Modulation of Klebsiella pneumoniae Outer Membrane Vesicle Protein Cargo under Antibiotic Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Curves
2.2. Genome Sequencing, Assembly, and Annotation
2.3. OMV Isolation
2.4. Nanoparticle Tracking Analysis
2.5. Transmission Electron Microscopy
2.6. Protein Extraction and Proteomic Analysis
3. Results
3.1. KpHCD1 Genome Assembly and Annotation
3.2. KpHCD1 Growth Curve in the Presence of Antibiotics
3.3. KpHCD1 OMV Characterization
3.4. Profiling KpHCD1 OMVs’ Proteome
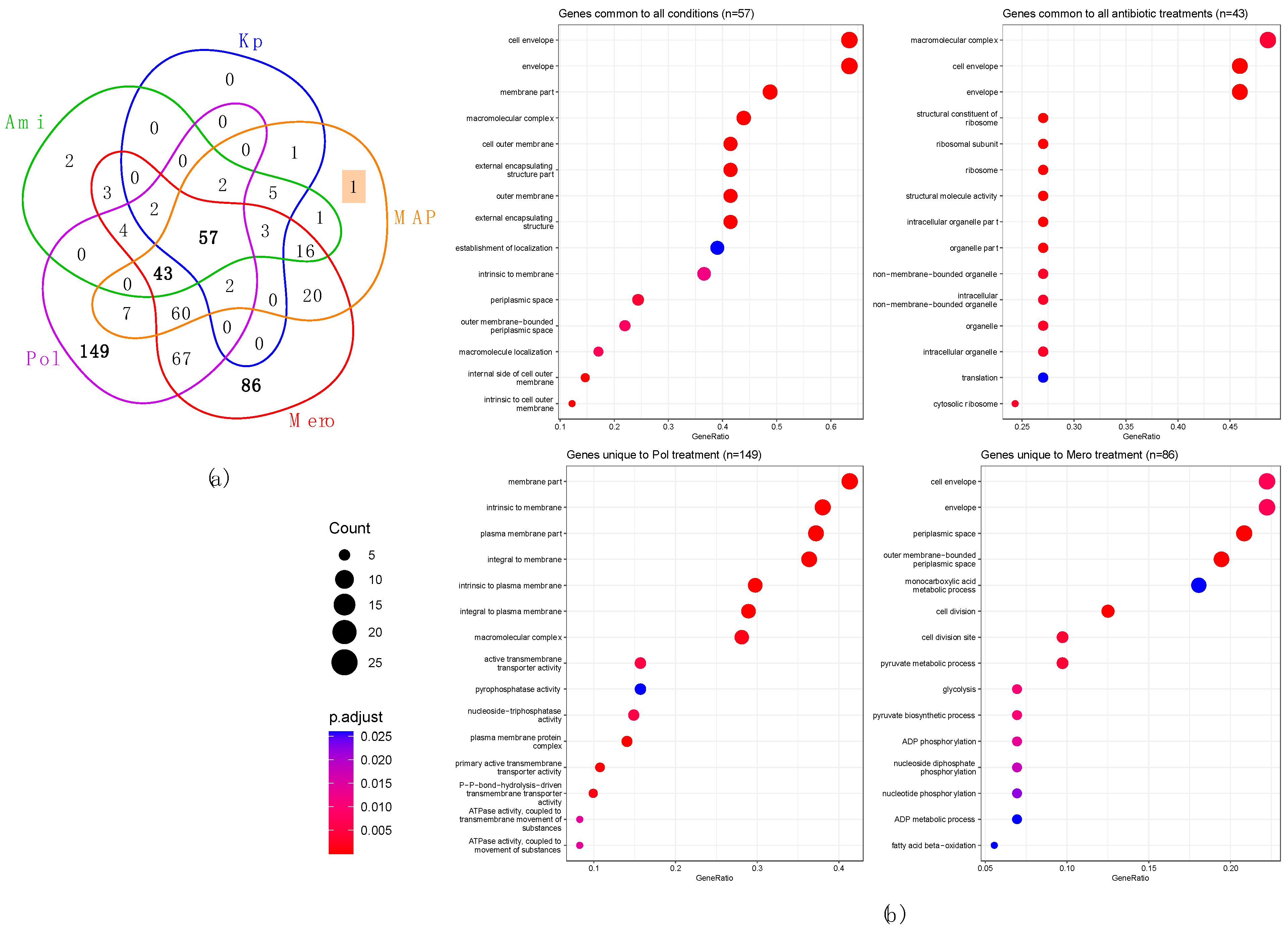
3.5. Perturbed Proteins in KpHCD1-OMVs after Treatments with Different Antibiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population Genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal Transfer of the OXA-24 Carbapenemase Gene via Outer Membrane Vesicles: A New Mechanism of Dissemination of Carbapenem Resistance Genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mondal, A.; Mitra, S.; Basu, S. Acinetobacter baumannii Transfers the BlaNDM-1 Gene via Outer Membrane Vesicles. J. Antimicrob. Chemother. 2017, 72, 2201–2207. [Google Scholar] [CrossRef]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-Mediated Transfer of Virulence Genes from Escherichia coli O157:H7 to Other Enteric Bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Daniel, O.; Karch, H.; Mellmann, A. Dissemination of the BlaCTX-M-15 Gene among Enterobacteriaceae via Outer Membrane Vesicles. J. Antimicrob. Chemother. 2020, 75, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Bitto, N.J.; Chapman, R.; Pidot, S.; Costin, A.; Lo, C.; Choi, J.; D’Cruze, T.; Reynolds, E.C.; Dashper, S.G.; Turnbull, L.; et al. Bacterial Membrane Vesicles Transport Their DNA Cargo into Host Cells. Sci. Rep. 2017, 7, 7072. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-H.; Chen, C.-H.; Goodwin, J.S.; Wang, B.-Y.; Xie, H. Functional Advantages of Porphyromonas Gingivalis Vesicles. PLoS ONE 2015, 10, e0123448. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jing, X.; Meng, D.; Wu, T.; Zhou, H.; Sun, R.; Min, X.; Liu, R.; Zeng, J. Newly Detected Transmission of BlaKPC-2 by Outer Membrane Vesicles in Klebsiella pneumoniae. Curr. Med. Sci. 2023, 43, 80–85. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Dell’Aversana, C.; Doti, N.; Donadio, G.; Dal Piaz, F.; Izzo, V.; De Filippis, A.; Galdiero, M.; Altucci, L.; Boccia, G.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Are a Driving Force for Horizontal Gene Transfer. Int. J. Mol. Sci. 2021, 22, 8732. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Collins, S.M.; Brown, A.C. Bacterial Outer Membrane Vesicles as Antibiotic Delivery Vehicles. Front. Immunol. 2021, 12, 733064. [Google Scholar] [CrossRef]
- Hua, Y.; Wang, J.; Huang, M.; Huang, Y.; Zhang, R.; Bu, F.; Yang, B.; Chen, J.; Lin, X.; Hu, X.; et al. Outer Membrane Vesicles-Transmitted Virulence Genes Mediate the Emergence of New Antimicrobial-Resistant Hypervirulent Klebsiella pneumoniae. Emerg. Microbes Infect. 2022, 11, 1281–1292. [Google Scholar] [CrossRef]
