Role of Artificial Intelligence for Autism Diagnosis Using DTI and fMRI: A Survey
Abstract
1. Introduction
2. MRI Findings for ASD
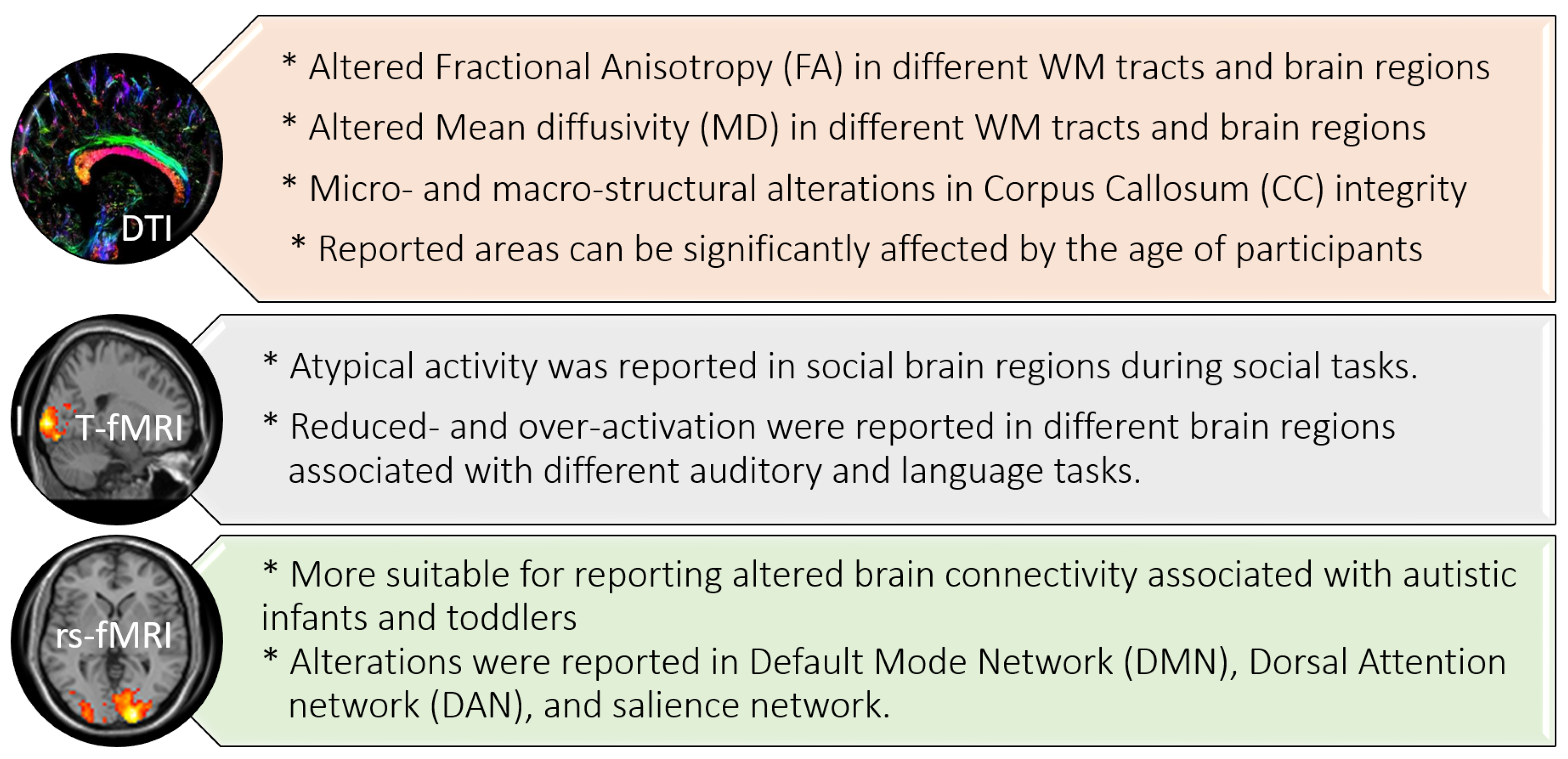
2.1. ASD Findings Using Diffusion Tensor Imaging (DTI)
2.2. ASD Findings Using fMRI
2.2.1. Task-Based (T-fMRI)
2.2.2. Resting-State (rs-fMRI)
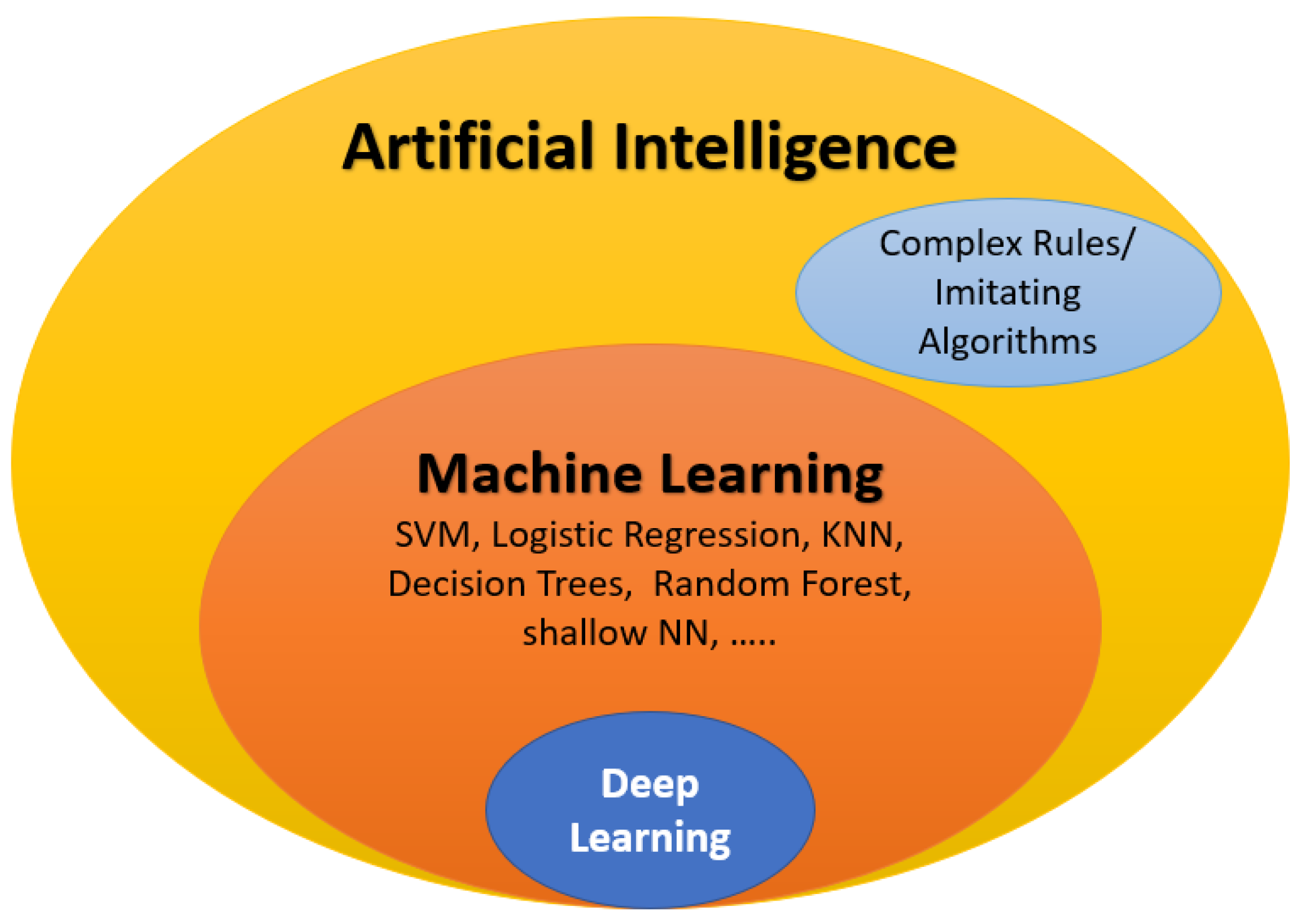
3. The Role of AI in ASD Diagnosis
- Specificity: ;
- Sensitivity (recall): ;
- Accuracy: ;
- Precision: ;
- F1-score: ;
- The AUC is the region beneath the receiver operating characteristics (ROC) curve that shows the relationship between the false positive rate (1-specificity, shown on the x-axis) and the true positive rate (sensitivity, on the y-axis). The AUC value ranges from 0 to 1, with a higher AUC value indicating better performance.
