State of the Art of microRNAs Signatures as Biomarkers and Therapeutic Targets in Parkinson’s and Alzheimer’s Diseases: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Instruments and Professionals Used for Study Eligibility
2.3. Eligibility Criteria, Study Quality, and Risk of Bias
2.4. Data Sources, Research Strategy, and Study Publication Date
2.5. Statistical Analysis—Meta-Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Titova, N.; Qamar, M.A.; Chaudhuri, K.R. The Nonmotor Features of Parkinson’s Disease. Int. Rev. Neurobiol. 2017, 132, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, M.; Junn, E.; Mochizuki, H.; Mouradian, M.M. Nucleic Acid-Based Therapeutics for Parkinson’s Disease. Neurotherapeutics 2019, 16, 287–298. [Google Scholar] [CrossRef]
- Braak, H.; Del Tredici, K.; Rub, U.; de Vos, R.A.; Jansen Steur, E.N.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211. [Google Scholar] [CrossRef]
- Hamza, T.H.; Zabetian, C.P.; Tenesa, A.; Laederach, A.; Montimurro, J.; Yearout, D.; Kay, D.M.; Doheny, K.F.; Paschall, J.; Pugh, E.; et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010, 42, 781–785. [Google Scholar] [CrossRef]
- Dawson, T.M.; Dawson, V.L. Molecular pathways of neurodegeneration in Parkinson’s disease. Science 2003, 302, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Gasser, T. Molecular pathogenesis of Parkinson disease: Insights from genetic studies. Expert Rev. Mol. Med. 2009, 11, e22. [Google Scholar] [CrossRef]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wei, L.; Wu, F.; Hu, Z.; Liu, Z.; Yuan, W. Advances with microRNAs in Parkinson’s disease research. Drug Des. Dev. Ther. 2013, 7, 1103–1113. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease results from the cerebral accumulation and cytotoxicity of amyloid beta-protein. J. Alzheimer’s Dis. 2001, 3, 75–80. [Google Scholar] [CrossRef]
- Mattson, M.P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 9–631. [Google Scholar] [CrossRef] [PubMed]
- LaFerla, F.M.; Green, K.M.; Oddo, S. Intracellular amyloid-beta in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H.; Burns, J.M.; Khan, S.M. The Alzheimer’s disease mitochondrial cascade hypothesis. J. Alzheimer’s Dis. 2010, 20 (Suppl. 2), S265–S279. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.H.; Manczak, M.; Mao, P.; Calkins, M.J.; Reddy, A.P.; Shirendeb, U. Amyloid-beta and mitochondria in aging and Alzheimer’s disease: Implications for synaptic damage and cognitive decline. J. Alzheimer’s Dis. 2010, 20 (Suppl. 2), S499–S512. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Manczak, M.; Calkins, M.J.; Truong, Q.; Reddy, T.P.; Reddy, A.P.; Shirendeb, U.; Lo, H.H.; Rabinovitch, P.S.; Reddy, P.H. Mitochondria-targeted catalase reduces abnormal APP processing, amyloid β production, and BACE1 in a mouse model of Alzheimer’s disease: Implications for neuroprotection and lifespan extension. Hum. Mol. Genet. 2012, 21, 2973–2990. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta 2016, 1862, 1617–1627. [Google Scholar] [CrossRef]
- Reddy, P.H. A critical assessment of research on neurotransmitters in Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 57, 969–974. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. MicroRNA-455-3p as a potential biomarker for Alzheimer’s disease: An update. Front. Aging Neurosci. 2018, 10, 41. [Google Scholar] [CrossRef]
- Abeliovich, A.; Flint, M. Beal, Parkinsonism genes: Culprits and clues. J. Neurochem. 2006, 99, 1062–1072. [Google Scholar] [CrossRef]
- Yaghoubi, Y.; Movassaghpour, A.; Zamani, M.; Talebi, M.; Mehdizadeh, A.; Yousefi, M. Human umbilical cord mesenchymal stem cells derived-exosomes in diseases treatment. Life Sci. 2019, 233, 116733. [Google Scholar] [CrossRef]
- Baharlooi, H.; Nouraei, Z.; Azimi, M.; Moghadasi, A.N.; Tavassolifar, M.J.; Moradi, B.; Sahraian, M.A.; Izad, M. Umbilical cord mesenchymal stem cells as well as their released exosomes suppress proliferation of activated PBMCs in multiple sclerosis. Scand. J. Immunol. 2020, 93, e13013. [Google Scholar] [CrossRef]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef]
- Mears, R.; Craven, R.A.; Hanrahan, S.; Totty, N.; Upton, C.; Young, S.L.; Patel, P.; Selby, P.J.; Banks, R.E. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 2004, 4, 4019–4031. [Google Scholar] [CrossRef]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef]
- Futter, C.E.; White, I.J. Annexins and endocytosis. Traffic 2007, 8, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Staubach, S.; Razawi, H.; Hanisch, F.G. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics 2009, 9, 2820–2835. [Google Scholar] [CrossRef]
- Yim, N.; Ryu, S.W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.-H.; et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, Z.; Lu, N. From pathogenesis to clinical application: Insights into exosomes as transfer vectors in cancer. J. Exp. Clin. Cancer Res. 2016, 35, 156. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, Y.; Zhang, Y.; Zhang, H.; Liu, W.; Zhang, N.; Zhang, X.; Zhou, G.; Wu, L.; Hua, K.; et al. Exosomes derived from human umbilical cord mesenchymal stem cells accelerate growth of VK2 vaginal epithelial cells through MicroRNAs in vitro. Hum. Reprod. 2019, 34, 248–260. [Google Scholar] [CrossRef]
- Zhang, B.; Shen, L.; Shi, H.; Pan, Z.; Wu, L.; Yan, Y.; Zhang, X.; Mao, F.; Qian, H.; Xu, W. Exosomes from Human Umbilical Cord Mesenchymal Stem Cells: Identification, Purification, and Biological Characteristics. Stem Cells Int. 2016, 2016, 1929536. [Google Scholar] [CrossRef]
- Pinnell, J.R.; Cui, M.; Tieu, K. Exosomes in Parkinson disease. J. Neurochem. 2020, 157, 413–428. [Google Scholar] [CrossRef]
- Vassileff, N.; Cheng, L.; Hill, A.F. Extracellular vesicles—Propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. J. Cell Sci. 2020, 133, jcs243139. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Surguchov, A. Biomarkers in Parkinson’s Disease. In Neurodegenerative Diseases Biomarkers; Peplow, P.V., Martinez, B., Gennarelli, T.A., Eds.; Neuromethods; Humana: New York, NY, USA, 2022; Volume 173. [Google Scholar] [CrossRef]
- Junn, E.; Mouradian, M.M. MicroRNAs in neurodegenerative disorders. Cell Cycle 2010, 9, 1717–1721. [Google Scholar] [CrossRef]
- Junn, E.; Mouradian, M.M. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 2012, 133, 142–150. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Balshem, H.; Helfand, M.; Schünemann, H.J.; Oxman, A.D.; Kunz, R.; Brozek, J.; Vist, G.E.; Falck-Ytter, Y.; Meerpohl, J.; Norris, S.; et al. Grade guidelines: 3 ratng the quality of evidence. J. Clin. Epidemiol. Md. Height 2011, 64, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Cochrane. Cochrane Handbook for Systematic Reviews of Interventions Version 6.3 (Updated February 2022). Higgins, J.P.T., Thomas, J., Chandler, J., Cumpston, M., Li, T., Page, M.J., Welch, V.A., Eds.; 2022. Available online: www.training.cochrane.org/handbook (accessed on 27 February 2022).