- Lan, Y.; Zhou, M.; Li, X.; Liu, X.; Li, J.; Liu, W. Preliminary Investigation of Iron Acquisition in Hypervirulent Klebsiella pneumoniae Mediated by Outer Membrane Vesicles. Infect. Drug Resist. 2022, 15, 311–320. [Google Scholar] [CrossRef]
- Guerrero-Mandujano, A.; Hernández-Cortez, C.; Ibarra, J.A.; Castro-Escarpulli, G. The Outer Membrane Vesicles: Secretion System Type Zero. Traffic 2017, 18, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Munhoz da Rocha, I.F.; Amatuzzi, R.F.; Lucena, A.C.R.; Faoro, H.; Alves, L.R. Cross-Kingdom Extracellular Vesicles EV-RNA Communication as a Mechanism for Host–Pathogen Interaction. Front. Cell. Infect. Microbiol. 2020, 10, 593160. [Google Scholar] [CrossRef]
- Martora, F.; Pinto, F.; Folliero, V.; Cammarota, M.; Dell’Annunziata, F.; Squillaci, G.; Galdiero, M.; Morana, A.; Schiraldi, C.; Giovane, A.; et al. Isolation, Characterization and Analysis of pro-Inflammatory Potential of Klebsiella pneumoniae Outer Membrane Vesicles. Microb. Pathog. 2019, 136, 103719. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Ilisso, C.P.; Dell’Aversana, C.; Greco, G.; Coppola, A.; Martora, F.; Dal Piaz, F.; Donadio, G.; Falanga, A.; Galdiero, M.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the MiRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020, 8, 1985. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Li, J.; Xia, Y.; Cao, J. Outer Membrane Vesicles Derived from Hypervirulent Klebsiella pneumoniae Stimulate the Inflammatory Response. Microb. Pathog. 2021, 154, 104841. [Google Scholar] [CrossRef]
- Orench-Rivera, N.; Kuehn, M.J. Differential Packaging into Outer Membrane Vesicles upon Oxidative Stress Reveals a General Mechanism for Cargo Selectivity. Front. Microbiol. 2021, 12, 561863. [Google Scholar] [CrossRef] [PubMed]
- Cahill, B.K.; Seeley, K.W.; Gutel, D.; Ellis, T.N. Klebsiella pneumoniae O Antigen Loss Alters the Outer Membrane Protein Composition and the Selective Packaging of Proteins into Secreted Outer Membrane Vesicles. Microbiol. Res. 2015, 180, 1–10. [Google Scholar] [CrossRef]
- Nevermann, J.; Silva, A.; Otero, C.; Oyarzún, D.P.; Barrera, B.; Gil, F.; Calderón, I.L.; Fuentes, J.A. Identification of Genes Involved in Biogenesis of Outer Membrane Vesicles (OMVs) in Salmonella enterica Serovar Typhi. Front. Microbiol. 2019, 10, 104. [Google Scholar] [CrossRef]
- Kulp, A.J.; Sun, B.; Ai, T.; Manning, A.J.; Orench-Rivera, N.; Schmid, A.K.; Kuehn, M.J. Genome-Wide Assessment of Outer Membrane Vesicle Production in Escherichia coli. PLoS ONE 2015, 10, e0139200. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Piro, V.C.; Faoro, H.; Weiss, V.A.; Steffens, M.B.; Pedrosa, F.O.; Souza, E.M.; Raittz, R.T. FGAP: An Automated Gap Closing Tool. BMC Res. Notes 2014, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. PlasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Reis, F.C.G.; Borges, B.S.; Jozefowicz, L.J.; Sena, B.A.G.; Garcia, A.W.A.; Medeiros, L.C.; Martins, S.T.; Honorato, L.; Schrank, A.; Vainstein, M.H.; et al. A Novel Protocol for the Isolation of Fungal Extracellular Vesicles Reveals the Participation of a Putative Scramblase in Polysaccharide Export and Capsule Construction in Cryptococcus gattii. Msphere 2019, 4, e00080-19. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-Wide Identification of Ubiquitin Interactions Using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 13.0; EUCAST: Växjö, Sweden, 2023; p. 12. [Google Scholar]
