A Proteome-Wide Effect of PHF8 Knockdown on Cortical Neurons Shows Downregulation of Parkinson’s Disease-Associated Protein Alpha-Synuclein and Its Interactors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Primary Culture of Cortical Neurons
2.3. Pharmacological Stimulation of Neural Network Activity
2.4. Cell Lysate Preparation and RNA/Protein Extraction
2.5. Isobaric Tag for Relative and Absolute Quantification of Proteins (iTRAQ)
2.6. Immunofluorescence
2.7. Widefield Imaging Microscopy
2.8. Overexpression of PHF8-YFP and Knockdown of PHF8 Levels Using shRNA and siRNA Transfection
3. Results
3.1. Relative Quantitation of the Activity-Regulated Cortical Neuronal Proteome
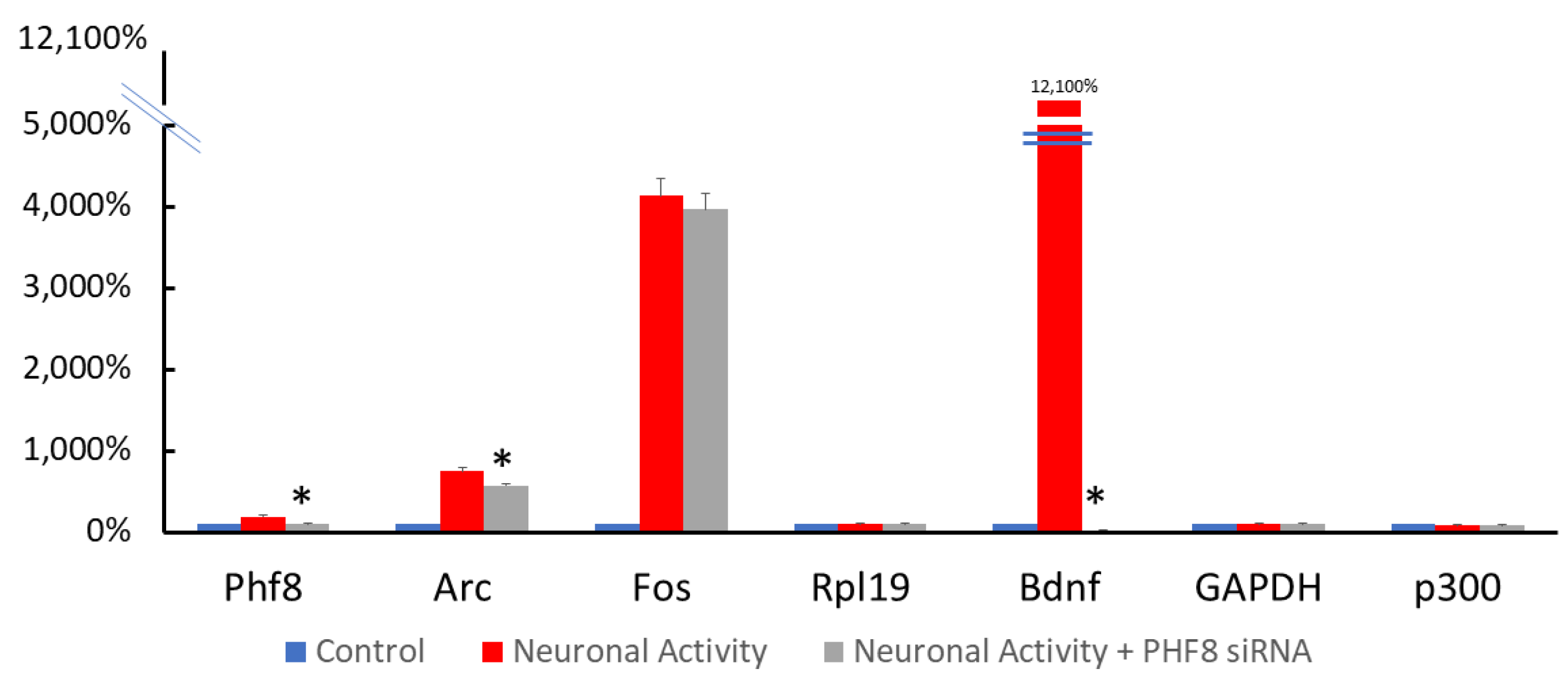
3.2. RNA Interference Specifically Depleted PHF8 in Primary Rat Cortical Neurons
3.3. Validation of Alpha-Synuclein as a Synaptic Protein Regulated by PHF8
3.4. Pathway Analysis Using DAVID Reveals Specific Downregulation of Proteins in PHF8 Knockdown That Are Involved in Synaptic Function
4. Discussion
4.1. The Potential Role of PHF8 in Synaptic Plasticity and PD Pathogenesis
4.2. Knockdown of PHF8 Reduces Levels of Alpha-Synuclein
4.3. Knockdown of PHF8 Reduces the Levels of Other Synaptic Plasticity-Related Proteins and SNCA Interactors
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Surguchov, A. Biomarkers in Parkinson’s Disease. In Neurodegenerative Diseases Biomarkers: Towards Translating Research to Clinical Practice; Peplow, P.V., Martinez, B., Gennarelli, T.A., Eds.; Springer: New York, NY, USA, 2022; pp. 155–180. [Google Scholar]
- Picconi, B.; Piccoli, G.; Calabresi, P. Synaptic Dysfunction in Parkinson’s Disease. In Synaptic Plasticity; part of the Advances in Experimental Medicine and Biology Book Series; Springer: Vienna, Austria, 2012; Volume 970, pp. 553–572. [Google Scholar] [CrossRef]
- Franklin, T.B.; Mansuy, I.M. The involvement of epigenetic defects in mental retardation. Neurobiol. Learn. Mem. 2011, 96, 61–67. [Google Scholar] [CrossRef] [PubMed]
- George, G.; Singh, S.; Lokappa, S.B.; Varkey, J. Gene co-expression network analysis for identifying genetic markers in Parkinson’s disease—A three-way comparative approach. Genomics 2018, 111, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Oey, N.E.; Leung, H.W.; Ezhilarasan, R.; Zhou, L.; Beuerman, R.W.; VanDongen, H.M.; VanDongen, A.M. A Neuronal Activity-Dependent Dual Function Chromatin-Modifying Complex Regulates Arc Expression. Eneuro 2015, 2. [Google Scholar] [CrossRef]
- Shepherd, J.; Bear, M.F. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011, 14, 279–284. [Google Scholar] [CrossRef]
- Janezic, S.; Threlfell, S.; Dodson, P.D.; Dowie, M.J.; Taylor, T.N.; Potgieter, D.; Parkkinen, L.; Senior, S.L.; Anwar, S.; Ryan, B.; et al. Deficits in dopaminergic transmission precede neuron loss and dysfunction in a new Parkinson model. Proc. Natl. Acad. Sci. USA 2013, 110, E4016–E4025. [Google Scholar] [CrossRef] [PubMed]
- Calo, L.; Węgrzynowicz, M.; Santivañez-Perez, J.; Spillantini, M.G. Synaptic failure and α-synuclein. Mov. Disord. 2016, 31, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Bridi, J.C.; Hirth, F. Mechanisms of α-Synuclein Induced Synaptopathy in Parkinson’s Disease. Front. Neurosci. 2018, 12, 80. [Google Scholar] [CrossRef]
- Nguyen, M.; Wong, Y.C.; Ysselstein, D.; Severino, A.; Krainc, D. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci. 2018, 42, 140–149. [Google Scholar] [CrossRef]