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Sun, Y.; Zhen, H.; Guo, M.; Ye, J.; Liu, Z.; Yang, Y.; Zhang, X. Differential Expression of Plasma Exo-miRNA in Neurodegenerative Diseases by Next-Generation Sequencing. Front. Neurosci. 2020, 14, 438. [Google Scholar] [CrossRef]
- Bekris, L.M.; Lutz, F.; Montine, T.J.; Yu, C.E.; Tsuang, D.; Peskind, E.R.; Leverenz, J.B. MicroRNA in Alzheimer’s disease: An exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers 2013, 18, 455–466. [Google Scholar] [CrossRef]
- Liu, C.G.; Meng, S.; Li, Y.; Lu, Y.; Zhao, Y.; Wang, P.C. MicroRNA-135a in ABCA1-labeled Exosome is a Serum Biomarker Candidate for Alzheimer’s Disease. Biomed. Environ. Sci. 2021, 34, 19–28. [Google Scholar] [CrossRef]
- De Felice, B.; Montanino, C.; Oliva, M.; Bonavita, S.; Di Onofrio, V.; Coppola, C. MicroRNA Expression Signature in Mild Cognitive Impairment Due to Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 4408–4416. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kang, J.; Svetnik, V.; Warden, D.; Wilcock, G.; David Smith, A.; Savage, M.J.; Laterza, O.F. A Machine Learning Approach to Identify a Circulating MicroRNA Signature for Alzheimer Disease. J. Appl. Lab. Med. 2020, 5, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Denk, J.; Oberhauser, F.; Kornhuber, J.; Wiltfang, J.; Fassbender, K.; Schroeter, M.L.; Volk, A.E.; Diehl-Schmid, J.; Prudlo, J.; Danek, A.; et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 2018, 13, e0197329. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, J.L.; Li, L.; Wang, P.C. MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 2014, 34, 160–166. [Google Scholar] [CrossRef]
- Galimberti, D.; Villa, C.; Fenoglio, C.; Serpente, M.; Ghezzi, L.; Cioffi, S.M.; Arighi, A.; Fumagalli, G.; Scarpini, E. Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimer’s Dis. 2014, 42, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Soleimani Zakeri, N.S.; Pashazadeh, S.; MotieGhader, H. Gene biomarker discovery at different stages of Alzheimer using gene co-expression network approach. Sci. Rep. 2020, 10, 12210. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Yu, M. MicroRNA-4722-5p and microRNA-615-3p serve as potential biomarkers for Alzheimer’s disease. Exp. Ther. Med. 2022, 23, 241. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef]
- Guévremont, D.; Tsui, H.; Knight, R.; Fowler, C.J.; Masters, C.L.; Martins, R.N.; Abraham, W.C.; Tate, W.P.; Cutfield, N.J.; Williams, J.M. Plasma microRNA vary in association with the progression of Alzheimer’s disease. Alzheimer’s Dement. (Amst.) 2022, 14, e12251. [Google Scholar] [CrossRef]
- Jia, L.; Zhu, M.; Yang, J.; Pang, Y.; Wang, Q.; Li, Y.; Li, T.; Li, F.; Wang, Q.; Li, Y.; et al. Prediction of P-tau/Aβ42 in the cerebrospinal fluid with blood microRNAs in Alzheimer’s disease. BMC Med. 2021, 19, 264. [Google Scholar] [CrossRef]
