4) |
| Metastasis | |
| No | 61 (17.7) |
| Yes | 149 (34.2) |
| Chemotherapy regimens | |
| Platinum/gemcitabine | 109 (31.6) |
| Platinum/paclitaxel | 59 (17.1) |
| Platinum/navelbine | 8 (2.3) |
| Platinum/etoposide | 77 (22.3) |
| Platinum/irinotecan | 8 (2.3) |
| Gene | Locus | dbSNP | Call Rate (%) | Polymorphism | MAF | Genotype | N (%) |
|---|---|---|---|---|---|---|---|
| ERCC1 | 19q13.32 | rs12984195 | 97.97 | T>C | 3.70 | TT | 325 (94.2) |
| TC | 13 (3.8) | ||||||
| CC | 0 | ||||||
| rs117128015 | 100.00 | C>T | 8.00 | CC | 315 (91.3) | ||
| CT | 30 (8.7) | ||||||
| TT | 0 | ||||||
| ERCC5 | 13q33.1 | rs873601 | 99.13 | G>A | 49.11 | GG | 84 (24.4) |
| GA | 165 (47.8) | ||||||
| AA | 93 (27.0) | ||||||
| PNKY | 6q16.1 | rs1869641 | 98.84 | G>A | 22.22 | GG | 247 (71.6) |
| GA | 82 (23.8) | ||||||
| AA | 12 (3.5) | ||||||
| rs1883306 | 93.91 | T>G | 38.53 | TT | 146 (42.3) | ||
| TG | 138 (40.0) | ||||||
| GG | 40 (11.6) | ||||||
| rs2444933 | 99.13 | A>G | 31.62 | AA | 194 (56.2) | ||
| AG | 126 (36.5) | ||||||
| GG | 22 (6.4) | ||||||
| SIRT1 | 10q21.3 | rs3758391 | 97.39 | T>C | 24.46 | TT | 248 (71.9) |
| TC | 74 (21.5) | ||||||
| CC | 14 (4.1) | ||||||
| rs3740051 | 96.52 | A>G | 33.41 | AA | 182 (52.8) | ||
| AG | 119 (34.5) | ||||||
| GG | 32 (9.3) | ||||||
| rs4746720 | 99.13 | T>C | 46.67 | TT | 118 (34.2) | ||
| TC | 138 (40.0) | ||||||
| CC | 86 (24.9) | ||||||
| rs12778366 | 95.65 | T>C | 22.09 | TT | 239 (69.3) | ||
| TC | 82 (23.8) | ||||||
| CC | 9 (2.6) | ||||||
| XPA | 9q22.33 | rs3176751 | 98.84 | G>C | 20.61 | GG | 2 (0.6) |
