Protein Kinase CK2 and Epstein–Barr Virus
Abstract
:1. Introduction
2. Epstein–Barr Virus
3. Phosphorylation of EBV Proteins by Protein Kinase CK2
4. CK2 and Cellular Proteins in the Balance between Lytic EBV Virus Replication and Cell Transformation
4.1. Ikaros and the Switch from EBV Latency to Lytic Replication
4.2. CK2 Binding Cellular Protein ARKL1 and the Regulation of EBV Replication
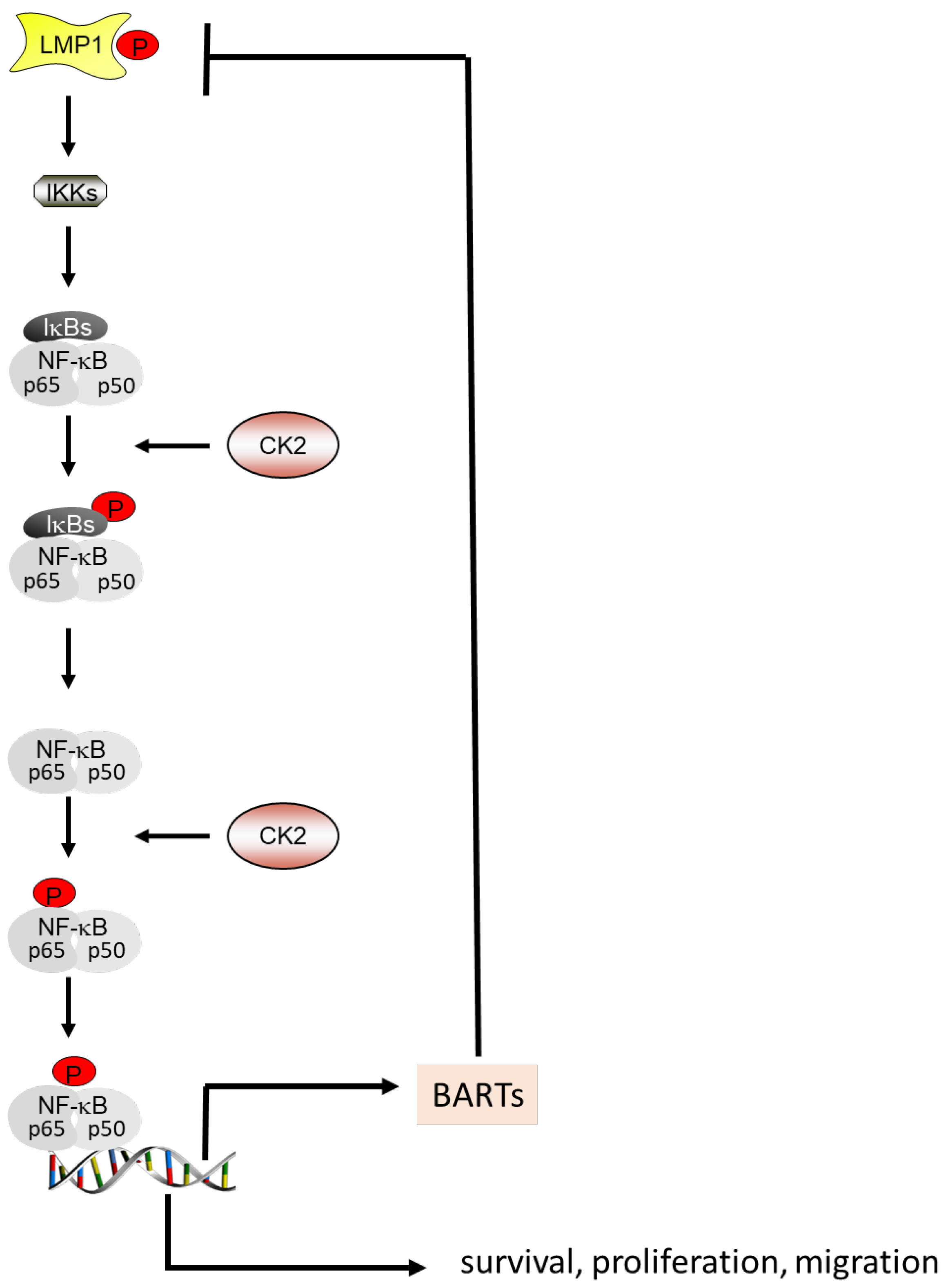
4.3. CK2 and the Autoregulatory Loop between NF-κB, BARTs and LMP1
5. Common Targets of EBV and CK2
5.1. EBV, CK2 and the NF-κB Pathway
5.2. EBV, CK2 and the PI3K/Akt Pathway
5.3. EBV, CK2 and the Wnt/β-Catenin Pathway
5.4. EBV, CK2 and the Janus Kinase/Signaling Transduction and Transcription Activator (JAK/STAT) Pathway
6. EBV-Encoded LMP1 Protein and p53
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Glossary
| BARF1 | BamHI-A rightward open reading frame 1 |
| BARTs | BamHI-A rightward transcripts |
| BZLF1 or ZEBRA or EB-1 or Zta | BamHI-Z EBV replication activator |
| BL | Burkitt’s lymphoma |
| DLBCL | Diffuse large B-cell lymphoma |
| PKR | Double-stranded RNA-dependent protein kinase |
| EBER | Epstein–Barr encoded small RNA |
| EBV | Epstein–Barr virus |
| EBNA | Epstein–Barr virus nuclear antigen |
| GC | Gastric carcinoma |
| HCV | Hepatitis C virus |
| HSV | Herpes simplex-1 virus |
| HD | Hodgkin’s disease |
| HCMV | Human cytomegalovirus |
| HIV | Human immunodeficiency virus |
| HTLV-1 | Human T-lymphotropic virus type 1 |
| JAK | Janus kinase |
| LMP | Latent membrane protein |
| MS | Multiple sclerosis |
| NKTL | Nasal NK/T-cell lymphoma |
| NPC | Nasopharyngeal carcinoma |
| NF-kB | Nuclear factor kappaB |
| PI3K | Phosphatidylinositol-3-kinase |
| PTLD | Post-transplant lymphoproliferative disease |
| Akt | Protein kinase B, also known as Akt |
| CK2 | Protein kinase CK2 |
| PKC | protein kinase C |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus type-2 |
| STAT | Signal transducer and activator of transcription protein |
References
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litchfield, D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef]
- Heriche, J.K.; Lebrin, F.; Rabilloud, T.; LeRoy, D.; Chambaz, E.M.; Goldberg, Y. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2alpha. Science 1997, 276, 952–955. [Google Scholar] [CrossRef]
- Stigare, J.; Buddelmeijer, N.; Pigon, A.; Egyhazi, E. A majority of CK2 alpha subunit is tightly bound to intranuclear compounds but not to the beta subunit. Mol. Cell. Biol. 1993, 129, 77–85. [Google Scholar]
- Lou, D.Y.; Dominguez, I.; Toselli, P.; Landesman-Bollag, E.; O’Brien, C.; Seldin, D.C. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell. Biol. 2008, 28, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Toselli, P.A.; Russell, L.D.; Seldin, D.C. Globozoospermia in mice lacking the casein kinase II α’ catalytic subunit. Nat. Genet. 1999, 23, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Vilk, G.; Saulnier, R.B.; St Pierre, R.; Litchfield, D.W. Inducible expression of protein kinase CK2 in mammalian cells-Evidence for functional specialization of CK2 isoforms. J. Biol. Chem. 1999, 274, 14406–14414. [Google Scholar] [CrossRef] [Green Version]
- Turowec, J.P.; Vilk, G.; Gabriel, M.; Litchfield, D.W. Characterizing the convergence of protein kinase CK2 and caspase-3 reveals isoform-specific phosphorylation of caspase-3 by CK2alpha’: Implications for pathological roles of CK2 in promoting cancer cell survival. Oncotarget 2013, 4, 560–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montenarh, M.; Götz, C. The interactome of protein kinase CK2. In Protein Kinase CK2; Pinna, L.A., Ed.; John Wiley & Sons, Inc.: Oxford, UK, 2013; pp. 76–116. [Google Scholar]
- Faust, M.; Montenarh, M. Subcellular localization of protein kinase CK2: A key to its function? Cell Tissue Res. 2000, 301, 329–340. [Google Scholar] [CrossRef]
- Guerra, B.; Issinger, O.G. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 1999, 20, 391–408. [Google Scholar] [CrossRef]
- Bren, G.D.; Pennington, K.N.; Paya, C.V. PKC-zeta-associated CK2 participates in the turnover of free IkappaBalpha. J. Mol. Biol. 2000, 297, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Issinger, O.G.; Wang, J.Y.J. Modulation of human checkpoint kinase Chk1 by the regulatory β-subunit of protein kinase CK2. Oncogene 2003, 22, 4933–4942. [Google Scholar] [CrossRef] [Green Version]
- Gray, G.K.; McFarland, B.C.; Rowse, A.L.; Gibson, S.A.; Benveniste, E.N. Therapeutic CK2 inhibition attenuates diverse prosurvival signaling cascades and decreases cell viability in human breast cancer cells. Oncotarget 2014, 5, 6484–6496. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Jones, K.A. CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr. Biol. 2006, 16, 2239–2244. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, H.Y. Casein kinase 2 Is activated and essential for Wnt/beta-catenin signaling. J. Biol. Chem. 2006, 281, 18394–18400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, B. Protein kinase CK2 subunits are positive regulators of AKT kinase. Int. J. Oncol. 2006, 28, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Ponce, D.P.; Yefi, R.; Cabello, P.; Maturana, J.L.; Niechi, I.; Silva, E.; Galindo, M.; Antonelli, M.; Marcelain, K.; Armisen, R.; et al. CK2 functionally interacts with AKT/PKB to promote the beta-catenin-dependent expression of survivin and enhance cell survival. Mol. Cell. Biochem. 2011, 356, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ponce, D.P.; Maturana, J.L.; Cabello, P.; Yefi, R.; Niechi, I.; Silva, E.; Armisen, R.; Galindo, M.; Antonelli, M.; Tapia, J.C. Phosphorylation of AKT/PKB by CK2 is necessary for the AKT-dependent up-regulation of beta-catenin transcriptional activity. J. Cell. Physiol. 2011, 226, 1953–1959. [Google Scholar] [CrossRef] [PubMed]