- Benskey, M.J.; Perez, R.G.; Manfredsson, F.P. The contribution of alpha synuclein to neuronal survival and function—Implications for Parkinson’s disease. J. Neurochem. 2016, 137, 331–359. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Xie, C.; Wang, Q.; Liu, Z. Interactions of CaMKII with dopamine D2 receptors: Roles in levodopa-induced dyskinesia in 6-hydroxydopamine lesioned Parkinson’s rats. Sci. Rep. 2014, 4, 6811. [Google Scholar] [CrossRef]
- Basso, M.; Giraudo, S.; Corpillo, D.; Bergamasco, B.; Lopiano, L.; Fasano, M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics 2004, 4, 3943–3952. [Google Scholar] [CrossRef] [PubMed]
- Gispert, S.; Kurz, A.; Brehm, N.; Rau, K.; Walter, M.; Riess, O.; Auburger, G. Complexin-1 and Foxp1 Expression Changes Are Novel Brain Effects of Alpha-Synuclein Pathology. Mol. Neurobiol. 2014, 52, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Hook, P.W.; McClymont, S.A.; Cannon, G.H.; Law, W.D.; Morton, A.J.; Goff, L.A.; McCallion, A.S. Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am. J. Hum. Genet. 2018, 102, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Zhang, X.; Zou, S.-C.; Zhu, Y.; Li, B.; Kuang, W.-P.; Guo, Y.; Li, X.-S.; Li, L.; Wang, X.-Y. A transcriptome-wide association study identifies susceptibility genes for Parkinson’s disease. npj Park. Dis. 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Roosen, D.A.; Blauwendraat, C.; Cookson, M.R.; Lewis, P.A. DNAJC proteins and pathways to parkinsonism. FEBS J. 2019, 286, 3080–3094. [Google Scholar] [CrossRef] [PubMed]
- Quadri, M.; Fang, M.; Picillo, M.; Olgiati, S.; Breedveld, G.J.; Graafland, J.; Wu, B.; Xu, F.; Erro, R.; Amboni, M.; et al. Mutation in the SYNJ1 Gene Associated with Autosomal Recessive, Early-Onset Parkinsonism. Hum. Mutat. 2013, 34, 1208–1215. [Google Scholar] [CrossRef]
- Zou, Z.; Ohta, T.; Miura, F.; Oki, S. ChIP-Atlas 2021 update: A data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022, 50, W175–W182. [Google Scholar] [CrossRef]
- Picconi, B.; Gardoni, F.; Centonze, D.; Mauceri, D.; Cenci, M.A.; Bernardi, G.; Calabresi, P.; Di Luca, M. Abnormal Ca2+-Calmodulin-Dependent Protein Kinase II Function Mediates Synaptic and Motor Deficits in Experimental Parkinsonism. J. Neurosci. 2004, 24, 5283–5291. [Google Scholar] [CrossRef]
- Peters, M.; Mizuno, K.; Ris, L.; Angelo, M.; Godaux, E.; Giese, K.P. Loss of Ca2+/calmodulin kinase kinase beta affects the formation of some, but not all, types of hippocampus-dependent long-term memory. J. Neurosci. 2003, 23, 9752–9760. [Google Scholar] [CrossRef] [Green Version]
- Benowitz, L.I.; Routtenberg, A. GAP-43: An intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997, 20, 84–91. [Google Scholar] [CrossRef]
- Van De Ven, T.J.; Vandongen, H.M.A.; Vandongen, A.M.J. The Nonkinase Phorbol Ester Receptor α1-Chimerin Binds the NMDA Receptor NR2A Subunit and Regulates Dendritic Spine Density. J. Neurosci. 2005, 25, 9488–9496. [Google Scholar] [CrossRef] [PubMed]
- Ecapurro, A.; Bodea, L.-G.; Eschaefer, P.; Eluthi-Carter, R.; Perreau, V.M. Computational deconvolution of genome wide expression data from Parkinson’s and Huntington’s disease brain tissues using population-specific expression analysis. Front. Neurosci. 2015, 8, 441. [Google Scholar] [CrossRef]
- Surmeier, D.J. Calcium, ageing, and neuronal vulnerability in Parkinson’s disease. Lancet Neurol. 2007, 6, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef]