- Grossi, I.; Radeghieri, A.; Paolini, L.; Porrini, V.; Pilotto, A.; Padovani, A.; Marengoni, A.; Barbon, A.; Bellucci, A.; Pizzi, M.; et al. MicroRNA 34a 5p expression in the plasma and in its extracellular vesicle fractions in subjects with Parkinson’s disease: An exploratory study. Int. J. Mol. Med. 2021, 47, 533–546. [Google Scholar] [CrossRef]
- Chen, Q.; Deng, N.; Lu, K.; Liao, Q.; Long, X.; Gou, D.; Bi, F.; Zhou, J. Elevated plasma miR-133b and miR-221-3p as biomarkers for early Parkinson’s disease. Sci. Rep. 2021, 11, 15268. [Google Scholar] [CrossRef]
- Manna, I.; Quattrone, A.; De Benedittis, S.; Vescio, B.; Iaccino, E.; Quattrone, A. Exosomal miRNA as peripheral biomarkers in Parkinson’s disease and progressive supranuclear palsy: A pilot study. Park. Relat. Disord. 2021, 93, 77–84. [Google Scholar] [CrossRef]
- Cai, M.; Chai, S.; Xiong, T.; Wei, J.; Mao, W.; Zhu, Y.; Li, X.; Wei, W.; Dai, X.; Yang, B.; et al. Aberrant Expression of Circulating MicroRNA Leads to the Dysregulation of Alpha-Synuclein and Other Pathogenic Genes in Parkinson’s Disease. Front. Cell Dev. Biol. 2021, 9, 695007. [Google Scholar] [CrossRef]
- He, S.; Huang, L.; Shao, C.; Nie, T.; Xia, L.; Cui, B.; Lu, F.; Zhu, L.; Chen, B.; Yang, Q. Several miRNAs derived from serum extracellular vesicles are potential biomarkers for early diagnosis and progression of Parkinson’s disease. Transl. Neurodegener. 2021, 10, 25. [Google Scholar] [CrossRef]
- Baghi, M.; Yadegari, E.; Rostamian Delavar, M.; Peymani, M.; Ganjalikhani-Hakemi, M.; Salari, M.; Nasr-Esfahani, M.H.; Megraw, T.L.; Ghaedi, K. MiR-193b deregulation is associated with Parkinson’s disease. J. Cell Mol. Med. 2021, 25, 6348–6360. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xiao, L.; Jiang, X.; Li, G.; Lu, Z. Screening of Parkinson’s Differential MicroRNA Based on GEO Database and Its Clinical Verification. Biomed. Res. Int. 2021, 2021, 8171236. [Google Scholar] [CrossRef]
- Lin, X.; Wang, R.; Li, R.; Tao, T.; Zhang, D.; Qi, Y. Diagnostic Performance of miR-485-3p in Patients with Parkinson’s Disease and its Relationship with Neuroinflammation. Neuromol. Med. 2021, 24, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed]
- Vallelunga, A.; Iannitti, T.; Dati, G.; Capece, S.; Maugeri, M.; Tocci, E.; Picillo, M.; Volpe, G.; Cozzolino, A.; Squillante, M.; et al. Serum miR-30c-5p is a potential biomarker for multiple system atrophy. Mol. Biol. Rep. 2019, 46, 1661–1666. [Google Scholar] [CrossRef]
- Starhof, C.; Hejl, A.M.; Heegaard, N.H.H.; Carlsen, A.L.; Burton, M.; Lilje, B.; Winge, K. The biomarker potential of cell-free microRNA from cerebrospinal fluid in Parkinsonian Syndromes. Mov. Disord. 2019, 34, 246–254. [Google Scholar] [CrossRef]
- Kingsbury, A.E.; Daniel, S.E.; Sangha, H.; Eisen, S.; Lees, A.J.; Foster, O.J. Alteration in alpha-synuclein mRNA expression in Parkinson’s disease. Mov. Disord. 2004, 19, 162–170. [Google Scholar] [CrossRef]
- Dachsel, J.C.; Lincoln, S.J.; Gonzalez, J.; Ross, O.A.; Dickson, D.W.; Farrer, M.J. The ups and downs of alpha-synuclein mRNA expression. Mov. Disord. 2007, 22, 293–295. [Google Scholar] [CrossRef]