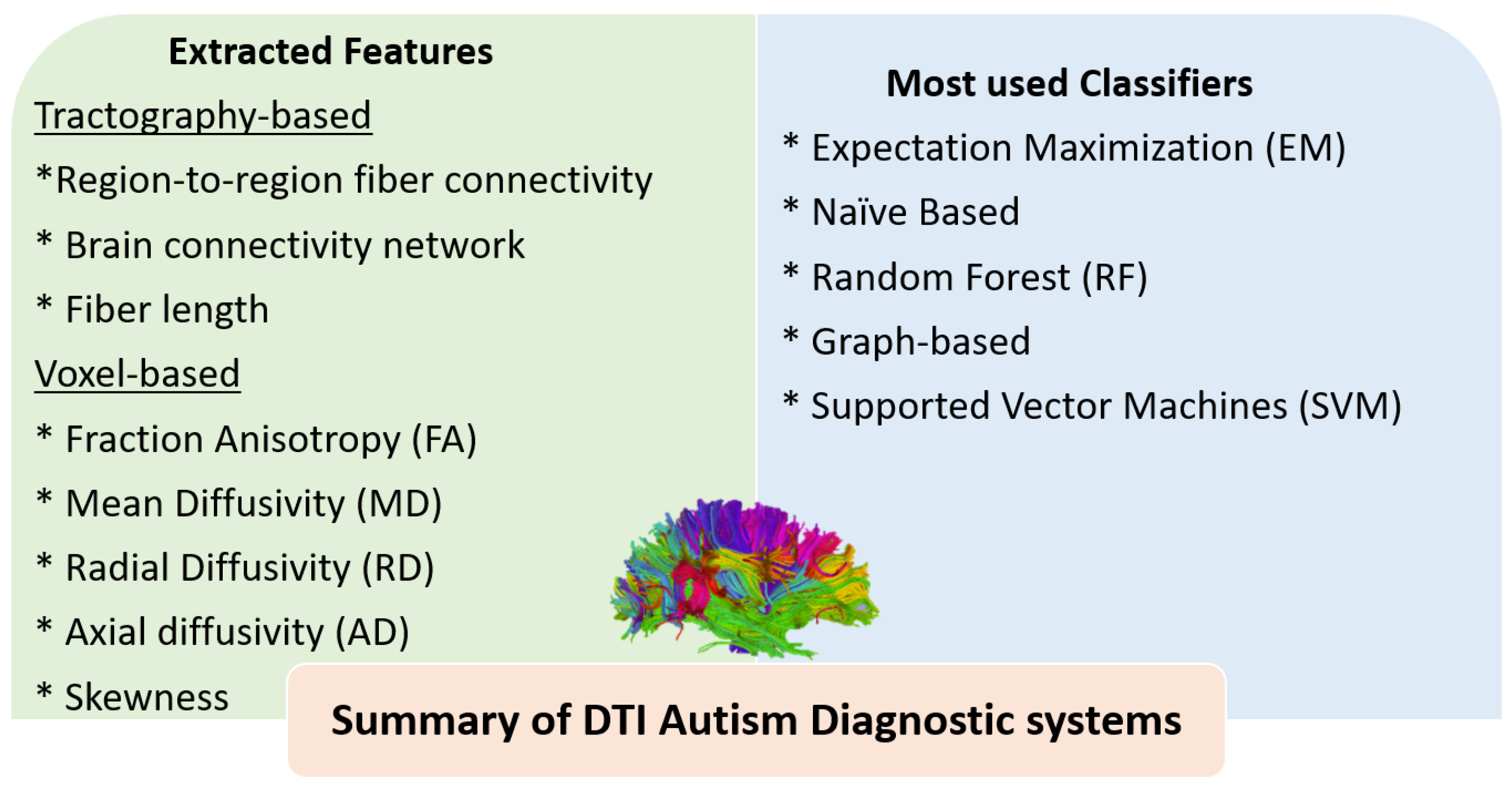
3.1. The Role of AI for ASD Diagnosis Using DTI
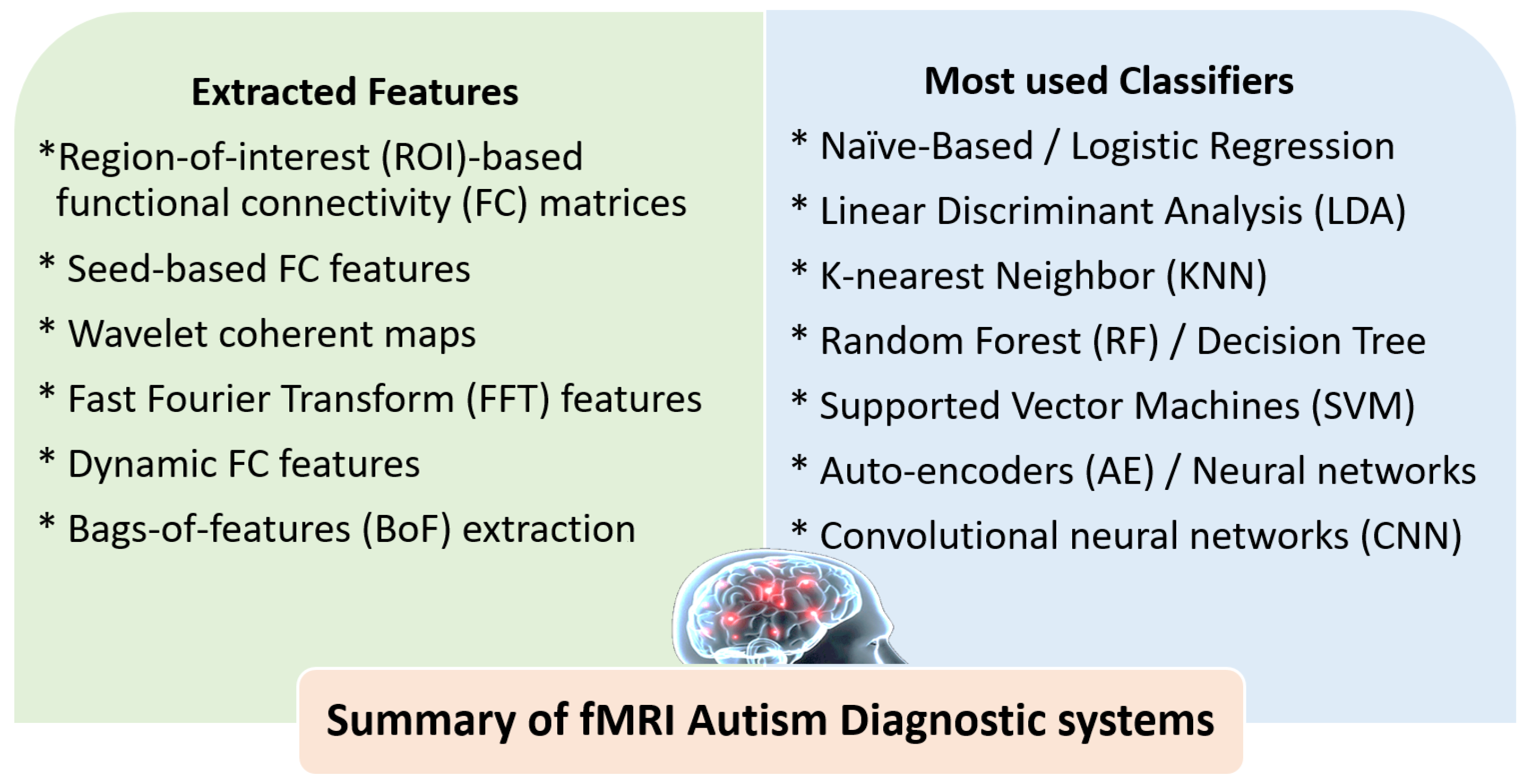
3.2. The Role of AI for ASD Diagnosis Using fMRI
4. Discussions and Future Trends
4.1. Summary of ASD Findings Using DTI and fMRI
4.2. Summary of ASD Diagnostic Systems Using DTI and fMRI
4.3. Strengths of Using DTI and fMRI for ASD Diagnosis
4.4. Limitations of Using DTI and fMRI for ASD Diagnosis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABIDE | Autism Brain Imaging Data Exchange |
| NDAR | National Database of Autism Research |
| ADI-R | Autism Diagnostic Interview-Revised |
| ADOS | Autism Diagnostic Observation Schedule |
| MRI | Magnetic resonance imaging |
| sMRI | Structural MRI |
| dwMRI | DWI | Diffusion-weighted MRI |
| DTI | Diffusion tensor imaging |
| fMRI | Functional MRI |
| rs-fMRI | Resting-state functional MRI |
| T-fMRI | Task-based functional MRI |
References
- Rafiee, F.; Rezvani Habibabadi, R.; Motaghi, M.; Yousem, D.M.; Yousem, I.J. Brain mri in autism spectrum disorder: Narrative review and recent advances. J. Magn. Reson. Imaging 2022, 55, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Joudar, S.S.; Albahri, A.; Hamid, R.A. Triage and priority-based healthcare diagnosis using artificial intelligence for autism spectrum disorder and Gene contribution: A systematic review. Comput. Biol. Med. 2022, 146, 105553. [Google Scholar] [CrossRef]
- van’t Hof, M.; Tisseur, C.; van Berckelear-Onnes, I.; van Nieuwenhuyzen, A.; Daniels, A.M.; Deen, M.; Hoek, H.W.; Ester, W.A. Age at autism spectrum disorder diagnosis: A systematic review and meta-analysis from 2012 to 2019. Autism 2021, 25, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Duda, M.; Zhang, H.; Li, H.D.; Wall, D.P.; Burmeister, M.; Guan, Y. Brain-specific functional relationship networks inform autism spectrum disorder gene prediction. Transl. Psychiatry 2018, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Song, D.Y.; Topriceanu, C.C.; Ilie-Ablachim, D.C.; Kinali, M.; Bisdas, S. Machine learning with neuroimaging data to identify autism spectrum disorder: A systematic review and meta-analysis. Neuroradiology 2021, 63, 2057–2072. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Calhoun, V.; Jiang, R.; Yan, W.; Sui, J. Brain imaging-based machine learning in autism spectrum disorder: Methods and applications. J. Neurosci. Methods 2021, 361, 109271. [Google Scholar] [CrossRef]