- Nuytemans, K.; Theuns, J.; Cruts, M.; Van Broeckhoven, C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Hum. Mutat. 2010, 31, 763–780. [Google Scholar] [CrossRef]
- Cesca, F.; Baldelli, P.; Valtorta, F.; Benfenati, F. The synapsins: Key actors of synapse function and plasticity. Prog. Neurobiol. 2010, 91, 313–348. [Google Scholar] [CrossRef]
- Lüthi, A.; Laurent, J.-P.; Figurovt, A.; Mullert, D.; Schachnert, M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature 1994, 372, 777–779. [Google Scholar] [CrossRef]
- Paratcha, G.; Ledda, F.; Ibáñez, C.F. The Neural Cell Adhesion Molecule NCAM Is an Alternative Signaling Receptor for GDNF Family Ligands. Cell 2003, 113, 867–879. [Google Scholar] [CrossRef]
- D’Andrea, M.R.; Ilyin, S.; Plata-Salaman, C.R. Abnormal patterns of microtubule-associated protein-2 (MAP-2) immuno-labeling in neuronal nuclei and Lewy bodies in Parkinson’s disease substantia nigra brain tissues. Neurosci. Lett. 2001, 306, 137–140. [Google Scholar] [CrossRef]
- Mandelkow, E.M.; Mandelkow, E. Tau in Alzheimer’s disease. Trends Cell Biol. 1998, 8, 425–427. [Google Scholar]
- Zhang, Y.Q.; Bailey, A.M.; Matthies, H.J.; Renden, R.B.; Smith, M.A.; Speese, S.D.; Rubin, G.M.; Broadie, K. Drosophila Fragile X-Related Gene Regulates the MAP1B Homolog Futsch to Control Synaptic Structure and Function. Cell 2001, 107, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Riederer, B.M. Microtubule-associated protein 1B, a growth-associated and phosphorylated scaffold protein. Brain Res. Bull. 2007, 71, 541–558. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, I.; Caroni, P.; Kolkova, K.; Berezin, V.; Bock, E.; Walmod, P.S. Characterization of BASP1-mediated neurite outgrowth. J. Neurosci. Res. 2008, 86, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Koch, I.; Schwarz, H.; Beuchle, D.; Goellner, B.; Langegger, M.; Aberle, H. Drosophila Ankyrin 2 Is Required for Synaptic Stability. Neuron 2008, 58, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Vits, L.; Van Camp, G.; Coucke, P.; Fransen, E.; De Boulle, K.; Reyniers, E.; Korn, B.; Poustka, A.; Wilson, G.; Schrander-Stumpel, C.; et al. MASA syndrome is due to mutations in the neural cell adhesion gene L1CAM. Nat. Genet. 1994, 7, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Umemori, H.; Sanes, J.R. Signal Regulatory Proteins (SIRPS) Are Secreted Presynaptic Organizing Molecules. J. Biol. Chem. 2008, 283, 34053–34061. [Google Scholar] [CrossRef]
- Toth, A.B.; Terauchi, A.; Zhang, L.Y.; Johnson-Venkatesh, E.M.; Larsen, D.J.; Sutton, M.; Umemori, H. Synapse maturation by activity-dependent ectodomain shedding of SIRPα. Nat. Neurosci. 2013, 16, 1417–1425. [Google Scholar] [CrossRef]
- Ting, J.T.; Peça, J.; Feng, G. Functional Consequences of Mutations in Postsynaptic Scaffolding Proteins and Relevance to Psychiatric Disorders. Annu. Rev. Neurosci. 2012, 35, 49–71. [Google Scholar] [CrossRef]
- Chang, D.; Nalls, M.A.; Hallgrímsdóttir, I.B.; Hunkapiller, J.; Van Der Brug, M.; Cai, F.; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team; Kerchner, G.A.; Ayalon, G.; et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017, 49, 1511–1516. [Google Scholar] [CrossRef]
- Song, L.; He, Y.; Ou, J.; Zhao, Y.; Li, R.; Cheng, J.; Lin, C.-H.; Ho, M.S. Auxilin Underlies Progressive Locomotor Deficits and Dopaminergic Neuron Loss in a Drosophila Model of Parkinson’s Disease. Cell Rep. 2017, 18, 1132–1143. [Google Scholar] [CrossRef] [Green Version]