- Grundemann, J.; Schlaudraff, F.; Haeckel, O.; Liss, B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson’s disease. Nucleic Acids Res. 2008, 36, e38. [Google Scholar] [CrossRef]
- Junn, E.; Lee, K.W.; Jeong, B.S.; Chan, T.W.; Im, J.Y.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. [Google Scholar] [CrossRef] [PubMed]
- Doxakis, E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 2010, 285, 12726–12734. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2013, 22, 608–620. [Google Scholar] [CrossRef]
- Ghanbari, M.; Darweesh, S.K.; de Looper, H.W.; Van Luijn, M.M.; Hofman, A.; Ikram, M.A.; Franco, O.; Erkeland, S.J.; Dehghan, A. Genetic Variants in MicroRNAs and Their Binding Sites Are Associated with the Risk of Parkinson Disease. Hum. Mutat. 2016, 37, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Sadlon, A.; Takousis, P.; Alexopoulos, P.; Evangelou, E.; Prokopenko, I.; Perneczky, R. miRNAs Identify Shared Pathways in Alzheimer’s and Parkinson’s Diseases. Trends Mol. Med. 2019, 25, 662–672. [Google Scholar] [CrossRef]
- Wen, M.M. Getting miRNA Therapeutics into the Target Cells for Neurodegenerative Diseases: A Mini-Review. Front. Mol. Neurosci. 2016, 9, 129. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.C.; Yoo, M.; Kabaria, S.; Junn, E. MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci. Lett. 2018, 678, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Dorsett, Y.; Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 2004, 10, 544–550. [Google Scholar] [CrossRef]
- Choi, D.C.; Chae, Y.J.; Kabaria, S.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Mouradian, M.M.; Junn, E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J. Neurosci. 2014, 34, 12725–12737. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.D.; Kabaria, S.; Choi, D.C.; Mouradian, M.M.; Junn, E. MicroRNA-7 Promotes Glycolysis to Protect against 1-Methyl-4-phenylpyridinium-induced Cell Death. J. Biol. Chem. 2015, 290, 12425–12434. [Google Scholar] [CrossRef]
- Kabaria, S.; Choi, D.C.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Junn, E. MicroRNA-7 activates Nrf2 pathway by targeting Keap1 expression. Free Radic Biol. Med. 2015, 89, 548–556. [Google Scholar] [CrossRef]
- Chaudhuri, A.D.; Choi, D.C.; Kabaria, S.; Tran, A.; Junn, E. MicroRNA-7 Regulates the Function of Mitochondrial Permeability Transition Pore by Targeting VDAC1 Expression. J. Biol. Chem. 2016, 291, 6483–6493. [Google Scholar] [CrossRef]
- Mushtaq, G.; Greig, N.H.; Anwar, F.; Zamzami, M.A.; Choudhry, H.; Shaik, M.M.; Tamargo, I.A.; Kamal, M.A. miRNAs as Circulating Biomarkers for Alzheimer’s Disease and Parkinson’s Disease. Med. Chem. 2016, 12, 217–225. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]

| PARTICIPANTS | INTERVENTION | CONTROL | OUTCOMES | STUDY DESIGN |
|---|---|---|---|---|
| Human serum, plasma, and cerebrospinal fluid samples | Peripheral blood and cerebrospinal fluid collection | Biological samples from healthy patients | Main microRNAs as biomarkers and therapeutic targets | In vitro clinical studies |