- Marchiori, F.; Meggio, F.; Marin, O.; Borin, G.; Calderan, A.; Ruzza, P.; Pinna, L.A. Synthetic peptide substrates for casein kinase 2. Assessment of minimum structural requirements for phosphorylation. Biochim. Biophys. Acta 1988, 971, 332–338. [Google Scholar] [CrossRef]
- de Villavicencio-Diaz, T.; Rabalski, A.J.; Litchfield, D.W. Protein Kinase CK2: Intricate Relationships within Regulatory Cellular Networks. Pharmaceuticals 2017, 10, E27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabjerg, M.; Bjerregaard, H.; Halekoh, U.; Jensen, B.L.; Walter, S.; Marcussen, N. Molecular characterization of clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1 as promising novel prognostic markers. APMIS 2016, 124, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Montenarh, M.; Götz, C. Protein kinase CK2 and ion channels. Biomed. Rep. 2020, 13, 55. [Google Scholar] [CrossRef]
- Al-Quobaili, F.; Montenarh, M. CK2 and the regulation of the carbohydrate metabolism. Metabolism 2012, 61, 1512–1517. [Google Scholar] [CrossRef]
- Ortega, C.E.; Seidner, Y.; Dominguez, I. Mining CK2 in cancer. PLoS ONE 2014, 9, e115609. [Google Scholar] [CrossRef]
- Seldin, D.C.; Leder, P. Casein kinase II alpha transgene-induced murine lymphoma: Relation to theileriosis in cattle. Science 1995, 267, 884–897. [Google Scholar] [CrossRef]
- Cozza, G.; Pinna, L.A.; Moro, S. Protein kinase CK2 inhibitors: A patent review. Expert Opin. Ther. Pat. 2012, 22, 1081–1097. [Google Scholar] [CrossRef] [PubMed]
- Sarno, S.; Salvi, M.; Battistutta, R.; Zanotti, G.; Pinna, L.A. Features and potentials of ATP-site directed CK2 inhibitors. Biochim. Biophys. Acta 2005, 1754, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G. The Development of CK2 Inhibitors: From Traditional Pharmacology to in Silico Rational Drug Design. Pharmaceuticals 2017, 10, E26. [Google Scholar] [CrossRef] [Green Version]
- Wells, C.I.; Drewry, D.H.; Pickett, J.E.; Tjaden, A.; Krämer, A.; Müller, S.; Gyenis, L.; Menyhart, D.; Litchfield, D.W.; Knapp, S.; et al. Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell Chem. Biol. 2021, 28, 546–558. [Google Scholar] [CrossRef]
- Prudent, R.; Cochet, C. New protein kinase CK2 inhibitors: Jumping out of the catalytic box. Chem. Biol. 2009, 16, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Pinna, L.A. Casein kinases as potential therapeutic targets. Expert Opin. Ther. Targets 2016, 20, 319–340. [Google Scholar] [CrossRef]
- Cozza, G.; Zanin, S.; Sarno, S.; Costa, E.; Girardi, C.; Ribaudo, G.; Salvi, M.; Zagotto, G.; Ruzzene, M.; Pinna, L.A. Design, validation and efficacy of bisubstrate inhibitors specifically affecting ecto-CK2 kinase activity. Biochem. J. 2015, 471, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; Cesaro, L.; Hirota, T.; Kuwata, K.; D’Amore, C.; Ruppert, T.; Blatnik, R.; Salvi, M.; Pinna, L.A. Comparing the efficacy and selectivity of CK2 inhibitors. A phosphoproteomics approach. Eur. J. Med. Chem. 2021, 214, 113217. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; Ruzzene, M. Protein kinase CK2 inhibition as a pharmacological strategy. Adv. Protein Chem. Struct. Biol. 2021, 124, 23–46. [Google Scholar] [PubMed]
- Firzlaff, J.M.; Galloway, D.A.; Eisenman, R.N.; Luscher, B. The E7 protein of human papillomavirus type 16 is phosphorylated by casein kinase II. New Biol. 1989, 1, 44–53. [Google Scholar] [PubMed]
- Ching, W.; Dobner, T.; Koyuncu, E. The human adenovirus type 5 E1B 55-kilodalton protein is phosphorylated by protein kinase CK2. J. Virol. 2012, 86, 2400–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franck, N.; Le Seyec, J.; Guguen-Guillouzo, C.; Erdtmann, L. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J. Virol. 2005, 79, 2700–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvisi, G.; Marin, O.; Pari, G.; Mancini, M.; Avanzi, S.; Loregian, A.; Jans, D.A.; Ripalti, A. Multiple phosphorylation sites at the C-terminus regulate nuclear import of HCMV DNA polymerase processivity factor ppUL44. Virology 2011, 417, 259–267. [Google Scholar] [CrossRef]
- Schubert, U.; Henklein, P.; Boldyreff, B.; Wingender, E.; Strebel, K.; Porstmann, T. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted alpha-helix-turn-alpha-helix-motif. J. Mol. Biol. 1994, 236, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Bidoia, C.; Mazzorana, M.; Pagano, M.A.; Arrigoni, G.; Meggio, F.; Pinna, L.A.; Bertazzoni, U. The pleiotropic protein kinase CK2 phosphorylates HTLV-1 Tax protein in vitro, targeting its PDZ-binding motif. Virus Genes 2010, 41, 149–157. [Google Scholar] [CrossRef]
- Piirsoo, A.; Piirsoo, M.; Kala, M.; Sankovski, E.; Lototskaja, E.; Levin, V.; Salvi, M.; Ustav, M. Activity of CK2alpha protein kinase is required for efficient replication of some HPV types. PLoS Pathog. 2019, 15, e1007788. [Google Scholar] [CrossRef] [PubMed]