- Chandra, S.; Fornai, F.; Kwon, H.-B.; Yazdani, U.; Atasoy, D.; Liu, X.; Hammer, R.E.; Battaglia, G.; German, D.C.; Castillo, P.E.; et al. Double-knockout mice for α- and β-synucleins: Effect on synaptic functions. Proc. Natl. Acad. Sci. USA 2004, 101, 14966–14971. [Google Scholar] [CrossRef] [PubMed]
- Dalfó, E.; Barrachina, M.; Rosa, J.; Ambrosio, S.; Ferrer, I. Abnormal α-synuclein interactions with rab3a and rabphilin in diffuse Lewy body disease. Neurobiol. Dis. 2004, 16, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Elferink, L.A.; Anzai, K.; Scheller, R.H. rab15, a novel low molecular weight GTP-binding protein specifically expressed in rat brain. J. Biol. Chem. 1992, 267, 5768–5775. [Google Scholar] [CrossRef]
- Van Vlijmen, T.; Vleugel, M.; Evers, M.; Mohammed, S.; Wulf, P.S.; Heck, A.J.; Hoogenraad, C.; Van Der Sluijs, P. A unique residue in rab3c determines the interaction with novel binding protein Zwint-1. FEBS Lett. 2008, 582, 2838–2842. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.M.; Khvotchev, M.; Trauterman, B.; Kavalali, E.T. Vti1a Identifies a Vesicle Pool that Preferentially Recycles at Rest and Maintains Spontaneous Neurotransmission. Neuron 2012, 73, 121–134. [Google Scholar] [CrossRef]
- Karim, S.; Mirza, Z.; Ansari, S.; Rasool, M.; Iqbal, Z.; Sohrab, S.; Kamal, M.; Abuzenadah, A.; Al-Qahtani, M. Transcriptomics Study of Neurodegenerative Disease: Emphasis on Synaptic Dysfunction Mechanism in Alzheimer’s Disease. CNS Neurol. Disord. Drug Targets 2014, 13, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Alldred, M.J.; Duff, K.E.; Ginsberg, S.D. Microarray analysis of CA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfunction. Neurobiol. Dis. 2012, 45, 751–762. [Google Scholar] [CrossRef]
- Ottone, C.; Galasso, A.; Gemei, M.; Pisa, V.; Gigliotti, S.; Piccioni, F.; Graziani, F.; di Pianella, A.V. Diminution of eIF4E activity suppresses parkin mutant phenotypes. Gene 2011, 470, 12–19. [Google Scholar] [CrossRef]
- Peters, M.M.; Hill, K.E.; Burk, R.F.; Weeber, E.J. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol. Neurodegener. 2006, 1, 12. [Google Scholar] [CrossRef]
- Li, S.; Overman, J.J.; Katsman, D.; Kozlov, S.V.; Donnelly, C.J.; Twiss, J.L.; Giger, R.J.; Coppola, G.; Geschwind, D.H.; Carmichael, S.T. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat. Neurosci. 2010, 13, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- International Parkinson Disease Genomics Consortium. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet 2011, 377, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Schreij, A.M.A.; Chaineau, M.; Ruan, W.; Lin, S.; Barker, P.A.; Fon, E.A.; McPherson, P.S. LRRK 2 localizes to endosomes and interacts with clathrin-light chains to limit Rac1 activation. EMBO Rep. 2014, 16, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.-Y.; Cai, Q.; Lin, L.; Lu, P.-H.; Duan, S.; Sheng, Z.-H. SNAP-29-mediated Modulation of Synaptic Transmission in Cultured Hippocampal Neurons. J. Biol. Chem. 2005, 280, 25769–25779. [Google Scholar] [CrossRef]
- Morgan, J.R.; Zhao, X.; Womack, M.; Prasad, K.; Augustine, G.J.; Lafer, E. A Role for the Clathrin Assembly Domain of AP180 in Synaptic Vesicle Endocytosis. J. Neurosci. 1999, 19, 10201–10212. [Google Scholar] [CrossRef]