| GC | 86 (24.9) | ||||||
| CC | 253 (73.3) | ||||||
| rs3176752 | 98.84 | C>A | 21.44 | CC | 249 (72.2) | ||
| CA | 88 (25.5) | ||||||
| AA | 4 (1.2) | ||||||
| XRCC3 | 14q32.33 | rs3212117 | 98.84 | C>A | 5.28 | CC | 322 (93.3) |
| CA | 19 (5.5) | ||||||
| AA | 0 | ||||||
| rs3212121 | 98.55 | A>G | 4.24 | AA | 325 (94.2) | ||
| AG | 14 (4.1) | ||||||
| GG | 1 (0.3) | ||||||
| XRCC5 | 2q35 | rs1051677 | 98.84 | T>C | 22.43 | TT | 245 (71.0) |
| TC | 87 (25.2) | ||||||
| CC | 9 (2.6) | ||||||
| rs2440 | 98.55 | C>T | 34.03 | CC | 24 (7.0) | ||
| CT | 139 (40.3) | ||||||
| TT | 177 (51.3) | ||||||
| rs1051685 | 99.13 | A>G | 12.14 | AA | 295 (85.5) | ||
| AG | 45 (13.0) | ||||||
| GG | 2 (0.6) |
| Variables | Patients N (%) | Death N (%) | MST-OS (Year) | MST-PFS (Year) |
|---|---|---|---|---|
| Lung cancer | 345 | 279 | 4.42 | 3.16 |
| NSCLC | 233 (67.5) | 188 (67.4) | 4.56 | 3.25 |
| SCLC | 99 (28.7) | 80 (28.7) | 4.17 | 3.10 |
| Age | ||||
| ≤56 | 172 (49.9) | 142 (50.9) | 4.48 | 2.95 |
| >56 | 173 (50.1) | 137 (49.1) | 4.36 | 3.80 |
| Clinical stage | ||||
| I/II/LD | 41 (11.9) | 31 (11.1) | 3.14 | 4.05 |
| III/IV/ED | 298 (86.4) | 243 (87.1) | 4.55 | 3.21 |
| Smoking status | ||||
| Non-smoker | 122 (35.4) | 97 (34.8) | 4.77 | 4.60 |
| Smoker | 223 (64.6) | 182 (65.2) | 4.26 | 2.61 |
| Gender | ||||
| male | 285 (82.6) | 229 (82.1) | 4.39 | 2.93 |
| female | 60 (17.4) | 50 (17.9) | 4.57 | 4.47 |
| Metastasis | ||||
| No | 61 (17.7) | 51 (18.3) | 3.84 | 2.28 |
| Yes | 149 (43.2) | 121 (43.4) | 4.53 | 3.94 |