- Koffa, M.D.; Kean, J.; Zachos, G.; Rice, S.A.; Clements, J.B. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J. Virol. 2003, 77, 4315–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Marrero, M.C.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; El-Guindy, A.; Countryman, J.; Ye, J.; Gradoville, L. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res.. 2007, 97, 81–109. [Google Scholar]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.; Gutkind, J.S. Human tumor-associated viruses and new insights into the molecular mechanisms of cancer. Oncogene 2008, 27 (Suppl. S2), S31–S42. [Google Scholar] [CrossRef] [Green Version]
- Ooka, T. The molecular biology of Epstein-Barr virus. Biomed. Pharm. 1985, 39, 59–66. [Google Scholar]
- Amon, W.; Farrell, P.J. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 2005, 15, 149–156. [Google Scholar] [CrossRef]
- Khan, G.; Miyashita, E.M.; Yang, B.; Babcock, G.J.; Thorley-Lawson, D.A. Is EBV persistence in vivo a model for B cell homeostasis? Immunity 1996, 5, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, J.B.; Weinberg, W.; Johnson, R.; Yuspa, S.; Levine, A.J. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 1990, 61, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Kulwichit, W.; Edwards, R.H.; Davenport, E.M.; Baskar, J.F.; Godfrey, V.; Raab-Traub, N. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 1998, 95, 11963–11968. [Google Scholar] [CrossRef] [Green Version]
- Curran, J.A.; Laverty, F.S.; Campbell, D.; Macdiarmid, J.; Wilson, J.B. Epstein-Barr virus encoded latent membrane protein-1 induces epithelial cell proliferation and sensitizes transgenic mice to chemical carcinogenesis. Cancer Res. 2001, 61, 6730–6738. [Google Scholar]
- Mainou, B.A.; Raab-Traub, N. LMP1 strain variants: Biological and molecular properties. J. Virol. 2006, 80, 6458–6468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, N.; Sugden, B. CD40 and its viral mimic, LMP1: Similar means to different ends. Cell. Signal. 2003, 15, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, R.G.; Wilson, J.B.; Anderson, S.J.; Longnecker, R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998, 9, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, R.G.; Brown, R.C.; Longnecker, R. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J. Virol. 2000, 74, 1101–1113. [Google Scholar] [CrossRef] [Green Version]
- Thornburg, N.J.; Kulwichit, W.; Edwards, R.H.; Shair, K.H.; Bendt, K.M.; Raab-Traub, N. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene 2006, 25, 288–297. [Google Scholar] [CrossRef] [Green Version]
- Everly, D.N., Jr.; Kusano, S.; Raab-Traub, N. Accumulation of cytoplasmic beta-catenin and nuclear glycogen synthase kinase 3beta in Epstein-Barr virus-infected cells. J. Virol. 2004, 78, 11648–11655. [Google Scholar] [CrossRef] [Green Version]
- Li, S.S.; Yang, S.; Wang, S.; Yang, X.M.; Tang, Q.L.; Wang, S.H. Latent membrane protein 1 mediates the resistance of nasopharyngeal carcinoma cells to TRAIL-induced apoptosis by activation of the PI3K/Akt signaling pathway. Oncol. Rep. 2011, 26, 1573–1579. [Google Scholar]
- Gires, O.; Kohlhuber, F.; Kilger, E.; Baumann, M.; Kieser, A.; Kaiser, C.; Zeidler, R.; Scheffer, B.; Ueffing, M.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999, 18, 3064–3073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.D.; Tsai, M.H.; Romero-Masters, J.C.; Ranheim, E.A.; Huebner, S.M.; Bristol, J.A.; Delecluse, H.J.; Kenney, S.C. Latent Membrane Protein 1 (LMP1) and LMP2A Collaborate to Promote Epstein-Barr Virus-Induced B Cell Lymphomas in a Cord Blood-Humanized Mouse Model but Are Not Essential. J. Virol. 2017, 91, e01928-16. [Google Scholar] [CrossRef] [Green Version]
- Skalsky, R.L.; Cullen, B.R. EBV Noncoding RNAs. Curr. Top Microbiol. Immunol. 2015, 391, 181–217. [Google Scholar] [PubMed] [Green Version]
- Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.A.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; Freeman, C.; Hunt, S.E.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Thorley-Lawson, D.A. EBV Persistence–Introducing the Virus. Curr. Top Microbiol. Immunol. 2015, 390, 151–209. [Google Scholar]
- Thacker, E.L.; Mirzaei, F.; Ascherio, A. Infectious mononucleosis and risk for multiple sclerosis: A meta-analysis. Ann. Neurol. 2006, 59, 499–503. [Google Scholar] [CrossRef]
- Wang, Z.; Kennedy, P.G.; Dupree, C.; Wang, M.; Lee, C.; Pointon, T.; Langford, T.D.; Graner, M.W.; Yu, X. Antibodies from Multiple Sclerosis Brain Identified Epstein-Barr Virus Nuclear Antigen 1 & 2 Epitopes which Are Recognized by Oligoclonal Bands. J. Neuroimmune Pharm. 2021, 16, 567–580. [Google Scholar]
- Lanz, T.V.; Brewer, R.C.; Ho, P.P.; Moon, J.-S.; Jude, K.M.; Fernandez, D.; Fernandes, R.A.; Gomez, A.M.; Nadj, G.-S.; Bartley, C.M.; et al. Clonally Expanded B Cells in Multiple Sclerosis Bind EBV EBNA1 and GlialCAM. Nature 2022, 603, 321–327. [Google Scholar] [CrossRef]
- Jacobs, B.M.; Giovannoni, G.; Cuzick, J.; Dobson, R. Systematic review and meta-analysis of the association between Epstein-Barr virus, multiple sclerosis and other risk factors. Mult. Scler. 2020, 26, 1281–1297. [Google Scholar] [CrossRef]
- Jeon, S.M.; Shin, E.A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lung, M.L.; Cheung, A.K.; Ko, J.M.; Lung, H.L.; Cheng, Y.; Dai, W. The interplay of host genetic factors and Epstein-Barr virus in the development of nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.J.; Feederle, R.; O’Sullivan, B.; Taniere, P. Epstein–Barr virus-associated tumours: An update for the attention of the working pathologist. J. Clin. Pathol. 2007, 60, 1358–1364. [Google Scholar] [CrossRef]