- Doi, Y. Treatment Options for Carbapenem-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2019, 69, S565–S575. [Google Scholar] [CrossRef]
- Ramirez, M.; Tolmasky, M. Amikacin: Uses, Resistance, and Prospects for Inhibition. Molecules 2017, 22, 2267. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. In Polymyxin Antibiotics: From Laboratory Bench to Bedside; Li, J., Nation, R.L., Kaye, K.S., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; Volume 1145, pp. 55–71. ISBN 978-3-030-16371-6. [Google Scholar]
- Zapun, A.; Contretras-Martel, C.; Vernet, T. Penicillin-Binding Proteins and β-Lactam Resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef] [PubMed]
- Bauwens, A.; Kunsmann, L.; Karch, H.; Mellmann, A.; Bielaszewska, M. Antibiotic-Mediated Modulations of Outer Membrane Vesicles in Enterohemorrhagic Escherichia Coli O104:H4 and O157:H7. Antimicrob. Agents Chemother. 2017, 61, e00937-17. [Google Scholar] [CrossRef] [PubMed]
- Hadadi-Fishani, M.; Najar-Peerayeh, S.; Siadat, S.D.; Sekhavati, M.; Mohabati Mobarez, A. Isolation and Immunogenicity of Extracted Outer Membrane Vesicles from Pseudomonas aeruginosa under Antibiotics Treatment Conditions. Iran. J. Microbiol. 2021, 13, 824. [Google Scholar] [CrossRef]
- Chiang, M.-H.; Chang, F.-J.; Kesavan, D.K.; Vasudevan, A.; Xu, H.; Lan, K.-L.; Huang, S.-W.; Shang, H.-S.; Chuang, Y.-P.; Yang, Y.-S.; et al. Proteomic Network of Antibiotic-Induced Outer Membrane Vesicles Released by Extensively Drug-Resistant Elizabethkingia anophelis. Microbiol. Spectr. 2022, 10, e00262-22. [Google Scholar] [CrossRef] [PubMed]
- Hennequin, C.; Robin, F. Correlation between Antimicrobial Resistance and Virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-K.; Fung, C.-P.; Lin, J.-C.; Chen, J.-H.; Chang, F.-Y.; Chen, T.-L.; Siu, L.K. Klebsiella pneumoniae Outer Membrane Porins OmpK35 and OmpK36 Play Roles in Both Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2011, 55, 1485–1493. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Kot, B.; Piechota, M.; Szweda, P.; Mitrus, J.; Wicha, J.; Grużewska, A.; Witeska, M. Virulence Analysis and Antibiotic Resistance of Klebsiella pneumoniae Isolates from Hospitalised Patients in Poland. Sci. Rep. 2023, 13, 4448. [Google Scholar] [CrossRef]
- Ballén, V.; Gabasa, Y.; Ratia, C.; Ortega, R.; Tejero, M.; Soto, S. Antibiotic Resistance and Virulence Profiles of Klebsiella pneumoniae Strains Isolated From Different Clinical Sources. Front. Cell. Infect. Microbiol. 2021, 11, 738223. [Google Scholar] [CrossRef]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Chng, S.-S.; Ruiz, N.; Chimalakonda, G.; Silhavy, T.J.; Kahne, D. Characterization of the Two-Protein Complex in Escherichia coli Responsible for Lipopolysaccharide Assembly at the Outer Membrane. Proc. Natl. Acad. Sci. USA 2010, 107, 5363–5368. [Google Scholar] [CrossRef]