| PUBMED | AND | PUBMED | AND | PUBMED | NOT | PUBMED |
| Parkinson’s disease OR Alzheimer’s disease | Alzheimer’s disease and microRNA and miRNA and human and serum and plasma and cerebrospinal fluid | Parkinson’s disease and microRNA and miRNA and human and serum and plasma and cerebrospinal fluid | Review study OR Editorials OR Short communications |
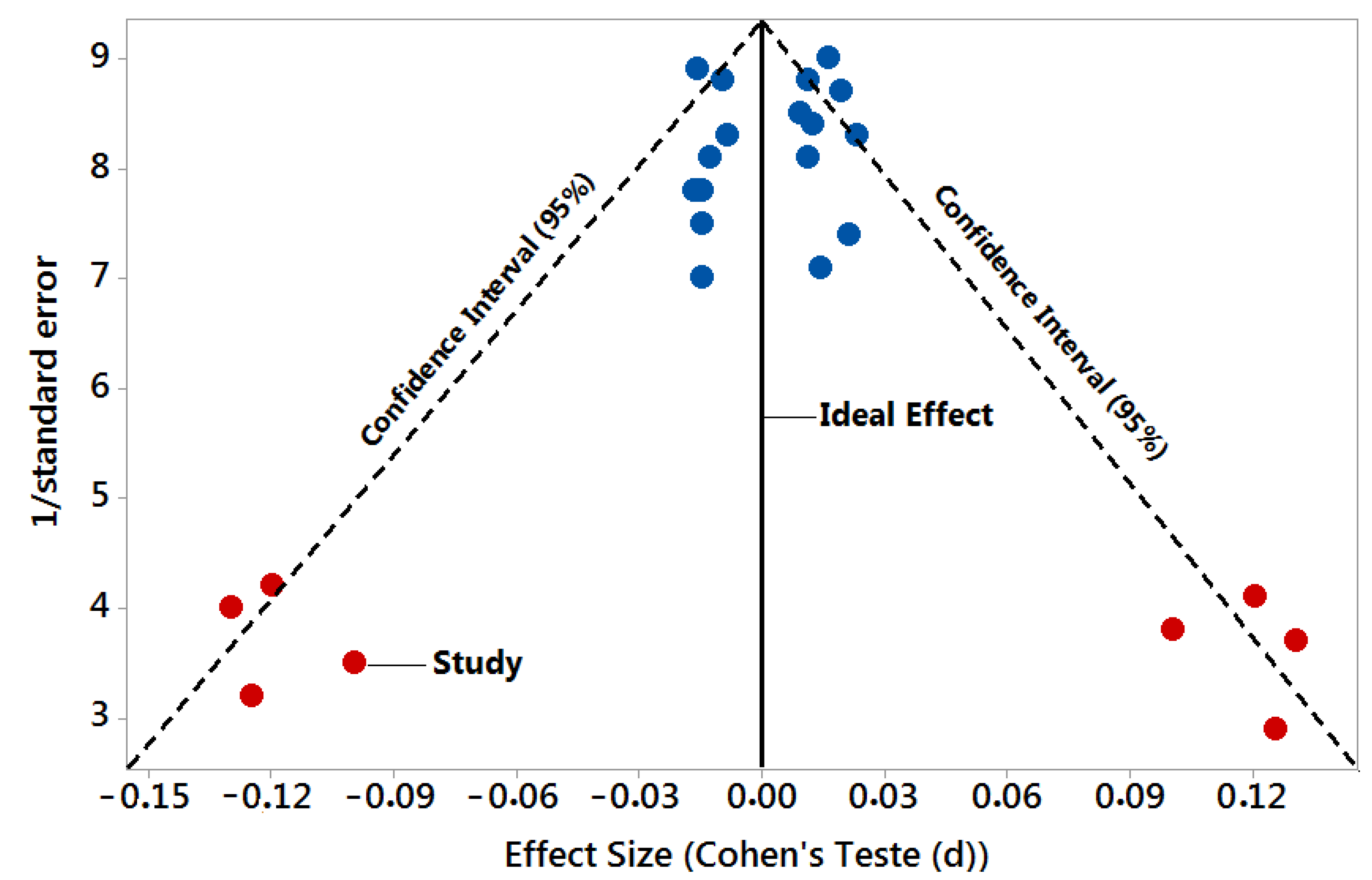
| Authors and Date/Variables | Detection Rate (Accuracy (%)) | p-Value | Effect Size | 1/Standard Error |
|---|---|---|---|---|
| N = 25 Studies | Test Group vs. Control | Reference < 0.05 | Cohen’s Test (d) | Precision or Sample Size |
| 1. Burgos et al. 2014 [39] | 73% AD 55% PD | >0.05 | 0.012 | 8.4 |
| 2. Nie et al. 2020 [40] | 84% AD 95% PD | >0.05 | −0.010 | 8.8 |
| 3. Bekris et al. 2013 [41] | 92% AD | >0.05 | −0.013 | 8.1 |
| 4. Liu et al. 2021 [42] | 95% AD | >0.05 | −0.017 | 7.8 |
| 5. De Felice et al. 2020 [43] | 85.7% AD | >0.05 | 0.021 | 7.4 |
| 6. Zhao et al. 2020 [44] | 76% AD | >0.05 | −0.016 | 8.9 |
| 7. Denk et al. 2018 [45] | 72% AD | >0.05 | 0.023 | 8.3 |
| 8. Liu et al. 2014 [46] | 96% AD | >0.05 | −0.100 | 3.5 |
| 9. Galimberti et al. 2014 [47] | 82% AD | >0.05 | 0.100 | 3.8 |
| 10. Soleimani, Pashazadeh, and MotieGhader 2020 [48] | 80% AD | >0.05 | −0.015 | 7.5 |
| 11. Liu, Xu, and Yu 2022 [49] | 87% AD | >0.05 | −0.015 | 7.8 |
| 12. Gámez-Valero et al. 2019 [50] | 90% AD | >0.05 | 0.120 | 4.1 |
| 13. Guévremont et al. 2022 [51] | 80% AD | >0.05 | −0.120 | 4.2 |
| 14. Jia et al. 2021 [52] | 90% AD | >0.05 | 0.130 | 3.7 |
| 15. Grossi et al. 2021 [53] | 73.8% PD | >0.05 | −0.130 | 4.0 |
| 16. Chen et al. 2021 [54] | 91.1% PD | >0.05 | 0.125 | 2.9 |
| 17. Manna et al. 2021 [55] | 75% PD | >0.05 | −0.125 | 3.2 |
| 18. Cai et al. 2021 [56] | 97% PD | >0.05 | 0.009 | 8.5 |
| 19. He et al. 2021 [57] | 79% PD | >0.05 | 0.011 | 8.8 |
| 20. Baghi et al. 2021 [58] | 79.3% PD | >0.05 | 0.011 | 8.1 |
| 21. Jiang et al. 2021 [59] | 88.6% PD | >0.05 | −0.009 | 8.3 |
| 22. Lin et al. 2021 [60] | 88.1% PD | >0.05 | 0.014 | 7.1 |
| 23. Gui et al. 2015 [61] | 85.6% PD | >0.05 | −0.015 | 7.0 |
| 24. Vallelunga et al. 2019 [62] | 82% PD | >0.05 | 0.019 | 8.7 |
| 25. Starhof et al. 2019 [63] | 88% PD | >0.05 | 0.016 | 9.0 |
| Authors/Study Data | Sample Size (n) (Human Participants) | Disease Type Alzheimer’ Disease (AD) and/or Parkinson’ Disease (PD) | Sample Type | Numbers and Types of miRNAs |
|---|---|---|---|---|
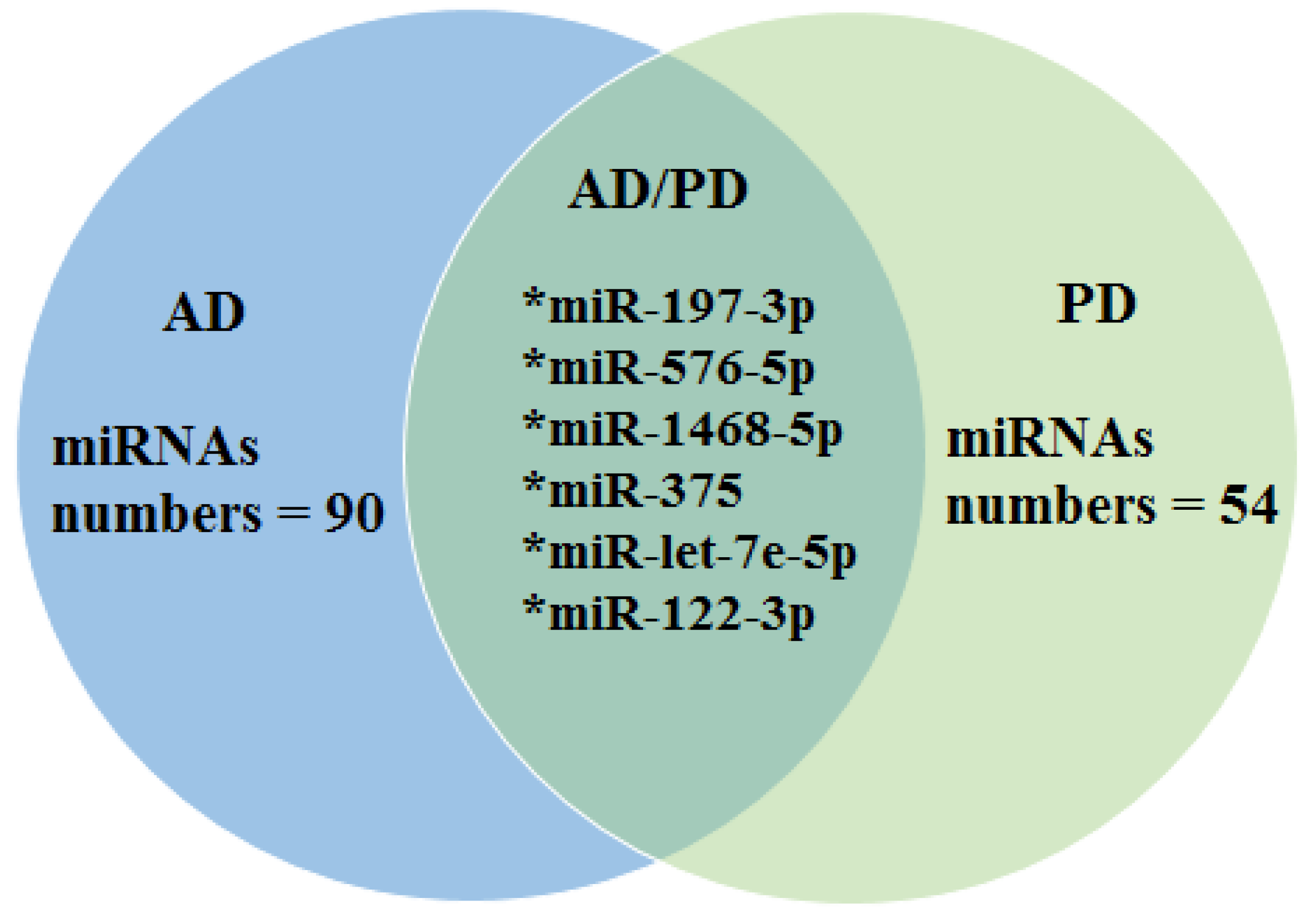
| 1. Burgos et al. 2014 [40] | 69 AD 67 PD 78 healthy controls | AD/PD | CSF and Serum (postmortem) | AD-Serum: Up-regulated: miR-34b-3p, miR-219-2-3p, miR-34c-5p, miR-34b-5p, miR-135a-5p Down-regulated: miR-182-5p, miR-21-5p, miR-375 AD-CSF: Down-regulated: N = 41 miRNAs (demonstrated in the supplementary material) PD-CSF: Up-regulated: miR-19a-3p, miR-19b-3p, let-7g-3p Down-regulated: miR-132-5p, miR-485-5p, miR-127-3p, miR-128, miR-409-3p, miR-433, miR-370, miR-431-3p, miR-873-3p, miR-136-3p, miR-212-3p, miR-10a-5p, miR-1224-5p, miR-4448 PD (Serum): Up-regulated: miR-338-3p, 30e-3p, 30a-3p Down-regulated: miR-16-2-3p, 1294 |