- Kramer, L.B.; Shim, J.; Previtera, M.L.; Isack, N.R.; Lee, M.-C.; Firestein, B.L.; Rongo, C. UEV-1 Is an Ubiquitin-Conjugating Enzyme Variant That Regulates Glutamate Receptor Trafficking in C. elegans Neurons. PLoS ONE 2010, 5, e14291. [Google Scholar] [CrossRef]
- Scott, D.A.; Tabarean, I.; Tang, Y.; Cartier, A.; Masliah, E.; Roy, S. A Pathologic Cascade Leading to Synaptic Dysfunction in Synuclein-Induced Neurodegeneration. J. Neurosci. 2010, 30, 8083–8095. [Google Scholar] [CrossRef]
- Watabe-Uchida, M.; John, K.A.; Janas, J.A.; Newey, S.E.; Van Aelst, L. The Rac Activator DOCK7 Regulates Neuronal Polarity through Local Phosphorylation of Stathmin/Op18. Neuron 2006, 51, 727–739. [Google Scholar] [CrossRef]
- Perrault, I.; Hamdan, F.F.; Rio, M.; Capo-Chichi, J.-M.; Boddaert, N.; Décarie, J.-C.; Maranda, B.; Nabbout, R.; Sylvain, M.; Lortie, A.; et al. Mutations in DOCK7 in Individuals with Epileptic Encephalopathy and Cortical Blindness. Am. J. Hum. Genet. 2014, 94, 891–897. [Google Scholar] [CrossRef]
- Nishimura-Akiyoshi, S.; Niimi, K.; Nakashiba, T.; Itohara, S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc. Natl. Acad. Sci. USA 2007, 104, 14801–14806. [Google Scholar] [CrossRef] [Green Version]
- Aoki-Suzuki, M.; Yamada, K.; Meerabux, J.; Iwayama-Shigeno, Y.; Ohba, H.; Iwamoto, K.; Takao, H.; Toyota, T.; Suto, Y.; Nakatani, N.; et al. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol. Psychiatry 2005, 57, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Kar, A.; Kuo, D.; Yu, B.; Havlioglu, N. SRp54 (SFRS11), a Regulator for tau Exon 10 Alternative Splicing Identified by an Expression Cloning Strategy. Mol. Cell. Biol. 2006, 26, 6739–6747. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, G.; Arnold, F.J.L.; Bading, H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 2001, 4, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Otmakhov, N.; Khibnik, L.; Otmakhova, N.; Carpenter, S.; Riahi, S.; Asrican, B.; Lisman, J. Forskolin-Induced LTP in the CA1 Hippocampal Region Is NMDA Receptor Dependent. J. Neurophysiol. 2004, 91, 1955–1962. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef]
- Rudrabhatla, P.; Grant, P.; Jaffe, H.; Strong, M.J.; Pant, H.C. Quantitative phosphoproteomic analysis of neuronal intermediate filament proteins (NF-M/H) in Alzheimer’s disease by iTRAQ. FASEB J. 2010, 24, 4396–4407. [Google Scholar] [CrossRef]
- Yu, P.; Pisitkun, T.; Wang, G.; Wang, R.; Katagiri, Y.; Gucek, M.; Knepper, M.A.; Geller, H.M. Global Analysis of Neuronal Phosphoproteome Regulation by Chondroitin Sulfate Proteoglycans. PLoS ONE 2013, 8, e59285. [Google Scholar] [CrossRef]
- Oki, S.; Ohta, T.; Shioi, G.; Hatanaka, H.; Ogasawara, O.; Okuda, Y.; Kawaji, H.; Nakaki, R.; Sese, J.; Meno, C. Ch IP-Atlas: A data-mining suite powered by full integration of public Ch IP-seq data. EMBO Rep. 2018, 19, e46255. [Google Scholar] [CrossRef]
- Walsh, R.M.; Shen, E.Y.; Bagot, R.C.; Anselmo, A.; Jiang, Y.; Javidfar, B.; Wojtkiewicz, G.J.; Cloutier, J.; Chen, J.W.; Sadreyev, R.; et al. Phf8 loss confers resistance to depression-like and anxiety-like behaviors in mice. Nat. Commun. 2017, 8, 15142. [Google Scholar] [CrossRef] [PubMed]
- Stanic, J.; Mellone, M.; Napolitano, F.; Racca, C.; Zianni, E.; Minocci, D.; Ghiglieri, V.; Thiolat, M.-L.; Li, Q.; Longhi, A.; et al. Rabphilin 3A: A novel target for the treatment of levodopa-induced dyskinesias. Neurobiol. Dis. 2017, 108, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Recabarren, D.; Alarcón, M. Gene networks in neurodegenerative disorders. Life Sci. 2017, 183, 83–97. [Google Scholar] [CrossRef] [PubMed]