- Wu, T.; McCandlish, A.C.; Gronenberg, L.S.; Chng, S.-S.; Silhavy, T.J.; Kahne, D. Identification of a Protein Complex That Assembles Lipopolysaccharide in the Outer Membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 11754–11759. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.M.; Holden, B.S.; Coburn, J.; Taylor, M.F.; Weber, S.; Hilton, B.; Zaugg, A.L.; McEwan, C.; Carson, R.; Andersen, J.L.; et al. Proteomic Analysis of Resistance of Gram-Negative Bacteria to Chlorhexidine and Impacts on Susceptibility to Colistin, Antimicrobial Peptides, and Ceragenins. Front. Microbiol. 2019, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.K.; Miu, A.; Oh, A.; Reichelt, M.; Ho, H.; Chalouni, C.; Labadie, S.; Wang, L.; Liang, J.; Nickerson, N.N.; et al. Disrupting Gram-Negative Bacterial Outer Membrane Biosynthesis through Inhibition of the Lipopolysaccharide Transporter MsbA. Antimicrob. Agents Chemother. 2018, 62, e01142-18. [Google Scholar] [CrossRef] [PubMed]
- Trent, M.S.; Ribeiro, A.A.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. An Inner Membrane Enzyme in Salmonella and Escherichia coli That Transfers 4-Amino-4-Deoxy-l-Arabinose to Lipid A. J. Biol. Chem. 2001, 276, 43122–43131. [Google Scholar] [CrossRef]
- Nang, S.C.; Han, M.-L.; Yu, H.H.; Wang, J.; Torres, V.V.L.; Dai, C.; Velkov, T.; Harper, M.; Li, J. Polymyxin Resistance in Klebsiella pneumoniae: Multifaceted Mechanisms Utilized in the Presence and Absence of the Plasmid-Encoded Phosphoethanolamine Transferase Gene Mcr-1. J. Antimicrob. Chemother. 2019, 74, 3190–3198. [Google Scholar] [CrossRef]
- Hussein, M.; Jasim, R.; Gocol, H.; Baker, M.; Thombare, V.J.; Ziogas, J.; Purohit, A.; Rao, G.G.; Li, J.; Velkov, T. Comparative Proteomics of Outer Membrane Vesicles from Polymyxin-Susceptible and Extremely Drug-Resistant Klebsiella pneumoniae. mSphere 2023, 8, e00537-22. [Google Scholar] [CrossRef]
- Smani, Y.; Fàbrega, A.; Roca, I.; Sánchez-Encinales, V.; Vila, J.; Pachón, J. Role of OmpA in the Multidrug Resistance Phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 1806–1808. [Google Scholar] [CrossRef]
- Choi, U.; Lee, C.-R. Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [CrossRef]
- Tiku, V.; Kofoed, E.M.; Yan, D.; Kang, J.; Xu, M.; Reichelt, M.; Dikic, I.; Tan, M.-W. Outer Membrane Vesicles Containing OmpA Induce Mitochondrial Fragmentation to Promote Pathogenesis of Acinetobacter baumannii. Sci. Rep. 2021, 11, 618. [Google Scholar] [CrossRef] [PubMed]