| Gene | Polymorphisms | Genotype | MST (Year) | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|---|
| BETA (95%CI) | p Value | BETA (95%CI) | p Value | BETA (95%CI) | p Value | ||||
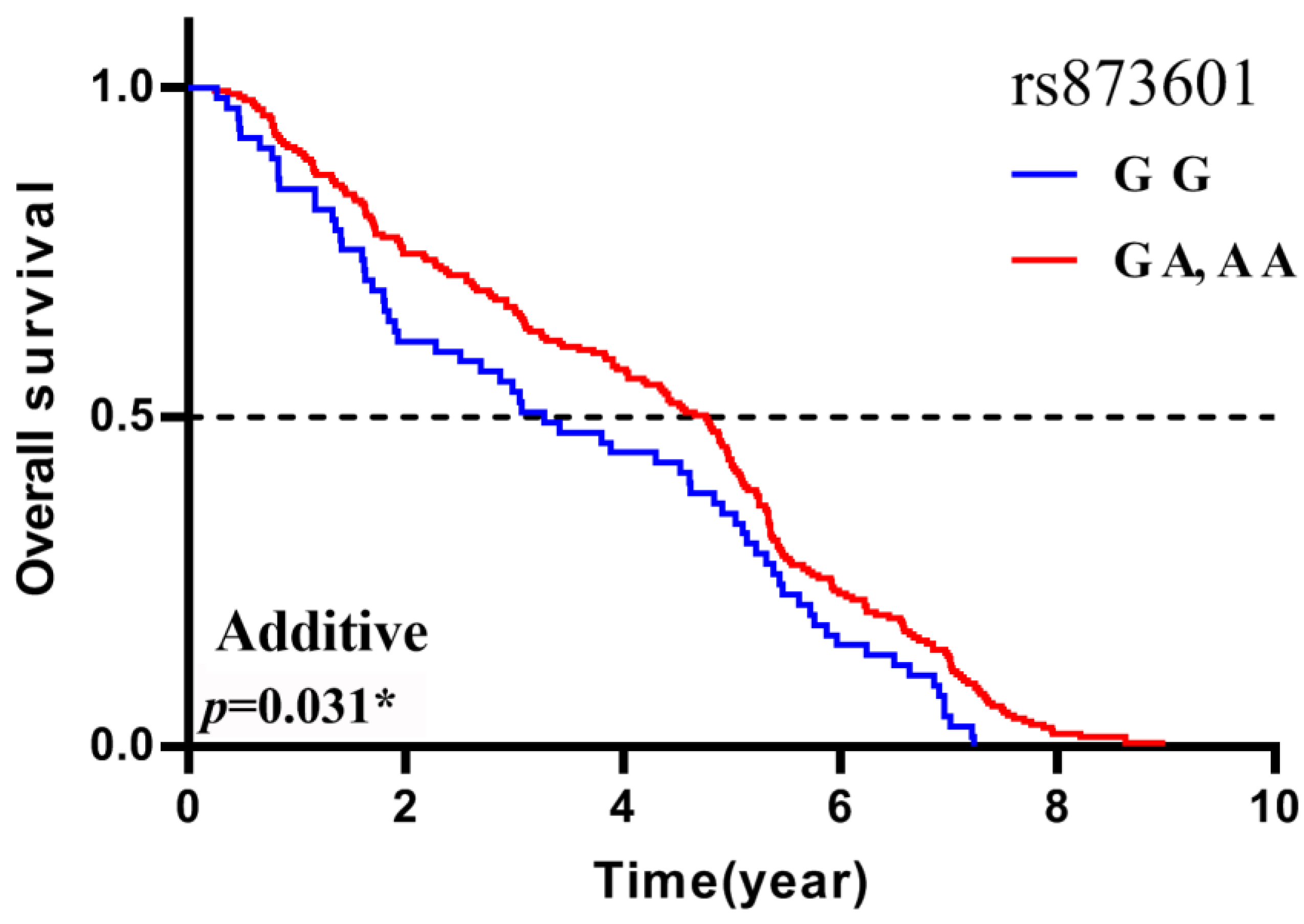
| ERCC5 | rs873601 | G G | 3.28 | −0.37 (−0.75, 0.01) | 0.061 | −0.30 (−0.91, 0.32) | 0.348 | −0.70 (−1.34, −0.07) | 0.031 * |
| G A | 4.88 | ||||||||
| A A | 4.02 |
| OS/PFS | Gene | Polymorphisms | Subgroup | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|---|
| BETA (95%CI) | p Value | BETA (95%CI) | p Value | BETA (95%CI) | p Value | ||||
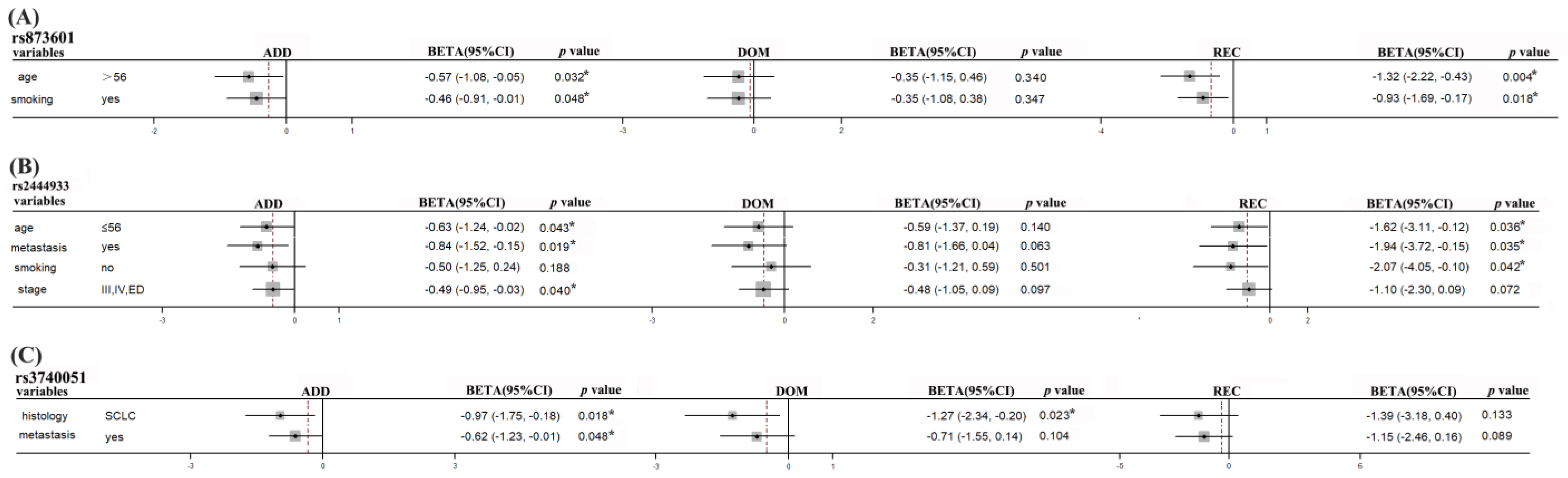
| OS | ERCC5 | rs873601 | age > 56 | −0.57 (−1.08, −0.05) | 0.032 * | −0.35 (−1.15, 0.46) | 0.399 | −1.32 (−2.22, −0.43) | 0.004 * |
| smokers | −0.46 (−0.91, −0.01) | 0.048 * | −0.35 (−1.08, 0.38) | 0.347 | −0.93 (−1.69, −0.17) | 0.018 * | |||
| PNKY | rs2444933 | age ≤ 56 | −0.63 (−1.24, −0.02) | 0.043 * | −0.59 (−1.37, 0.19) | 0.140 | −1.62 (−3.11, −0.12) | 0.036 * | |
| metastasis | −0.84 (−1.53, −0.15) | 0.019 * | −0.81 (−1.66, 0.04) | 0.063 | −1.94 (−3.72, −0.15) | 0.035 * | |||
| non-smoker | −0.51 (−1.25, 0.24) | 0.188 | −0.31 (−1.21, 0.59) | 0.501 | −2.07 (−4.05, −0.10) | 0.042 * | |||
| III/IV/ED | −0.49 (−0.95, −0.03) | 0.040 * | −0.48 (−1.05, 0.09) | 0.097 | −1.10 (−2.30, 0.09) | 0.072 | |||
| STIR1 | rs3740051 | SCLC | −0.97 (−1.75, −0.18) | 0.018 * | −1.27 (−2.34, −0.20) | 0.023 * | −1.39 (−3.18, 0.40) | 0.133 | |
| metastasis | −0.62 (−1.23, −0.01) | 0.048 * | −0.71 (−1.55, 0.14) | 0.104 | −1.15 (−2.46, 0.16) | 0.089 | |||