| Proteins downregulated by PHF8 knockdown | ||||
|---|---|---|---|---|
| Common Name | ID | Ratio | CV | Role in Human Diseases |
| Long-Term Potentiation (LTP) and Synaptic Plasticity | ||||
| Calcium/Calmodulin Dependent Protein Kinase II Alpha | CaMKIIa | 0.239 | 0.352 | Master regulator of synaptic plasticity; may be overactive in Parkinson’s disease [12,20] |
| Calcium/Calmodulin-dependent Protein Kinase Kinase 2 | CaMKK2 | 0.251 | 0.795 | Regulates LTP in the hippocampus [21] |
| Growth-associated Protein 43/Neuromodulin | GAP43 | 0.306 | 0.044 | Enriched in the substantia nigra; regulates axonal regeneration in Parkinson’s disease [22] |
| Alpha-Chimerin | CHN1 | 0.275 | 0.01 | Regulates dendritic spines [23]; downregulated in Parkinson’s disease [24] |
| Calbindin | CALB1 | 0.457 | 0.259 | Physically sequesters SNCA; mutation protects against aggregation in Parkinson’s disease [25] |
| Alpha-Synuclein | SNCA | 0.329 | 0.01 | First gene identified to be causative for Parkinson’s disease [26]; regulates pre-synaptic vesicle pool [27] |
| Synapsin 1 | SYN1 | 0.667 | 0.381 | Modulates neurotransmitter release, vesicular fusion and recycling [28] |
| Synaptic Structure | ||||
| Neural Cell Adhesion Molecule 1 | NCAM1 | 0.256 | 0.05 | Mediates long-term potentiation [29], serves as receptor for GDNF ligands [30] |
| Microtubule-Associated Protein 2 | MAP2 | 0.348 | 0.052 | Major component of neuronal dendrites, colocalizes with SNCA in Lewy bodies [31] |
| Microtubule-Associated Protein 4 | MAP4 | 0.310 | 0.156 | Regulates synaptic vesicles along microtubular tracks, implicated in Alzheimer’s disease [32] |
| Microtubule-associated protein 1B | MAP1B | 0.661 | 0.169 | Controls dendritic structure [33], implicated in intellectual disability, schizophrenia, and neurodegeneration [34] |
| Brain Acid Soluble Protein 1 | BASP1 | 0.468 | 0.006 | Regulates morphology of neuronal membranes [35] |
| Ankyrin 2 | ANK2 | 0.375 | 0.454 | Regulates synaptic stability, implicated in autism spectrum disorder [36] |
| L1 Cell Adhesion Molecule | L1CAM | 0.472 | 0.527 | Organizes the neuronal ankryin–spectrin interactions, implicated in MASA syndrome characterized by intellectual disability [37] |
| Signal Regulatory Protein α | SIRPA | 0.515 | 0.64 | Organizes pre-synaptic vesicle clusters [38], underlies synaptic maturation [39] |
| A-kinase anchoring protein 5 | AKAP5 | 0.53 | 0.521 | Scaffolding for PSD95 and SAP97, implicated in schizophrenia [40] |
| Synaptic Vesicle Endocytosis/Release | ||||
| Synaptojanin 1 | SYNJ1 | 0.525 | 0.162 | Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism |
| Endophilin A1 | SH3GL2 | 0.685 | 0.026 | Together with SNCA, drives membrane bending and clathrin pit formation [41] |
| Auxilin | DNAJC6 | 0.182 | 0.307 | A major presynaptic endocytic protein linked to PD [42] |
| Complexin-1/Synaphin | CPLX1 | 0.435 | 0.046 | Interacts with SNCA; critical in the regulation of neurotransmitter release [43] |
| Rabphilin 3A | RPH3A | 0.299 | 0.01 | Interacts with SNCA; involved in neurotransmitter release in Parkinson’s disease [44] |
| Rab-like protein 6 | RABL6 | 0.388 | 0.01 | Analogue of Rab3, involved in neurotransmitter release [45] |
| ZW10-interacting protein | ZWINT | 0.487 | 0.855 | Binds to Rab3C, regulates synaptic vesicle release [46] |