- Vockerodt, M.; Yap, L.F.; Shannon-Lowe, C.; Curley, H.; Wei, W.; Vrzalikova, K.; Murray, P.G. The Epstein-Barr virus and the pathogenesis of lymphoma. J. Pathol. 2015, 235, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Kempkes, B.; Robertson, E.S. Epstein-Barr virus latency: Current and future perspectives. Curr. Opin. Virol. 2015, 14, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, J.B.; Bell, J.L.; Levine, A.J. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 1996, 15, 3117–3126. [Google Scholar] [CrossRef]
- Kang, M.S.; Soni, V.; Bronson, R.; Kieff, E. Epstein-Barr virus nuclear antigen 1 does not cause lymphoma in C57BL/6J mice. J. Virol. 2008, 82, 4180–4183. [Google Scholar] [CrossRef] [Green Version]
- Nanbo, A.; Inoue, K.; Adachi-Takasawa, K.; Takada, K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002, 21, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Ruf, I.K.; Lackey, K.A.; Warudkar, S.; Sample, J.T. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J. Virol. 2005, 79, 14562–14569. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Feederle, R.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Bencun, M.; Cullen, B.R.; Delecluse, H.J. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011, 7, e1001294. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yu, F.; Wu, W.; Wang, Y.; Ding, H.; Qian, L. Epstein-Barr virus-encoded microRNAs as regulators in host immune responses. Int. J. Biol. Sci. 2018, 14, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Pang, M.F.; Lin, K.W.; Peh, S.C. The signaling pathways of Epstein-Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell. Mol. Biol. Lett. 2009, 14, 222–247. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Han, C.; Wang, L.; Zhang, X.; He, X.; Lu, X. Targeting tumor suppressor genes for cancer therapy. BioEssays 2015, 37, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Leight, E.R.; Sugden, B. EBNA-1: A protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 2000, 10, 83–100. [Google Scholar] [CrossRef]
- Raab-Traub, N. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 2002, 12, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Sivachandran, N.; Dawson, C.W.; Young, L.S.; Liu, F.F.; Middeldorp, J.; Frappier, L. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J. Virol. 2012, 86, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Sivachandran, N.; Sarkari, F.; Frappier, L. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008, 4, e1000170. [Google Scholar] [CrossRef] [Green Version]
- Holowaty, M.N.; Zeghouf, M.; Wu, H.; Tellam, J.; Athanasopoulos, V.; Greenblatt, J.; Frappier, L. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 2003, 278, 29987–29994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, L.M.; Yu, J.S.; Chang, Y.S. Identification of protein kinase CK2 as a potent kinase of Epstein-Barr virus latent membrane protein 1. Biochem. Biophys. Res. Commun. 2002, 294, 586–591. [Google Scholar] [CrossRef]
- Cao, J.Y.; Shire, K.; Landry, C.; Gish, G.D.; Pawson, T.; Frappier, L. Identification of a novel protein interaction motif in the regulatory subunit of casein kinase 2. Mol. Cell. Biol. 2014, 34, 246–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frappier, L.; O’Donnell, M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J. Biol. Chem. 1991, 266, 7819–7826. [Google Scholar] [CrossRef]
- Duellman, S.J.; Thompson, K.L.; Coon, J.J.; Burgess, R.R. Phosphorylation sites of Epstein-Barr virus EBNA1 regulate its function. J. Gen. Virol. 2009, 90, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Frappier, L. Viral disruption of promyelocytic leukemia (PML) nuclear bodies by hijacking host PML regulators. Virulence 2011, 2, 58–62. [Google Scholar] [CrossRef] [Green Version]
- Scaglioni, P.P.; Yung, T.M.; Cai, L.F.; Erdjument-Bromage, H.; Kaufman, A.J.; Singh, B.; Teruya-Feldstein, J.; Tempst, P.; Pandolfi, P.P. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 2006, 126, 269–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaglioni, P.P.; Yung, T.M.; Choi, S.C.; Baldini, C.; Konstantinidou, G.; Pandolfi, P.P. CK2 mediates phosphorylation and ubiquitin-mediated degradation of the PML tumor suppressor. Mol. Cell. Biochem. 2008, 316, 149–154. [Google Scholar] [CrossRef]
- Kang, M.S.; Lee, E.K.; Soni, V.; Lewis, T.A.; Koehler, A.N.; Srinivasan, V.; Kieff, E. Roscovitine inhibits EBNA1 serine 393 phosphorylation, nuclear localization, transcription, and episome maintenance. J. Virol. 2011, 85, 2859–2868. [Google Scholar] [CrossRef] [Green Version]
- Shire, K.; Kapoor, P.; Jiang, K.; Hing, M.N.; Sivachandran, N.; Nguyen, T.; Frappier, L. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 2006, 80, 5261–5272. [Google Scholar] [CrossRef] [Green Version]
- Grässer, F.A.; Gottel, S.; Haiss, P.; Boldyreff, B.; Issinger, O.G.; Mueller-Lantzsch, N. Phosphorylation of the Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 1992, 186, 1694–1701. [Google Scholar] [CrossRef]
- Gross, H.; Hennard, C.; Masouris, I.; Cassel, C.; Barth, S.; Stober-Grässer, U.; Mamiani, A.; Moritz, B.; Ostareck, D.; Ostareck-Lederer, A.; et al. Binding of the heterogeneous ribonucleoprotein K (hnRNP K) to the Epstein-Barr virus nuclear antigen 2 (EBNA2) enhances viral LMP2A expression. PLoS ONE 2012, 7, e42106. [Google Scholar] [CrossRef]