| 2. Nie et al. 2020 [41] | 34 healthy controls, 5 AD donors, and 7 PD donors | AD and PD | Plasma | AD: Up-regulated: miR-423-5p, miR369-5p, miR-23a-3p Down-regulated: miR-204-5p, miR125a-5p, miR-1468-5p, miR-375, let-7e-5p PD: Up-regulated: let-7e-5p, let-7i-5p miR-652-3p, miR-4732-3p, miR-6131, miR-3184-3p, miR-378g Down-regulated: miR-197-3p, miR-576-5p, miR-1468-5p, miR-375, miR-211-5p, let-7e-3p, miR-122-3p, miR-941, miR-30d-5p, miR-192-5p, miR-93-5p, miR-425-5p, miR-99b-5p |
| 3. Bekris et al. 2013 [42] | 21 AD 21 healthy controls | AD | CSF, Plasma (during life); Cerebellum and Hippocampus were obtainedat autopsy. | Up-regulated: miR-15a (Plasma high levels) |
| 4. Liu et al. 2021 [43] | 198 AD 30 healthy controls | AD | LCR Serum | Up-regulated: miR-135a |
| 5. De Felice et al. 2020 [44] | 18 AD 18 mild cognitive impairment | AD | LCR | Up-regulated: hsa-mir-5588-5p, hsa-mir-3658, hsa-mir-567 e hsa-mir-3908 Highlight: hsa-mir-567 (Blood, LCR, and Serum) |
| 6. Zhao et al. 2020 [45] | 32 AD 51 healthy controls 13 mild cognitive impairment | AD | Serum | Up-regulated: mir-346, mir-345-5p, mir-122-3p, mir-1291, mir-640, mir-650, mir-1285-3p, mir-1299, mir-1267 Down-regulated: mir-208b-3p, mir-499a-5p, mir-206 |
| 7. Denk et al. 2018 [46] | 48 AD 44 healthy controls 48 frontotemporal lobar degeneration | AD | LCR Serum | Up-regulated: miR-320a and miR-26b-5p |
| 8. Liu et al. 2014 [47] | 45 AD 22 MCI 50 healthy controls | AD | LCR Serum | Down-regulated: miR-384 |
| 9. GalimberTi et al. 2014 [48] | 10 AD 8 healthy controls | AD | LCR Serum | Down-regulated: miR-125b, miR-23a, miR-26b |
| 10. Soleiman, Pashazadeh, and MotieGhader 2020 [49] | 145 AD 80 mild cognitive impairment (MCI) 104 healthy controls | AD | LCR Serum | Up-regulated: miR-615-3p, miR-4722-5p, miR-4768-3p, miR-1827, miR-940 e miR-30b-3p |
| 11. Liu, Xu and Yu 2022 [50] | 33 AD 33 healthy controls | AD | Serum | Up-regulated: miR-4722-5p e miR-615-3p |
| 12. Gámez-Valero et al. 2019 [51] | 10 AD 18 DLB (dementia with Lewy bodies) 15 healthy controls | AD | Plasma | Down-regulated: hsa-miR-451a e hsa-miR-21-5p, hsa-miR-23a-3p, hsa- miR-126-3p, hsa-let-7i-5p e hsa-miR-151a-3p |
| 13. Guévremont et al. 2022 [52] | 65 AD 74 MCI 89 healthy controls | AD | Plasma | Down-regulated: miR-27a-3p, miR-27b-3p e miR-324-5p Up-regulated: miR-122-5p, miR-132-3p, miR-193b-3p, miR-320a-3p, miR-365-3p, miR-885-5p |
| 14. Jia et al. 2021 [53] | Pilot study (21 controls; 23 AD3), followed by the second (216 controls; 190 AD) and third groups (153 controls; 151 AD). (139 controls; 155 AD; Amnestic mild cognitive impairment, 55 (aMCI); 51 VaD; 53 PDD; 53 bvFTD; 52 DLB) | AD | Serum | Down-regulated: miR-139-3p, miR-143-3p, miR-146a-5p, miR-485-5p Up-regulated: miR-10a-5P, miR-26b-5p e miR-451a-5p |