| Vesicle transport through interaction with T-SNAREs 1A | VTI1A | 0.525 | 0.021 | Mediates spontaneous neurotransmitter release [47], implicated in Alzheimer’s disease [48] |
| Syntaxin 7 | STX7 | 0.611 | 0.045 | Downregulated in aging mice in a Tau-dependent manner [49] |
| Synaptic Translation | ||||
| Eukaryotic Initiation Factor 4E | EIF4E | 0.637 | 0.032 | Interacts with Parkin; regulates synaptic protein translation [50] |
| Eukaryotic Elongation Factor, Selenocysteine-TRNA-Specific | EEFSEC | 0.402 | 0.045 | Regulates selenium protein synthesis [51] |
| Synaptic Membrane Recycling/Trafficking | ||||
| Ras-related protein 2B | RAB2B | 0.603 | 0.287 | Involved in Golgi body fragmentation; downregulated in aging [52] |
| N-ethylmaleimide sensitive fusion | NSF | 0.573 | 0.097 | Interacts with SNCA; an independent risk locus for Parkinson’s disease; downregulated in Parkinson’s disease [53] |
| Clathrin, light-chain | CLTA | 0.446 | 0.219 | Directly binds to LRRK2, regulates dendritic spine morphology [54] |
| Phosphatidylinositide phosphatase SAC2 | INPP5F | 0.525 | 0.045 | Involved in clathrin-mediated endocytosis, identified in multiple GWAS studies as a validated independent risk locus for Parkinson’s disease [55] |
| Soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein 29 | SNAP29 | 0.335 | 1.143 | Negative regulator of neurotransmitter release [56] |
| Clathrin Coat Assembly Protein AP180 | SNAP91 | 0.581 | 0.078 | Mediates clathrin-mediated vesicle recycling [57] |
| Ubiquitin-conjugating enzyme E2 variant 2 | UBE2V2 | 0.628 | 0.277 | Regulates glutamate receptor trafficking [58] |
| Amphiphysin | AMPH | 0.631 | 0.345 | Links Dynamin with Clathrin to mediate endocytosis, mediated in SCNA-related neurodegeneration [59] |
| Axonal Proteins | ||||
| Dedicator Of Cytokinesis 7 | DOCK7 | 0.394 | 0.034 | Regulates axonal polarity [60], implicated in epileptic encephalopathy [61] |
| Netrin G2 | NTNG2 | 0.616 | 0.175 | Regulates axonal pathfinding [62], implicated in schizophrenia [63] |
| Proteins with Nuclear Roles/Transcriptional Modulation | ||||
| DNA Methyltransferase 1 | DNMT1 | 0.353 | 0.032 | Critical DNA methyltransferase for activity-regulated synaptic function [64] |
| DNA Methyltransferase 3A | DNMT3A | 0.752 | 0.328 | Critical DNA methyltransferase for activity-regulated synaptic function [64] |
| Serine/Arginine-Rich Splicing Factor 11 | SRSF11 | 0.51 | 0.173 | Regulates Tau gene splicing [65] |
| RNA binding motif protein 17 | RBM17 | 0.649 | 0.189 | RNA metabolism and splicing, implicated in spinocerebellar ataxia |
| GO TERM | Biological Process | % | p-Value | Fold Enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|
| GO:0006886 | Intracellular protein transport | 4.13257 | 7.81 × 10−25 | 2.964188 | 4.93 × 10−21 | 4.74 × 10−21 |
| GO:0006457 | Protein folding | 2.536825 | 1.04 × 10−22 | 3.917003 | 3.29 × 10−19 | 3.16 × 10−19 |
| GO:0015031 | Protein transport | 4.13257 | 8.30 × 10−22 | 2.731703 | 1.75 × 10−18 | 1.68 × 10−18 |
| GO:0007420 | Brain development | 4.500818 | 4.41 × 10−18 | 2.346359 | 6.96 × 10−15 | 6.69 × 10−15 |
| GO:0007409 | Axonogenesis | 2.12766 | 6.74 × 10−18 | 3.710042 | 8.51 × 10−15 | 8.19 × 10−15 |
| GO:0006888 | ER to Golgi vesicle transport | 2.00491 | 6.12 × 10−16 | 3.557335 | 6.44 × 10−13 | 6.19 × 10−13 |
| GO:0016192 | Vesicle-mediated transport | 2.782324 | 3.34 × 10−15 | 2.772339 | 3.01 × 10−12 | 2.90 × 10−12 |