- Moridian, P.; Ghassemi, N.; Jafari, M.; Salloum-Asfar, S.; Sadeghi, D.; Khodatars, M.; Shoeibi, A.; Khosravi, A.; Ling, S.H.; Subasi, A.; et al. Automatic autism spectrum disorder detection using artificial intelligence methods with MRI neuroimaging: A review. arXiv 2022, arXiv:2206.11233. [Google Scholar] [CrossRef]
- Shoeibi, A.; Ghassemi, N.; Khodatars, M.; Moridian, P.; Khosravi, A.; Zare, A.; Gorriz, J.M.; Chale-Chale, A.H.; Khadem, A.; Acharya, U.R. Automatic Diagnosis of Schizophrenia and Attention Deficit Hyperactivity Disorder in rs-fMRI Modality using Convolutional Autoencoder Model and Interval Type-2 Fuzzy Regression. arXiv 2022, arXiv:2205.15858. [Google Scholar] [CrossRef]
- Li, D.; Karnath, H.O.; Xu, X. Candidate biomarkers in children with autism spectrum disorder: A review of MRI studies. Neurosci. Bull. 2017, 33, 219–237. [Google Scholar] [CrossRef]
- Sui, J.; Jiang, R.; Bustillo, J.; Calhoun, V. Neuroimaging-based individualized prediction of cognition and behavior for mental disorders and health: Methods and promises. Biol. Psychiatry 2020, 88, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Nogay, H.S.; Adeli, H. Machine learning (ML) for the diagnosis of autism spectrum disorder (ASD) using brain imaging. Rev. Neurosci. 2020, 31, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Parikh, M.N.; Li, H.; He, L. Enhancing diagnosis of autism with optimized machine learning models and personal characteristic data. Front. Comput. Neurosci. 2019, 13, 9. [Google Scholar] [CrossRef]
- Rajagopalan, V.; Jiang, Z.; Stojanovic-Radic, J.; Yue, G.; Pioro, E.P.; Wylie, G.; Das, A. A basic introduction to diffusion tensor imaging mathematics and image processing steps. Brain Disord. Ther. 2017, 6, 2. [Google Scholar] [CrossRef]
- Razek, A.A.K.A.; Baky, K.A.; Helmy, E. Diffusion Tensor Imaging in Characterization of Mediastinal Lymphadenopathy. Acad. Radiol. 2022, 29, S165–S172. [Google Scholar] [CrossRef]
- Razek, A.A.K.A.; Helmy, E.M.; Maher, H.; Kasem, M.A. Diffusion tensor imaging of the lateral rectus muscle in Duane retraction syndrome. J. Comput. Assist. Tomogr. 2019, 43, 467–471. [Google Scholar] [CrossRef]
- Lazar, M.; Miles, L.M.; Babb, J.S.; Donaldson, J.B. Axonal deficits in young adults with High Functioning Autism and their impact on processing speed. NeuroImage Clin. 2014, 4, 417–425. [Google Scholar] [CrossRef]
- Hrdlicka, M.; Sanda, J.; Urbanek, T.; Kudr, M.; Dudova, I.; Kickova, S.; Pospisilova, L.; Mohaplova, M.; Maulisova, A.; Krsek, P.; et al. Diffusion tensor imaging and tractography in autistic, dysphasic, and healthy control children. Neuropsychiatr. Dis. Treat. 2019, 15, 2843. [Google Scholar] [CrossRef]