| 15. Grossi et al. 2021 [54] | 15 PD 14 healthy controls | PD | Plasma | Up-regulated: miR-34a-5p |
| 16. Chen et al. 2021 [55] | 151 PD 21 Patients with multiple system atrophy 138 healthy controls | PD | Plasma | Up-regulated: miR-133b, miR-221-3p e miR-4454 |
| 17. Manna et al. 2021 [56] | 40 PD 20 Progressive Supranuclear Palsy 33 healthy controls | PD | Serum | Up-regulated: miR-21-3p, miR-22-3p e miR-223-5p |
| 18. Cai et al. 2021 [57] | 5 PD 7 healthy controls | PD | Plasma | Down-regulated: miR-23b3p, miR-30b-5p, miR-342-3p Up-regulated: miR-195-3p and miR-195-5p |
| 19. He et al. 2021 [58] | 72 PD 31 healthy controls | PD | Serum | Up-regulated: hsa-miR-374a-5p, hsa-miR-374b-5p, hsa-miR-199a-3p, hsa-miR-28-5p, hsa-miR-22-5p e hsa-miR-151a-5p |
| 20. Baghi et al. 2021 [59] | 20 PD 20 healthy controls | PD | Serum | Up-regulated: miR-193b |
| 21. Jiang et al. 2021 [60] | 68 PD 50 healthy controls | PD | Serum | Down-regulated: miR-374a-5p |
| 22. Lin et al. 2021 [61] | 92 PD 64 healthy controls | PD | Serum | Up-regulated: miR-485-3p |
| 23. Gui et al. 2015 [62] | 47 PD 27 healthy controls | PD | LCR | Down-regulated: miR-1 e miR-19b-3p Up-regulated: miR-153, miR-409-3p, miR-10a-5p e let-7g-3p |
| 24. Vallelunga et al. 2019 [63] | 56 PD 49 Multiple System Atrophy 50 healthy controls | PD | Plasma; Serum; LCR | Up-regulated: miR-30c-5p and miR148b-5p |
| 25. Starhof et al. 2019 [64] | 37 PD; 29 atypical Parkinson’sdisorder; 32 atypical Parkinson’s (AP) spectrum; 23 healthy controls. | PD | LCR | Up-regulated: miR-7-5p Down-regulated: miR-331-5p e miR-145-5p, miR-9-3p, miR-106b-5p |
| Studies (AD) | Accuracy (%) Mean | StDev | Mean = 84.37 ± 7.94% | 95% CI |
|---|---|---|---|---|
| 1 [39] | 71.667 | 1.528 |  | (70.276; 73.058) |
| 2 [40] | 83.667 | 1.528 | (82.276; 85.058) | |
| 3 [41] | 91.500 | 0.500 | (90.109; 92.891) | |
| 4 [42] | 94.500 | 0.500 | (93.109; 95.891) | |
| 5 [43] | 85.467 | 1.365 | (84.076; 86.858) | |
| 6 [44] | 75.500 | 0.500 | (74.109; 76.891) | |
| 7 [45] | 72.167 | 0.764 | (70.776; 73.558) | |
| 8 [46] | 96.333 | 1.528 | (94.942; 97.724) | |
| 9 [47] | 81.167 | 0.764 | (79.776; 82.558) | |
| 10 [48] | 80.167 | 0.764 | (78.776; 81.558) | |
| 11 [49] | 87.333 | 1.528 | (85.942; 88.724) | |
| 12 [50] | 91.000 | 1.000 | (89.609; 92.391) | |
| 13 [51] | 80.333 | 1.528 | (78.942; 81.724) | |
| 14 [52] | 90.333 | 1.528 | (88.942; 91.724) |
| Studies | Grouping | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 8 | A | ||||||||
| 4 | A | B | |||||||
| 3 | B | C | |||||||
| 12 | B | C | |||||||
| 14 | C | D | |||||||
| 11 | D | E | |||||||
| 5 | E | F | |||||||
| 2 | F | G | |||||||
| 9 | G | ||||||||
| 13 | G | ||||||||
| 10 | G | ||||||||
| 6 | H | ||||||||
| 7 | H | I | |||||||
| 1 | I | ||||||||
| Studies (PD) | Mean | StDev | Mean = 84.32 ± 7.15% | 95% CI |
|---|---|---|---|---|
| 15 [53] | 73.867 | 1.102 | 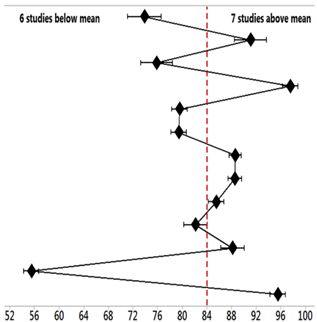 | (73.030; 74.703) |
| 16 [54] | 91.067 | 1.050 | (90.230; 91.903) | |