- Pichavant, M.; Delneste, Y.; Jeannin, P.; Fourneau, C.; Brichet, A.; Tonnel, A.-B.; Gosset, P. Outer Membrane Protein A from Klebsiella pneumoniae Activates Bronchial Epithelial Cells: Implication in Neutrophil Recruitment. J. Immunol. 2003, 171, 6697–6705. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Lee, J.S.; Park, S.B.; Lee, A.R.; Jung, J.W.; Chun, J.H.; Lazarte, J.M.S.; Kim, J.; Seo, J.-S.; Kim, J.-H.; et al. The Importance of Porins and β-Lactamase in Outer Membrane Vesicles on the Hydrolysis of β-Lactam Antibiotics. Int. J. Mol. Sci. 2020, 21, 2822. [Google Scholar] [CrossRef] [PubMed]
- Kaczanowska, M.; Rydén-Aulin, M. Ribosome Biogenesis and the Translation Process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef]
- Pütsep, K.; Brändén, C.; Boman, H. Antibacterial Peptide from H. pylori. Nature 1999, 398, 671–672. [Google Scholar] [CrossRef] [PubMed]
- Pidutti, P.; Federici, F.; Brandi, J.; Manna, L.; Rizzi, E.; Marini, U.; Cecconi, D. Purification and Characterization of Ribosomal Proteins L27 and L30 Having Antimicrobial Activity Produced by the Lactobacillus salivarius SGL 03. J. Appl. Microbiol. 2018, 124, 398–407. [Google Scholar] [CrossRef]
- Hurtado-Rios, J.J.; Carrasco-Navarro, U.; Almanza-Pérez, J.C.; Ponce-Alquicira, E. Ribosomes: The New Role of Ribosomal Proteins as Natural Antimicrobials. Int. J. Mol. Sci. 2022, 23, 9123. [Google Scholar] [CrossRef]
- McCoy, L.S.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Velkov, T.; Li, J.; Tor, Y. Polymyxins and Analogues Bind to Ribosomal RNA and Interfere with Eukaryotic Translation in Vitro. ChemBioChem 2013, 14, 2083–2086. [Google Scholar] [CrossRef]
- Andrade, J.M.; dos Santos, R.F.; Chelysheva, I.; Ignatova, Z.; Arraiano, C.M. The RNA-binding Protein Hfq Is Important for Ribosome Biogenesis and Affects Translation Fidelity. EMBO J. 2018, 37, e97631. [Google Scholar] [CrossRef]
- Chiang, M.-K.; Lu, M.-C.; Liu, L.-C.; Lin, C.-T.; Lai, Y.-C. Impact of Hfq on Global Gene Expression and Virulence in Klebsiella pneumoniae. PLoS ONE 2011, 6, e22248. [Google Scholar] [CrossRef]
- Ge, X.; Lyu, Z.-X.; Liu, Y.; Wang, R.; Zhao, X.S.; Fu, X.; Chang, Z. Identification of FkpA as a Key Quality Control Factor for the Biogenesis of Outer Membrane Proteins under Heat Shock Conditions. J. Bacteriol. 2014, 196, 672–680. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Yu, G.; Li, T.; Bai, L.; Yang, Y.; Xue, Z.; Pang, Y.; Reichmann, D.; Hiller, S.; He, L.; et al. Chaperone Spy Protects Outer Membrane Proteins from Folding Stress via Dynamic Complex Formation. mBio 2021, 12, e02130-21. [Google Scholar] [CrossRef] [PubMed]
- Sklar, J.G.; Wu, T.; Kahne, D.; Silhavy, T.J. Defining the Roles of the Periplasmic Chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Onishi, M.; Nishi, K.; Kawasaki, K.; Watanabe, K. Enhancement of Membrane Vesicle Production by Disrupting the DegP Gene in Meiothermus ruber H328. AMB Express 2021, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.M.; Lyseng-Williamson, K.A.; Keam, S.J. Meropenem: A Review of Its Use in the Treatment of Serious Bacterial Infections. Drugs 2008, 68, 803–838. [Google Scholar] [CrossRef]