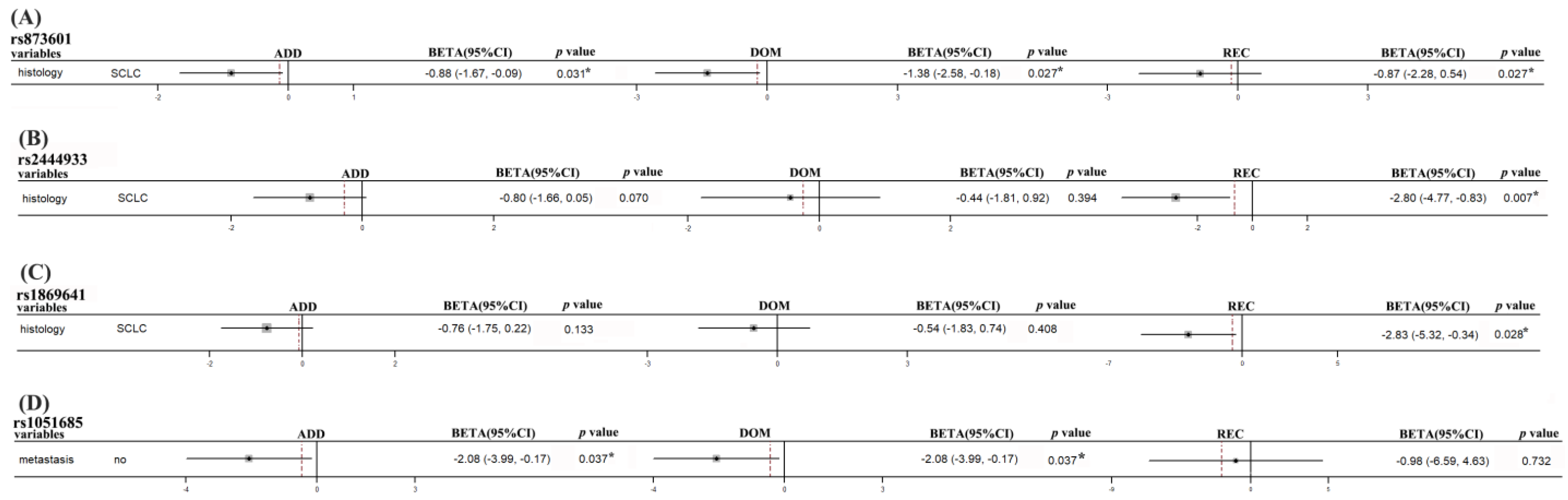
| PFS | ERCC5 | rs873601 | SCLC | −0.88 (−1.67, −0.09) | 0.031 * | −1.38 (−2.58, −0.18) | 0.027 * | −0.87 (−2.28, 0.54) | 0.230 |
| PNKY | rs2444933 | SCLC | −0.80 (−1.66, 0.05) | 0.070 | −0.49 (−1.60, 0.63) | 0.394 | −2.80 (−4.77, −0.83) | 0.007 * | |
| rs1869641 | SCLC | −0.76 (−1.75, 0.22) | 0.133 | −0.54 (−1.83, 0.74) | 0.408 | −2.83 (−5.32, −0.34) | 0.028 * | ||
| XRCC5 | rs1051685 | Non-metastasis | −2.08 (−3.99, −0.17) | 0.037 * | −2.08 (−3.99, −0.17) | 0.037 * | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, T.; Liu, J.-Y.; Qin, Q.; Guo, J.; Zhou, W.-Z.; Li, X.-P.; Zhou, H.-H.; Chen, J.; Liu, Z.-Q. Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Biomedicines 2023, 11, 3133. https://doi.org/10.3390/biomedicines11123133
Zou T, Liu J-Y, Qin Q, Guo J, Zhou W-Z, Li X-P, Zhou H-H, Chen J, Liu Z-Q. Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Biomedicines. 2023; 11(12):3133. https://doi.org/10.3390/biomedicines11123133
Chicago/Turabian StyleZou, Ting, Jun-Yan Liu, Qun Qin, Jie Guo, Wen-Zhi Zhou, Xiang-Ping Li, Hong-Hao Zhou, Juan Chen, and Zhao-Qian Liu. 2023. "Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy" Biomedicines 11, no. 12: 3133. https://doi.org/10.3390/biomedicines11123133
APA StyleZou, T., Liu, J.-Y., Qin, Q., Guo, J., Zhou, W.-Z., Li, X.-P., Zhou, H.-H., Chen, J., & Liu, Z.-Q. (2023). Role of rs873601 Polymorphisms in Prognosis of Lung Cancer Patients Treated with Platinum-Based Chemotherapy. Biomedicines, 11(12), 3133. https://doi.org/10.3390/biomedicines11123133