| 17 [55] | 75.833 | 1.041 | (74.997; 76.670) | |
| 18 [56] | 97.533 | 0.503 | (96.697; 98.370) | |
| 19 [57] | 79.567 | 0.513 | (78.730; 80.403) | |
| 20 [58] | 79.433 | 0.513 | (78.597; 80.270) | |
| 21 [59] | 88.600 | 0.400 | (87.763; 89.437) | |
| 22 [60] | 88.533 | 0.451 | (87.697; 89.370) | |
| 23 [61] | 85.533 | 0.503 | (84.697; 86.370) | |
| 24 [62] | 82.167 | 0.764 | (81.330; 83.003) | |
| 25 [63] | 88.167 | 0.764 | (87.330; 89.003) | |
| 1 [39] | 55.500 | 0.500 | (54.663; 56.337) | |
| 2 [40] | 95.500 | 0.500 | (94.663; 96.337) |
| Studies | Grouping | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 18 | A | ||||||||
| 2 | A | ||||||||
| 16 | B | ||||||||
| 21 | C | ||||||||
| 22 | C | ||||||||
| 25 | C | ||||||||
| 23 | D | ||||||||
| 24 | E | ||||||||
| 19 | F | ||||||||
| 20 | F | ||||||||
| 17 | G | ||||||||
| 15 | G | ||||||||
| 1 | H | ||||||||
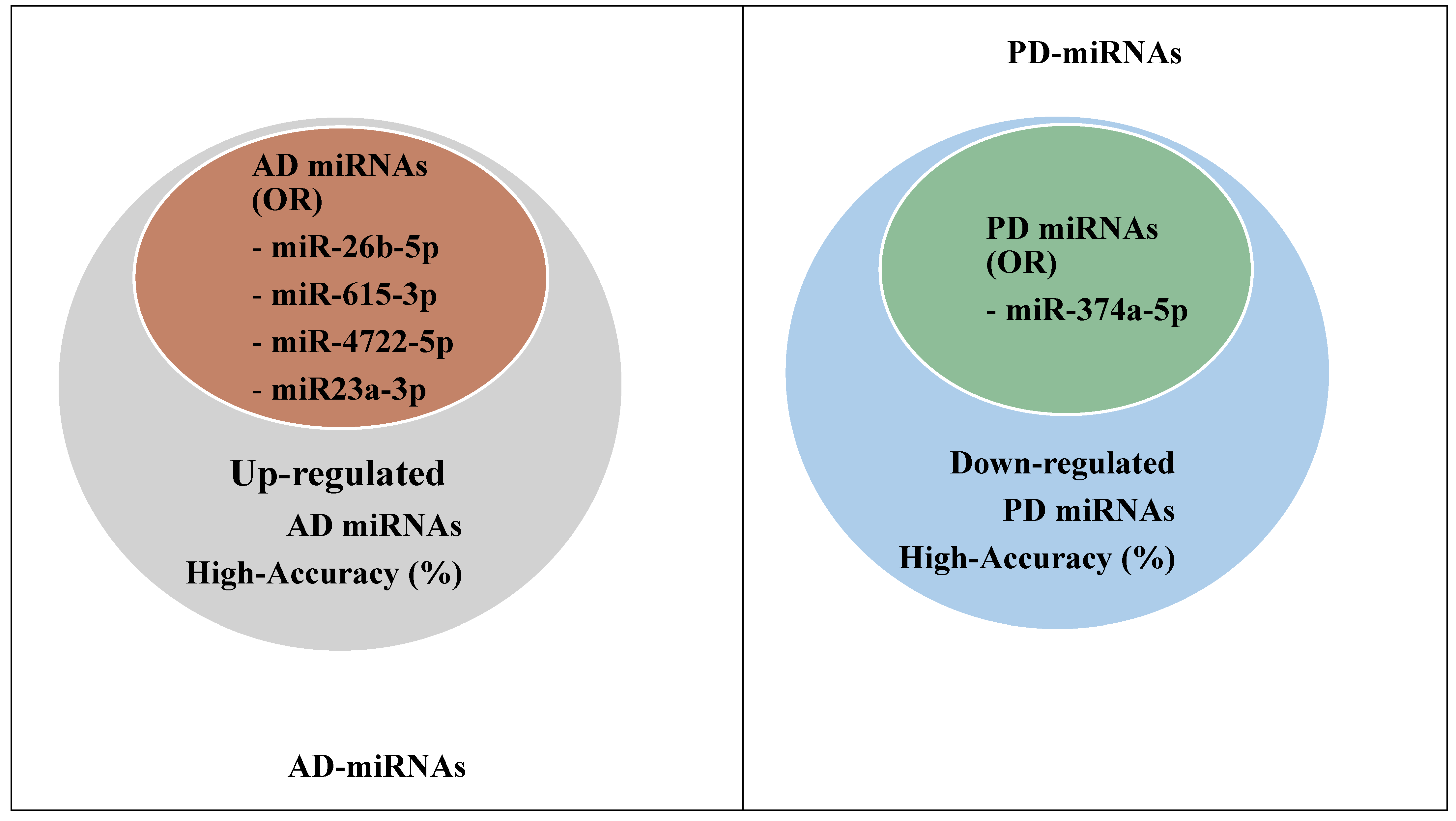
| AD/PD | miRNAs | Odds Ratio (OR)/ p-Value (95% CI) |
|---|---|---|
| AD | miR-26b-5p miR-615-3p miR-4722-5p miR23a-3p miR-27b-3p | OR = 2.55 (1.023–3.432); p = 0.004 < 0.05 |
| PD | miR-374a-5p | OR = 2.16 (0.087–3.567); p = 0.0035 < 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zotarelli-Filho, I.J.; Mogharbel, B.F.; Irioda, A.C.; Stricker, P.E.F.; de Oliveira, N.B.; Saçaki, C.S.; Perussolo, M.C.; da Rosa, N.N.; Lührs, L.; Dziedzic, D.S.M.; et al. State of the Art of microRNAs Signatures as Biomarkers and Therapeutic Targets in Parkinson’s and Alzheimer’s Diseases: A Systematic Review and Meta-Analysis. Biomedicines 2023, 11, 1113. https://doi.org/10.3390/biomedicines11041113
Zotarelli-Filho IJ, Mogharbel BF, Irioda AC, Stricker PEF, de Oliveira NB, Saçaki CS, Perussolo MC, da Rosa NN, Lührs L, Dziedzic DSM, et al. State of the Art of microRNAs Signatures as Biomarkers and Therapeutic Targets in Parkinson’s and Alzheimer’s Diseases: A Systematic Review and Meta-Analysis. Biomedicines. 2023; 11(4):1113. https://doi.org/10.3390/biomedicines11041113
Chicago/Turabian StyleZotarelli-Filho, Idiberto José, Bassam Felipe Mogharbel, Ana Carolina Irioda, Priscila Elias Ferreira Stricker, Nathalia Barth de Oliveira, Claudia Sayuri Saçaki, Maiara Carolina Perussolo, Nádia Nascimento da Rosa, Larissa Lührs, Dilcele Silva Moreira Dziedzic, and et al. 2023. "State of the Art of microRNAs Signatures as Biomarkers and Therapeutic Targets in Parkinson’s and Alzheimer’s Diseases: A Systematic Review and Meta-Analysis" Biomedicines 11, no. 4: 1113. https://doi.org/10.3390/biomedicines11041113
APA StyleZotarelli-Filho, I. J., Mogharbel, B. F., Irioda, A. C., Stricker, P. E. F., de Oliveira, N. B., Saçaki, C. S., Perussolo, M. C., da Rosa, N. N., Lührs, L., Dziedzic, D. S. M., Vaz, R. S., & de Carvalho, K. A. T. (2023). State of the Art of microRNAs Signatures as Biomarkers and Therapeutic Targets in Parkinson’s and Alzheimer’s Diseases: A Systematic Review and Meta-Analysis. Biomedicines, 11(4), 1113. https://doi.org/10.3390/biomedicines11041113