- Reimer, S.L.; Beniac, D.R.; Hiebert, S.L.; Booth, T.F.; Chong, P.M.; Westmacott, G.R.; Zhanel, G.G.; Bay, D.C. Comparative Analysis of Outer Membrane Vesicle Isolation Methods with an Escherichia coli TolA Mutant Reveals a Hypervesiculating Phenotype with Outer-Inner Membrane Vesicle Content. Front. Microbiol. 2021, 12, 628801. [Google Scholar] [CrossRef]
- Tsang, M.-J.; Yakhnina, A.A.; Bernhardt, T.G. NlpD Links Cell Wall Remodeling and Outer Membrane Invagination during Cytokinesis in Escherichia coli. PLOS Genet. 2017, 13, e1006888. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Han, M.-L.; Zhao, J.; Zhu, Y.; Rao, G.; Forrest, A.; Song, J.; Kaye, K.S.; Hertzog, P.; Purcell, A.; et al. Synergistic Combination of Polymyxin B and Enrofloxacin Induced Metabolic Perturbations in Extensive Drug-Resistant Pseudomonas aeruginosa. Front. Pharmacol. 2019, 10, 1146. [Google Scholar] [CrossRef]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-Negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Warner, D.M.; Levy, S.B. Different Effects of Transcriptional Regulators MarA, SoxS and Rob on Susceptibility of Escherichia coli to Cationic Antimicrobial Peptides (CAMPs): Rob-Dependent CAMP Induction of the MarRAB Operon. Microbiology 2010, 156, 570–578. [Google Scholar] [CrossRef]




| Protein IDs | Protein Name | ARGs | Intensity 1 | Treatment 2 |
|---|---|---|---|---|
| Kpim_31130 | Outer membrane protein A | OmpA | 3.25 × 1011 | All |
| Kpim_36000 | Outer membrane protein N | OmpK37 | 1.38 × 1011 | All |
| Kpim_15180; Kpim_08710 | Elongation factor Tu 1 | EF-Tu | 1.96 × 1010 | All |
| Kpim_53330 | Carbapenem-hydrolyzing beta-lactamase KPC | KPC-2 | 5.39 ×109 | All |
| Kpim_20790 | LPS-assembly protein LptD | LptD | 3.67 × 109 | MERO, AMI, POL, MAP |
| Kpim_00390 | Efflux pump periplasmic linker BepF | oqxA | 1.69 × 109 | MERO, AMI, POL, MAP |
| Kpim_00400 | Efflux pump membrane transporter BepE | oqxB | 1.13 × 109 | MERO, POL, MAP |
| Kpim_30630 | Lipid A export ATP-binding/permease protein MsbA | msbA | 8.35 × 108 | MERO, POL, MAP |
| Kpim_08900 | cAMP-activated global transcriptional regulator CRP | CRP | 6.86 × 108 | MERO |
| Kpim_43440 | DNA-binding protein H-NS | H-NS | 3.00 × 108 | POL, MAP |
| Kpim_09910 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | ArnT | 1.75 × 108 | POL |
| Kpim_51580 | Beta-lactamase TEM | TEM-1 | 1.50 × 108 | AMI |
| Kpim_30850 | Beta-lactamase CTX-M-1 | CTX-M-15 | 1.17 × 108 | POL |
| Kpim_21190 | Peptidoglycan D, D-transpeptidase Ftsl | PBP3 | 1.13 × 108 | MERO |
| Kpim_05620 | DNA topoisomerase 4 subunit A | parC | 1.50 × 107 | MERO |
| VF * Category | Protein Ids | VF * Name | Protein Name | Intensity 1 | Treatment 2 |
|---|---|---|---|---|---|
| Adherence | Kpim_08900 | Type IV pili | cAMP activated global transcriptional regulator CRP | 6.86 × 108 | MERO |
| Kpim_15180 | EF-Tu | Elongation factor Tu-1 | 1.96 × 1010 | All | |
| Kpim_16990 | Hsp60 | 60 kDa chaperonin | 4.09 × 109 | All | |
| Kpim_22480 | IlpA | D methionine binding lipoprotein MetQ | 5.29 × 109 | MAP, MERO, POL | |
| Antimicrobial activity/Competitive advantage | Kpim_08190 | AcrAB | Multidrug efflux pump subunit AcrA | 2.33 × 1010 | All |