- Jung, M.; Tu, Y.; Lang, C.A.; Ortiz, A.; Park, J.; Jorgenson, K.; Kong, X.J.; Kong, J. Decreased structural connectivity and resting-state brain activity in the lateral occipital cortex is associated with social communication deficits in boys with autism spectrum disorder. Neuroimage 2019, 190, 205–212. [Google Scholar] [CrossRef]
- Yamasaki, T.; Maekawa, T.; Fujita, T.; Tobimatsu, S. Connectopathy in autism spectrum disorders: A review of evidence from visual evoked potentials and diffusion magnetic resonance imaging. Front. Neurosci. 2017, 11, 627. [Google Scholar] [CrossRef]
- Rane, P.; Cochran, D.; Hodge, S.M.; Haselgrove, C.; Kennedy, D.; Frazier, J.A. Connectivity in autism: A review of MRI connectivity studies. Harv. Rev. Psychiatry 2015, 23, 223. [Google Scholar] [CrossRef] [PubMed]
- Valenti, M.; Pino, M.C.; Mazza, M.; Panzarino, G.; Di Paolantonio, C.; Verrotti, A. Abnormal structural and functional connectivity of the corpus callosum in autism spectrum disorders: A review. Rev. J. Autism Dev. Disord. 2020, 7, 46–62. [Google Scholar] [CrossRef]
- Shukla, D.K.; Keehn, B.; Lincoln, A.J.; Müller, R.A. White matter compromise of callosal and subcortical fiber tracts in children with autism spectrum disorder: A diffusion tensor imaging study. J. Am. Acad. Child Adolesc. Psychiatry 2010, 49, 1269–1278. [Google Scholar] [PubMed]
- McLaughlin, K.; Travers, B.G.; Dadalko, O.I.; Dean III, D.C.; Tromp, D.; Adluru, N.; Destiche, D.; Freeman, A.; Prigge, M.D.; Froehlich, A.; et al. Longitudinal development of thalamic and internal capsule microstructure in autism spectrum disorder. Autism Res. 2018, 11, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Saaybi, S.; AlArab, N.; Hannoun, S.; Saade, M.; Tutunji, R.; Zeeni, C.; Shbarou, R.; Hourani, R.; Boustany, R.M. Pre-and post-therapy assessment of clinical outcomes and white matter integrity in autism Spectrum disorder: Pilot study. Front. Neurol. 2019, 10, 877. [Google Scholar] [CrossRef]
- Vogan, V.; Morgan, B.; Leung, R.; Anagnostou, E.; Doyle-Thomas, K.; Taylor, M. Widespread white matter differences in children and adolescents with autism spectrum disorder. J. Autism Dev. Disord. 2016, 46, 2138–2147. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, R.; Yuan, Y.; Lian, D.; Qi, X.; Zheng, N.; Li, K. The value of diffusion tensor imaging for differentiating autism spectrum disorder with language delay from developmental language disorder among toddlers. Medicine 2019, 98, e15058. [Google Scholar] [CrossRef]