- Shimada, K.; Kondo, K.; Yamanishi, K. Human herpesvirus 6 immediate-early 2 protein interacts with heterogeneous ribonucleoprotein K and casein kinase 2. Microbiol. Immunol. 2004, 48, 205–210. [Google Scholar] [CrossRef]
- Wadd, S.; Bryant, H.; Filhol, O.; Scott, J.E.; Hsieh, T.Y.; Everett, R.D.; Clements, J.B. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 1999, 274, 28991–28998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, H.E.; Matthews, D.A.; Wadd, S.; Scott, J.E.; Kean, J.; Graham, S.; Russell, W.C.; Clements, J.B. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J. Virol. 2000, 74, 11322–11328. [Google Scholar] [CrossRef] [Green Version]
- Barth, S.; Liss, M.; Voss, M.D.; Dobner, T.; Fischer, U.; Meister, G.; Grasser, F.A. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine-glycine repeat to the survival motor neuron protein. J. Virol. 2003, 77, 5008–5013. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.D.; Cheng, C.P.; Fang, J.S.; Chen, L.C.; Zhao, B.; Kieff, E.; Peng, C.W. Modulation of Epstein-Barr virus nuclear antigen 2-dependent transcription by protein arginine methyltransferase 5. Biochem. Biophys. Res. Commun. 2013, 430, 1097–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, H.; Barth, S.; Palermo, R.D.; Mamiani, A.; Hennard, C.; Zimber-Strobl, U.; West, M.J.; Kremmer, E.; Grasser, F.A. Asymmetric arginine dimethylation of Epstein-Barr virus nuclear antigen 2 promotes DNA targeting. Virology 2010, 397, 299–310. [Google Scholar] [CrossRef] [Green Version]
- Kaye, K.M.; Izumi, K.M.; Kieff, E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 1993, 90, 9150–9154. [Google Scholar] [CrossRef] [Green Version]
- Kieser, A.; Sterz, K.R. The Latent Membrane Protein 1 (LMP1). Curr. Top Microbiol. Immunol. 2015, 391, 119–149. [Google Scholar]
- Baichwal, V.R.; Sugden, B. Posttranslational processing of an Epstein-Barr virus-encoded membrane protein expressed in cells transformed by Epstein-Barr virus. J. Virol. 1987, 61, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Diduk, S.V.; Smirnova, K.V.; Pavlish, O.A.; Gurtsevitch, V.E. Functionally significant mutations in the Epstein-Barr virus LMP1 gene and their role in activation of cell signaling pathways. Biochemistry 2008, 73, 1134–1139. [Google Scholar] [CrossRef]
- Mainou, B.A.; Everly, D.N., Jr.; Raab-Traub, N. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J. Virol. 2007, 81, 9680–9692. [Google Scholar] [CrossRef] [Green Version]
- Chien, K.Y.; Chang, Y.S.; Yu, J.S.; Fan, L.W.; Lee, C.W.; Chi, L.M. Identification of a new in vivo phosphorylation site in the cytoplasmic carboxyl terminus of EBV-LMP1 by tandem mass spectrometry. Biochem. Biophys. Res. Commun. 2006, 348, 47–55. [Google Scholar] [CrossRef] [PubMed]
- St Denis, N.; Gabriel, M.; Turowec, J.P.; Gloor, G.B.; Li, S.S.; Gingras, A.C.; Litchfield, D.W. Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J. Proteom. 2014, 118, 49–62. [Google Scholar] [CrossRef] [Green Version]
- El-Guindy, A.S.; Miller, G. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 2004, 78, 7634–7644. [Google Scholar] [CrossRef] [Green Version]
- El-Guindy, A.S.; Paek, S.Y.; Countryman, J.; Miller, G. Identification of constitutive phosphorylation sites on the Epstein-Barr virus ZEBRA protein. J. Biol. Chem. 2006, 281, 3085–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolman, J.L.; Taylor, N.; Marshak, D.R.; Miller, G. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc. Natl. Acad. Sci. USA 1993, 90, 10115–10119. [Google Scholar] [CrossRef] [Green Version]
- Gruffat, H.; Batisse, J.; Pich, D.; Neuhierl, B.; Manet, E.; Hammerschmidt, W.; Sergeant, A. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 2002, 76, 9635–9644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, I.D.; Shanahan, F.; Farrell, P.J. Epstein-Barr virus SM protein. Virology 1994, 205, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Medina-Palazon, C.; Gruffat, H.; Mure, F.; Filhol, O.; Vingtdeux-Didier, V.; Drobecq, H.; Cochet, C.; Sergeant, N.; Sergeant, A.; Manet, E. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J. Virol. 2007, 81, 11850–11860. [Google Scholar] [CrossRef] [Green Version]
- Sergeant, A.; Gruffat, H.; Manet, E. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 2008, 13, 3798–3813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, L.B.; Ward, A.C. The Ikaros gene family: Transcriptional regulators of hematopoiesis and immunity. Mol. Immunol. 2011, 48, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Dovat, E.; Song, C.; Hu, T.; Rahman, M.; Dhanyamraju, P.; Klink, M.; Bogush, D.; Soliman, M.; Kane, S.; McGrath, M.; et al. Transcriptional Regulation of PIK3CD and PIKFYVE in T-Cell Acute Lymphoblastic Leukemia by IKAROS and Protein Kinase CK2. Int. J. Mol. Sci. 2021, 22, 819. [Google Scholar] [CrossRef]
- Klink, M.; Rahman, M.; Song, C.; Dhanyamraju, P.; Ehudin, M.; Ding, Y.; Steffens, S.; Bhadauria, P.; Iyer, S.; Aliaga, C.; et al. Mechanistic Basis for In Vivo Therapeutic Efficacy of CK2 Inhibitor CX-4945 in Acute Myeloid Leukemia. Cancers 2021, 13, 1127. [Google Scholar] [CrossRef] [PubMed]
- Uckun, F.M.; Ma, H.; Zhang, J.; Ozer, Z.; Dovat, S.; Mao, C.; Ishkhanian, R.; Goodman, P.; Qazi, S. Serine phosphorylation by SYK is critical for nuclear localization and transcription factor function of Ikaros. Proc. Natl. Acad. Sci. USA 2012, 109, 18072–18077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-del, A.P.; Koipally, J.; Georgopoulos, K. Ikaros SUMOylation: Switching out of repression. Mol. Cell. Biol. 2005, 25, 2688–2697. [Google Scholar] [CrossRef] [Green Version]