| Kpim_08200 | AcrAB | Multidrug efflux pump subunit AcrB | 1.26 × 1010 | All | |
| Biofilm | Kpim_00400 | AdeFGH efflux pump | Efflux pump membrane transporter BepE | 1.13 × 109 | MAP, MERO, POL |
| Kpim_03480 | Type 3 fimbriae | Outer membrane usher protein HtrE | 8.66 × 107 | MAP, MERO | |
| Kpim_03500 | Type 3 fimbriae | Hypothetical protein | 6.30 × 108 | AMI, MAP, MERO, POL | |
| Immune modulation | Kpim_11280 | LOS | 3 deoxy D manno octulosonic acid transferase | 1.10 × 108 | POL |
| Kpim_30630 | LOS | Lipid A export ATP binding/permease protein MsbA | 8.35 × 108 | MAP, MERO, POL | |
| Kpim_31890 | LOS | Lipid A biosynthesis lauroyltransferase | 1.21 × 108 | POL | |
| Kpim_32150 | LPS | Acyl carrier protein | 4.38 × 108 | KP, MAP | |
| Kpim_43450 | LOS | UTP glucose 1 phosphate uridylyltransferase | 1.59 × 108 | MAP, MERO | |
| Kpim_46450 | Capsule | 6 phosphogluconate dehydrogenase, decarboxylating | 2.12 × 107 | MERO | |
| Kpim_46600 | Capsule | Putative tyrosine protein kinase in cps region | 2.87 × 109 | MAP, MERO, POL | |
| Kpim_46610 | Capsule | Hypothetical protein | 6.01 × 109 | AMI, MAP, MERO, POL | |
| Kpim_46620 | Capsule | Hypothetical protein | 5.28 × 109 | All | |
| Invasion | Kpim_31130 | OmpA | Outer membrane protein A | 3.25 × 1011 | All |
| Nutritional/ Metabolic factor | Kpim_09560 | GGT | Glutathione hydrolase proenzyme | 9.62 × 108 | AMI, MERO |
| Kpim_18230 | MgtBC | Magnesium transporting ATPase, P type 1 | 4.29 × 108 | MAP, MERO, POL | |
| Kpim_20690 | Pyrimidine biosynthesis | Carbamoyl phosphate synthase large chain | 7.81 × 108 | POL | |
| Kpim_26680 | Ent | Ferrienterobactin receptor | 1.42 × 1010 | AMI, MAP, MERO, POL | |
| Kpim_32310 | Aerobactin | Ferric aerobactin receptor | 2.93 × 1010 | All | |
| Regulation | Kpim_01780 | RpoS | RNA polymerase sigma factor RpoD | 8.44 × 108 | POL |
| Kpim_32500 | PhoPQ | Sensor protein PhoQ | 2.14 × 108 | POL | |
| Stress survival | Kpim_51330 | ClpC | ATP-dependent Clp protease ATP-binding subunit ClpC | 4.07 × 108 | MAP, MERO, POL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucena, A.C.R.; Ferrarini, M.G.; de Oliveira, W.K.; Marcon, B.H.; Morello, L.G.; Alves, L.R.; Faoro, H. Modulation of Klebsiella pneumoniae Outer Membrane Vesicle Protein Cargo under Antibiotic Treatment. Biomedicines 2023, 11, 1515. https://doi.org/10.3390/biomedicines11061515
Lucena ACR, Ferrarini MG, de Oliveira WK, Marcon BH, Morello LG, Alves LR, Faoro H. Modulation of Klebsiella pneumoniae Outer Membrane Vesicle Protein Cargo under Antibiotic Treatment. Biomedicines. 2023; 11(6):1515. https://doi.org/10.3390/biomedicines11061515
Chicago/Turabian StyleLucena, Aline Castro Rodrigues, Mariana Galvão Ferrarini, Willian Klassen de Oliveira, Bruna Hilzendeger Marcon, Luis Gustavo Morello, Lysangela Ronalte Alves, and Helisson Faoro. 2023. "Modulation of Klebsiella pneumoniae Outer Membrane Vesicle Protein Cargo under Antibiotic Treatment" Biomedicines 11, no. 6: 1515. https://doi.org/10.3390/biomedicines11061515