| GO:0050808 | Synapse organization | 1.595745 | 2.46 × 10−14 | 3.888839 | 1.94 × 10−11 | 1.87 × 10−11 |
| GO:0031175 | Neuron projection development | 2.536825 | 7.92 × 10−14 | 2.758749 | 5.56 × 10−11 | 5.35 × 10−11 |
| GO:0010976 | Neuron projection regulation | 2.454992 | 1.20 × 10−13 | 2.789746 | 7.57 × 10−11 | 7.28 × 10−11 |
| GO:0050804 | Modulation of synaptic transmission | 2.00491 | 4.79 × 10−13 | 3.072244 | 2.75 × 10−10 | 2.65 × 10−10 |
| GO:0000226 | Microtubule cytoskeleton organization | 2.045827 | 5.66 × 10−13 | 3.020528 | 2.98 × 10−10 | 2.87 × 10−10 |
| GO:0050821 | Protein stabilization | 2.495908 | 1.05 × 10−12 | 2.6432 | 5.08 × 10−10 | 4.89 × 10−10 |
| GO:1990090 | Response to nerve growth factor stimulus | 1.309329 | 4.20 × 10−12 | 3.952835 | 1.89 × 10−9 | 1.82 × 10−9 |
| GO:0050790 | Regulation of catalytic activity | 4.255319 | 6.06 × 10−12 | 1.987829 | 2.55 × 10−9 | 2.46 × 10−9 |
| GO:0006099 | Tricarboxylic acid cycle | 0.859247 | 9.07 × 10−12 | 5.60649 | 3.58 × 10−9 | 3.45 × 10−9 |
| GO:0048488 | Synaptic vesicle endocytosis | 1.104746 | 7.36 × 10−11 | 4.138124 | 2.73 × 10−8 | 2.63 × 10−8 |
| GO:0007005 | Mitochondrion organization | 1.595745 | 2.05 × 10−10 | 3.045035 | 6.80 × 10−8 | 6.55 × 10−8 |
| GO:0032981 | Mitochondrial respiratory chain complex I | 1.186579 | 2.34 × 10−10 | 3.750175 | 7.39 × 10−8 | 7.11 × 10−8 |
| GO:0016310 | Phosphorylation | 2.086743 | 6.63 × 10−10 | 2.512432 | 1.99 × 10−7 | 1.92 × 10−7 |
| GO:0007269 | Neurotransmitter secretion | 0.859247 | 2.81 × 10−9 | 4.456441 | 8.07 × 10−7 | 7.77 × 10−7 |
| GO:0021987 | Cerebral cortex development | 1.636661 | 3.15 × 10−9 | 2.758749 | 8.65 × 10−7 | 8.32 × 10−7 |
| GO:0007030 | Golgi organization | 1.554828 | 3.63 × 10−9 | 2.83331 | 9.54 × 10−7 | 9.18 × 10−7 |
| GO:0072659 | Protein localization to plasma membrane | 2.00491 | 5.14 × 10−9 | 2.42836 | 1.30 × 10−6 | 1.25 × 10−6 |
| GO:0061077 | Chaperone-mediated protein folding | 0.818331 | 6.82 × 10−9 | 4.473647 | 1.60 × 10−6 | 1.54 × 10−6 |
| GO:0007612 | Learning | 1.186579 | 8.56 × 10−9 | 3.287824 | 1.93 × 10−6 | 1.86 × 10−6 |
| GO:0021766 | Hippocampus development | 1.513912 | 2.92 × 10−8 | 2.686151 | 6.37 × 10−6 | 6.13 × 10−6 |
| GO:0032456 | Endocytic recycling | 1.06383 | 3.47 × 10−8 | 3.362226 | 7.30 × 10−6 | 7.03 × 10−6 |
| GO:0030036 | Actin cytoskeleton organization | 2.045827 | 4.41 × 10−8 | 2.26127 | 8.73 × 10−6 | 8.40 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oey, N.E.; Zhou, L.; Chan, C.H.S.; VanDongen, A.M.J.; Tan, E.K. A Proteome-Wide Effect of PHF8 Knockdown on Cortical Neurons Shows Downregulation of Parkinson’s Disease-Associated Protein Alpha-Synuclein and Its Interactors. Biomedicines 2023, 11, 486. https://doi.org/10.3390/biomedicines11020486
Oey NE, Zhou L, Chan CHS, VanDongen AMJ, Tan EK. A Proteome-Wide Effect of PHF8 Knockdown on Cortical Neurons Shows Downregulation of Parkinson’s Disease-Associated Protein Alpha-Synuclein and Its Interactors. Biomedicines. 2023; 11(2):486. https://doi.org/10.3390/biomedicines11020486
Chicago/Turabian StyleOey, Nicodemus E., Lei Zhou, Christine Hui Shan Chan, Antonius M. J. VanDongen, and Eng King Tan. 2023. "A Proteome-Wide Effect of PHF8 Knockdown on Cortical Neurons Shows Downregulation of Parkinson’s Disease-Associated Protein Alpha-Synuclein and Its Interactors" Biomedicines 11, no. 2: 486. https://doi.org/10.3390/biomedicines11020486