- Popescu, M.; Gurel, Z.; Ronni, T.; Song, C.; Hung, K.Y.; Payne, K.J.; Dovat, S. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J. Biol. Chem. 2009, 284, 13869–13880. [Google Scholar] [CrossRef] [Green Version]
- Dovat, S.; Song, C.; Payne, K.J.; Li, Z. Ikaros, CK2 kinase, and the road to leukemia. Mol. Cell. Biochem. 2011, 356, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Li, Z.; Erbe, A.K.; Savic, A.; Dovat, S. Regulation of Ikaros function by casein kinase 2 and protein phosphatase 1. World J. Biol. Chem. 2011, 2, 126–131. [Google Scholar] [CrossRef]
- Gurel, Z.; Ronni, T.; Ho, S.; Kuchar, J.; Payne, K.J.; Turk, C.W.; Dovat, S. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J. Biol. Chem. 2008, 283, 8291–8300. [Google Scholar] [CrossRef] [Green Version]
- Bogush, D.; Schramm, J.; Ding, Y.; He, B.; Singh, C.; Sharma, A.; Tukaramrao, D.B.; Iyer, S.; Desai, D.; Nalesnik, G.; et al. Signaling pathways and regulation of gene expression in hematopoietic cells. Adv. Biol. Regul. 2022, 88, 100942. [Google Scholar] [CrossRef]
- Iempridee, T.; Reusch, J.A.; Riching, A.; Johannsen, E.C.; Dovat, S.; Kenney, S.C.; Mertz, J.E. Epstein-Barr virus utilizes Ikaros in regulating its latent-lytic switch in B cells. J. Virol. 2014, 88, 4811–4827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqi, U.Z.; Vaidya, A.S.; Li, X.; Marcon, E.; Tsao, S.W.; Greenblatt, J.; Frappier, L. Identification of ARKL1 as a Negative Regulator of Epstein-Barr Virus Reactivation. J. Virol. 2019, 93, e00989-19. [Google Scholar] [CrossRef] [Green Version]
- Domingues, P.; Golebiowski, F.; Tatham, M.H.; Lopes, A.M.; Taggart, A.; Hay, R.T.; Hale, B.G. Global Reprogramming of Host SUMOylation during Influenza Virus Infection. Cell Rep. 2015, 13, 1467–1480. [Google Scholar] [CrossRef] [Green Version]
- Vernin, C.; Thenoz, M.; Pinatel, C.; Gessain, A.; Gout, O.; Delfau-Larue, M.-H.; Nazaret, N.; Legras-Lachuer, C.; Wattel, E.; Mortreux, F. HTLV-1 bZIP factor HBZ promotes cell proliferation and genetic instability by activating OncomiRs. Cancer Res. 2014, 74, 6082–6093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.L.; Lung, M.M.; Sham, J.S.; Choy, D.T.; Griffin, B.E.; Ng, M.H. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology 1992, 191, 193–201. [Google Scholar] [CrossRef]
- Hitt, M.M.; Allday, M.J.; Hara, T.; Karran, L.; Jones, M.D.; Busson, P.; Tursz, T.; Ernberg, I.; Griffin, B.E. EBV gene expression in an NPC-related tumour. EMBO J. 1989, 8, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Hoesel, B.; Schmid, J.A. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, I.; Sonenshein, G.E.; Seldin, D.C. CK2 and its role in Wnt and NF-kappaB signaling: Linking development and cancer. Cell Mol. Life Sci. 2009, 66, 1850–1857. [Google Scholar] [CrossRef]
- Verhoeven, R.J.; Tong, S.; Zhang, G.; Zong, J.; Chen, Y.; Jin, D.Y.; Chen, M.R.; Pan, J.; Chen, H. NF-kappaB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J. Virol. 2016, 90, 6475–6488. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Liu, Y.; Wang, C.; Gan, R. Signaling pathways of EBV-induced oncogenesis. Cancer Cell Int. 2021, 21, 93. [Google Scholar] [CrossRef]
- Romieu-Mourez, R.; Landesman-Bollag, E.; Seldin, D.C.; Sonenshein, G.E. Protein kinase CK2 promotes aberrant activation of nuclear factor-kappaB, transformed phenotype, and survival of breast cancer cells. Cancer Res. 2002, 62, 6770–6778. [Google Scholar] [PubMed]
- Kato, T., Jr.; Delhase, M.; Hoffmann, A.; Karin, M. CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol. Cell 2003, 12, 829–839. [Google Scholar] [CrossRef]
- Eddy, S.F.; Guo, S.; Demicco, E.G.; Romieu-Mourez, R.; Landesman-Bollag, E.; Seldin, D.C.; Sonenshein, G.E. Inducible IkappaB kinase/IkappaB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor-kappaB activation in breast cancer cells. Cancer Res. 2005, 65, 11375–11383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantome, A.; Pance, A.; Gauthier, N.; Vandroux, D.; Chenu, J.; Solary, E.; Jeannin, J.F.; Reveneau, S. Casein kinase II-mediated phosphorylation of NF-kappaB p65 subunit enhances inducible nitric-oxide synthase gene transcription in vivo. J. Biol. Chem. 2004, 279, 23953–23960. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Westerheide, S.D.; Hanson, J.L.; Baldwin, A.S., Jr. Tumor necrosis factor alpha-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 2000, 275, 32592–32597. [Google Scholar] [CrossRef] [Green Version]
- Parhar, K.; Morse, J.; Salh, B. The role of protein kinase CK2 in intestinal epithelial cell inflammatory signaling. Int. J. Color. Dis. 2007, 22, 601–609. [Google Scholar] [CrossRef]
- Yu, M.; Yeh, J.; Van, W.C. Protein kinase casein kinase 2 mediates inhibitor-kappaB kinase and aberrant nuclear factor-kappaB activation by serum factor(s) in head and neck squamous carcinoma cells. Cancer Res. 2006, 66, 6722–6731. [Google Scholar] [CrossRef] [Green Version]
- Lambert, S.L.; Martinez, O.M. Latent membrane protein 1 of EBV activates phosphatidylinositol 3-kinase to induce production of IL-10. J. Immunol. 2007, 179, 8225–8234. [Google Scholar] [CrossRef] [Green Version]