- Solso, S.; Xu, R.; Proudfoot, J.; Hagler, D.J., Jr.; Campbell, K.; Venkatraman, V.; Barnes, C.C.; Ahrens-Barbeau, C.; Pierce, K.; Dale, A.; et al. Diffusion tensor imaging provides evidence of possible axonal overconnectivity in frontal lobes in autism spectrum disorder toddlers. Biol. Psychiatry 2016, 79, 676–684. [Google Scholar] [CrossRef]
- Ouyang, M.; Cheng, H.; Mishra, V.; Gong, G.; Mosconi, M.W.; Sweeney, J.; Peng, Y.; Huang, H. Atypical age-dependent effects of autism on white matter microstructure in children of 2–7 years. Hum. Brain Mapp. 2016, 37, 819–832. [Google Scholar] [CrossRef]
- Thompson, A.; Shahidiani, A.; Fritz, A.; O’Muircheartaigh, J.; Walker, L.; D’Almeida, V.; Murphy, C.; Daly, E.; Murphy, D.; Williams, S.; et al. Age-related differences in white matter diffusion measures in autism spectrum condition. Mol. Autism 2020, 11, 36. [Google Scholar] [CrossRef]
- Schmithorst, V.J.; Wilke, M.; Dardzinski, B.J.; Holland, S.K. Correlation of white matter diffusivity and anisotropy with age during childhood and adolescence: A cross-sectional diffusion-tensor MR imaging study. Radiology 2002, 222, 212. [Google Scholar] [CrossRef] [PubMed]
- Payabvash, S.; Palacios, E.M.; Owen, J.P.; Wang, M.B.; Tavassoli, T.; Gerdes, M.; Brandes-Aitken, A.; Cuneo, D.; Marco, E.J.; Mukherjee, P. White matter connectome edge density in children with autism spectrum disorders: Potential imaging biomarkers using machine-learning models. Brain Connect. 2019, 9, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.H.; Vicnesh, J.; Wei, J.K.E.; Oh, S.L.; Arunkumar, N.; Abdulhay, E.W.; Ciaccio, E.J.; Acharya, U.R. Autism spectrum disorder diagnostic system using HOS bispectrum with EEG signals. Int. J. Environ. Res. Public Health 2020, 17, 971. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.M.; Smith, S.M.; Beckmann, C.F. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front. Syst. Neurosci. 2010, 4, 8. [Google Scholar] [CrossRef]
- Haweel, R.; Shalaby, A.; Mahmoud, A.; Seada, N.; Ghoniemy, S.; Ghazal, M.; Casanova, M.F.; Barnes, G.N.; El-Baz, A. A robust DWT–CNN-based CAD system for early diagnosis of autism using task-based fMRI. Med. Phys. 2021, 48, 2315–2326. [Google Scholar] [CrossRef]
- Khodatars, M.; Shoeibi, A.; Sadeghi, D.; Ghaasemi, N.; Jafari, M.; Moridian, P.; Khadem, A.; Alizadehsani, R.; Zare, A.; Kong, Y.; et al. Deep learning for neuroimaging-based diagnosis and rehabilitation of autism spectrum disorder: A review. Comput. Biol. Med. 2021, 139, 104949. [Google Scholar] [CrossRef]
- Müller, R.A.; Fishman, I. Brain connectivity and neuroimaging of social networks in autism. Trends Cogn. Sci. 2018, 22, 1103–1116. [Google Scholar] [CrossRef]
- Dichter, G.S. Functional magnetic resonance imaging of autism spectrum disorders. Dialogues Clin. Neurosci. 2022, 14, 319–351. [Google Scholar] [CrossRef]
- Pierce, K.; Redcay, E. Fusiform function in children with an autism spectrum disorder is a matter of “who”. Biol. Psychiatry 2008, 64, 552–560. [Google Scholar] [CrossRef]
- Herrington, J.D.; Nymberg, C.; Schultz, R.T. Biological motion task performance predicts superior temporal sulcus activity. Brain Cogn. 2011, 77, 372–381. [Google Scholar] [CrossRef]
- Weng, S.J.; Carrasco, M.; Swartz, J.R.; Wiggins, J.L.; Kurapati, N.; Liberzon, I.; Risi, S.; Lord, C.; Monk, C.S. Neural activation to emotional faces in adolescents with autism spectrum disorders. J. Child Psychol. Psychiatry 2011, 52, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, K.E.; Hernandez, L.M.; Eilbott, J.; Jack, A.; Aylward, E.; Gaab, N.; Van Horn, J.D.; Bernier, R.A.; Geschwind, D.H.; McPartland, J.C.; et al. Neural responsivity to social rewards in autistic female youth. Transl. Psychiatry 2020, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Philip, R.C.; Dauvermann, M.R.; Whalley, H.C.; Baynham, K.; Lawrie, S.M.; Stanfield, A.C. A systematic review and meta-analysis of the fMRI investigation of autism spectrum disorders. Neurosci. Biobehav. Rev. 2012, 36, 901–942. [Google Scholar] [CrossRef]
- Lau, W.K.; Leung, M.K.; Lau, B.W. Resting-state abnormalities in autism spectrum disorders: A meta-analysis. Sci. Rep. 2019, 9, 3892. [Google Scholar] [CrossRef]
- Sun, J.W.; Fan, R.; Wang, Q.; Wang, Q.Q.; Jia, X.Z.; Ma, H.B. Identify abnormal functional connectivity of resting state networks in Autism spectrum disorder and apply to machine learning-based classification. Brain Res. 2021, 1757, 147299. [Google Scholar] [CrossRef]
- He, C.; Chen, Y.; Jian, T.; Chen, H.; Guo, X.; Wang, J.; Wu, L.; Chen, H.; Duan, X. Dynamic functional connectivity analysis reveals decreased variability of the default-mode network in developing autistic brain. Autism Res. 2018, 11, 1479–1493. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.C.; Wang, N.Y.; Li, C.J.; Wang, H.L.S. Resting-state functional connectivity within default mode network in Chinese-speaking children with specific learning disabilities. Neuropsychiatry 2018, 8, 873–880. [Google Scholar] [CrossRef]