- Chen, J. Roles of the PI3K/Akt pathway in Epstein-Barr virus-induced cancers and therapeutic implications. World J. Virol. 2012, 1, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Fresno Vara, J.A.; Casado, E.; De Castro, J.; Cejas, P.; Belda-Iniesta, C.; Gonzalez-Baron, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Pulido, R. The Tumor Suppressor PTEN Is Phosphorylated by the Protein Kinase CK2 at Its C Terminus. Implications for pten stability to proteasome- mediated degradation. J. Biol. Chem. 2001, 276, 993–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.J.; Lou, D.Y.; Seldin, D.C.; Lane, W.S.; Neel, B.G. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002, 528, 145–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez, F.; Grossman, S.R.; Takahashi, Y.; Rokas, M.V.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001, 276, 48627–48630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Khouri, A.M.; Ma, Y.; Togo, S.H.; Williams, S.; Mustelin, T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 2005, 280, 35195–35202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccario, H.; Perera, N.M.; Davidson, L.; Downes, C.P.; Leslie, N.R. PTEN is destabilized by phosphorylation on Thr366. Biochem. J. 2007, 405, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Cordier, F.; Chaffotte, A.; Terrien, E.; Prehaud, C.; Theillet, F.X.; Delepierre, M.; Lafon, M.; Buc, H.; Wolff, N. Ordered phosphorylation events in two independent cascades of the PTEN C-tail revealed by NMR. J. Am. Chem. Soc. 2012, 134, 20533–20543. [Google Scholar] [CrossRef]
- Cai, L.M.; Lyu, X.M.; Luo, W.R.; Cui, X.F.; Ye, Y.F.; Yuan, C.C.; Peng, Q.X.; Wu, D.H.; Liu, T.-F.; Wang, E.; et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015, 34, 2156–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacPherson, M.R.; Molina, P.; Souchelnytskyi, S.; Wernstedt, C.; Martin-Perez, J.; Portillo, F.; Cano, A. Phosphorylation of serine 11 and serine 92 as new positive regulators of human Snail1 function: Potential involvement of casein kinase-2 and the cAMP-activated kinase protein kinase A. Mol. Biol. Cell 2010, 21, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.F.; Yang, G.-D.; Huang, T.-J.; Li, R.; Chu, Q.-Q.; Xu, L.; Wang, M.-S.; Cai, M.-D.; Zhong, L.; Wei, H.-J.; et al. EB-virus latent membrane protein 1 potentiates the stemness of nasopharyngeal carcinoma via preferential activation of PI3K/AKT pathway by a positive feedback loop. Oncogene 2016, 35, 3419–3431. [Google Scholar] [CrossRef]
- Ma, W.; Feng, L.; Zhang, S.; Zhang, H.; Zhang, X.; Qi, X.; Zhang, Y.; Feng, Q.; Xiang, T.; Zeng, Y. Induction of chemokine (C-C motif) ligand 5 by Epstein-Barr virus infection enhances tumor angiogenesis in nasopharyngeal carcinoma. Cancer Sci. 2018, 109, 1710–1722. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ren, J.; Li, G.; Moorman, J.P.; Yao, Z.Q.; Ning, S. LMP1 signaling pathway activates IRF4 in latent EBV infection and a positive circuit between PI3K and Src is required. Oncogene 2017, 36, 2265–2274. [Google Scholar] [CrossRef] [Green Version]
- Qing, L.Z.; Li, N.Y.; Li, L.; Shuang, W.; Yu, F.Y.; Yi, D.; Divakaran, J.; Xin, L.; Yan, Q.D. LMP1 antagonizes WNT/beta-catenin signalling through inhibition of WTX and promotes nasopharyngeal dysplasia but not tumourigenesis in LMP1(B95-8) transgenic mice. J. Pathol. 2011, 223, 574–583. [Google Scholar] [CrossRef]
- Song, D.H.; Dominguez, I.; Mizuno, J.; Kaut, M.; Mohr, S.C.; Seldin, D.C. CK2 phosphorylation of the armadillo repeat region of b-catenin potentiates Wnt signaling. J. Biol. Chem. 2003, 278, 24018–24025. [Google Scholar] [CrossRef] [Green Version]
- Song, D.H.; Sussman, D.J.; Seldin, D.C. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J. Biol. Chem. 2000, 275, 23790–23797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernatik, O.; Ganji, R.S.; Dijksterhuis, J.P.; Konik, P.; Cervenka, I.; Polonio, T.; Krejci, P.; Schulte, G.; Bryja, V. Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. J. Biol. Chem. 2011, 286, 10396–10410. [Google Scholar] [CrossRef] [Green Version]
- DiMaira, G.; Salvi, M.; Arrigoni, G.; Marin, O.; Sarno, S.; Brustolon, F.; Pinna, L.A.; Ruzzene, M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005, 12, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Ruzzene, M.; Bertacchini, J.; Toker, A.; Marmiroli, S. Cross-talk between the CK2 and AKT signaling pathways in cancer. Adv. Biol. Regul. 2017, 64, 1–8. [Google Scholar] [CrossRef]
- Zheng, Y.; Qin, H.; Frank, S.J.; Deng, L.; Litchfield, D.W.; Tefferi, A.; Pardanani, A.; Lin, F.-T.; Li, J.; Sha, B.; et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood 2011, 118, 156–166. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Karube, K.; Yamamoto, K.; Takizawa, J.; Tsuzuki, S.; Yatabe, Y.; Kanda, T.; Katayama, M.; Ozawa, Y.; Ishitsuka, K.; et al. Gene expression profiling of Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly reveals alterations of characteristic oncogenetic pathways. Cancer Sci. 2014, 105, 537–544. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.L.; Hu, D.S.; Deng, X.Y.; Cao, Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol. Immunol. 2007, 4, 185–196. [Google Scholar]
- Timofeeva, O.A.; Plisov, S.; Evseev, A.A.; Peng, S.; Jose-Kampfner, M.; Lovvorn, H.N.; Dome, J.S.; Perantoni, A.O. Serine-phosphorylated STAT1 is a prosurvival factor in Wilms’ tumor pathogenesis. Oncogene 2006, 25, 7555–7564. [Google Scholar] [CrossRef] [Green Version]
- Mandato, E.; Manni, S.; Zaffino, F.; Semenzato, G.; Piazza, F. Targeting CK2-driven non-oncogene addiction in B-cell tumors. Oncogene 2016, 35, 6045–6052. [Google Scholar] [CrossRef] [PubMed]
- Manni, S.; Brancalion, A.; Mandato, E.; Tubi, L.Q.; Colpo, A.; Pizzi, M.; Cappellesso, R.; Zaffino, F.; Di Maggio, S.A.; Cabrelle, A.; et al. Protein Kinase CK2 Inhibition Down Modulates the NF-kappaB and STAT3 Survival Pathways, Enhances the Cellular Proteotoxic Stress and Synergistically Boosts the Cytotoxic Effect of Bortezomib on Multiple Myeloma and Mantle Cell Lymphoma Cells. PLoS ONE 2013, 8, e75280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalathur, M.; Toso, A.; Chen, J.; Revandkar, A.; Danzer-Baltzer, C.; Guccini, I.; Alajati, A.; Sarti, M.; Pinton, S.; Brambilla, L.; et al. A chemogenomic screening identifies CK2 as a target for pro-senescence therapy in PTEN-deficient tumours. Nat. Commun. 2015, 6, 7227. [Google Scholar] [CrossRef]
- Jiang, L.; Sheikh, M.S.; Huang, Y. Decision Making by p53: Life versus Death. Mol. Cell Pharm. 2010, 2, 69–77. [Google Scholar]
- Michael-Michalovitz, D.; Yehiely, F.; Gottlieb, E.; Oren, M. Simian virus 40 can overcome the antiproliferative effect of wild-type p53 in the absence of stable large T antigen- p53 binding. J. Virol. 1991, 65, 4160–4168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Tsurumi, T. Genome guardian p53 and viral infections. Rev. Med. Virol. 2012, 23, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Krauer, K.G.; Burgess, A.; Buck, M.; Flanagan, J.; Sculley, T.B.; Gabrielli, B. The EBNA-3 gene family proteins disrupt the G2/M checkpoint. Oncogene 2004, 23, 1342–1353. [Google Scholar] [CrossRef] [Green Version]
- Wade, M.; Allday, M.J. Epstein-Barr virus suppresses a G(2)/M checkpoint activated by genotoxins. Mol. Cell. Biol. 2000, 20, 1344–1360. [Google Scholar] [CrossRef] [Green Version]
- Saridakis, V.; Sheng, Y.; Sarkari, F.; Holowaty, M.N.; Shire, K.; Nguyen, T.; Zhang, R.G.; Liao, J.; Lee, W.; Edwards, A.M.; et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 2005, 18, 25–36. [Google Scholar] [CrossRef]
- Liu, M.T.; Chang, Y.T.; Chen, S.C.; Chuang, Y.C.; Chen, Y.R.; Lin, C.S.; Chen, J.Y. Epstein-Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene 2005, 24, 2635–2646. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Guo, L.; Tao, Y.; Zhou, S.; Wang, Z.; Luo, W.; Hu, D.; Li, Z.; Xiao, L.; Tang, M.; et al. Latent membrane protein 1 of Epstein-Barr virus regulates p53 phosphorylation through MAP kinases. Cancer Lett. 2007, 255, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, S.; Chen, X.; Guo, L.; Li, Z.; Hu, D.; Luo, X.; Ma, X.; Tang, M.; Yi, W.; et al. The activation of p53 mediated by Epstein-Barr virus latent membrane protein 1 in SV40 large T-antigen transformed cells. FEBS Lett. 2008, 582, 755–762. [Google Scholar] [CrossRef] [Green Version]
- Husaini, R.; Ahmad, M.; Soo-Beng, K.A. Epstein-Barr virus Latent Membrane Protein LMP1 reduces p53 protein levels independent of the PI3K-Akt pathway. BMC Res. Notes 2011, 4, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maclaine, N.J.; Hupp, T.R. How phosphorylation controls p53. Cell Cycle 2011, 10, 916–921. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, A.; Herrmann, C.P.E.; Issinger, O.-G.; Montenarh, M. Phosphorylation of wild-type and mutant phenotypes of p53 by an associated protein kinase. Int. J. Oncol. 1992, 1, 571–580. [Google Scholar] [CrossRef]
- Herrmann, C.P.E.; Kraiss, S.; Montenarh, M. Association of casein kinase II with immunopurified p53. Oncogene 1991, 6, 877–884. [Google Scholar]
- Kraiss, S.; Barnekow, A.; Montenarh, M. Protein kinase activity associated with immunopurified p53 protein. Oncogene 1990, 5, 845–855. [Google Scholar] [PubMed]
- Achison, M.; Hupp, T.R. Hypoxia attenuates the p53 response to cellular damage. Oncogene 2003, 22, 3431–3440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, L.R.; Baladrón, I.; Rittoles, A.; Díaz, P.A.; Valenzuela, C.; Santana, R.; Vázquez, M.M.; García, A.; Chacón, D.; Thompson, D.; et al. Treatment with an Anti-CK2 Synthetic Peptide Improves Clinical Response in COVID-19 Patients with Pneumonia. A Randomized and Controlled Clinical Trial. ACS Pharm. Transl. Sci. 2021, 4, 206–212. [Google Scholar] [CrossRef] [PubMed]



| Gene(s)/Protein(s) Expressed in EBV Latency Patterns | ||||||||
|---|---|---|---|---|---|---|---|---|
| Latency Type | EBERs | BART miRNAs | EBNA1 | LMP1/2 | EBNA2-6 | BHRF1 BHRF1-miRNAs | v-snoRNA | Cell Type/ Tumor Type |
| 0 | + | + | Memory B-cell | |||||
| I | + | + | + | BL/GC B-cell | ||||
| II | + | + | + | + | HD/NPC/DLBCL | |||
| III | + | + | + | + | + | + | + | PTLD/LCLs |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montenarh, M.; Grässer, F.A.; Götz, C. Protein Kinase CK2 and Epstein–Barr Virus. Biomedicines 2023, 11, 358. https://doi.org/10.3390/biomedicines11020358
Montenarh M, Grässer FA, Götz C. Protein Kinase CK2 and Epstein–Barr Virus. Biomedicines. 2023; 11(2):358. https://doi.org/10.3390/biomedicines11020358
Chicago/Turabian StyleMontenarh, Mathias, Friedrich A. Grässer, and Claudia Götz. 2023. "Protein Kinase CK2 and Epstein–Barr Virus" Biomedicines 11, no. 2: 358. https://doi.org/10.3390/biomedicines11020358
APA StyleMontenarh, M., Grässer, F. A., & Götz, C. (2023). Protein Kinase CK2 and Epstein–Barr Virus. Biomedicines, 11(2), 358. https://doi.org/10.3390/biomedicines11020358