- Nair, S.; Jao Keehn, R.J.; Berkebile, M.M.; Maximo, J.O.; Witkowska, N.; Müller, R.A. Local resting state functional connectivity in autism: Site and cohort variability and the effect of eye status. Brain Imaging Behav. 2018, 12, 168–179. [Google Scholar] [CrossRef]
- Uddin, L.Q.; Supekar, K.; Lynch, C.J.; Khouzam, A.; Phillips, J.; Feinstein, C.; Ryali, S.; Menon, V. Salience network–based classification and prediction of symptom severity in children with autism. JAMA Psychiatry 2013, 70, 869–879. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Wang, R.; Duan, X.; Chen, H.; He, C.; Zhai, J.; Wu, L.; Chen, H. Atypical resting-state functional connectivity of intra/inter-sensory networks is related to symptom severity in young boys with autism spectrum disorder. Front. Physiol. 2021, 12, 626338. [Google Scholar] [CrossRef]
- Paakki, J.J.; Rahko, J.; Long, X.; Moilanen, I.; Tervonen, O.; Nikkinen, J.; Starck, T.; Remes, J.; Hurtig, T.; Haapsamo, H.; et al. Alterations in regional homogeneity of resting-state brain activity in autism spectrum disorders. Brain Res. 2010, 1321, 169–179. [Google Scholar] [CrossRef]
- Rausch, A.; Zhang, W.; Haak, K.V.; Mennes, M.; Hermans, E.J.; van Oort, E.; van Wingen, G.; Beckmann, C.F.; Buitelaar, J.K.; Groen, W.B. Altered functional connectivity of the amygdaloid input nuclei in adolescents and young adults with autism spectrum disorder: A resting state fMRI study. Mol. Autism 2016, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Maximo, J.O.; Keown, C.L.; Nair, A.; Müller, R.A. Approaches to local connectivity in autism using resting state functional connectivity MRI. Front. Hum. Neurosci. 2013, 7, 605. [Google Scholar] [CrossRef] [PubMed]
- Jann, K.; Hernandez, L.M.; Beck-Pancer, D.; McCarron, R.; Smith, R.X.; Dapretto, M.; Wang, D.J. Altered resting perfusion and functional connectivity of default mode network in youth with autism spectrum disorder. Brain Behav. 2015, 5, e00358. [Google Scholar] [CrossRef] [PubMed]
- Kleinhans, N.M.; Reiter, M.A.; Neuhaus, E.; Pauley, G.; Martin, N.; Dager, S.; Estes, A. Subregional differences in intrinsic amygdala hyperconnectivity and hypoconnectivity in autism spectrum disorder. Autism Res. 2016, 9, 760–772. [Google Scholar] [CrossRef]
- Ingalhalikar, M.; Parker, D.; Bloy, L.; Roberts, T.P.; Verma, R. Diffusion based abnormality markers of pathology: Toward learned diagnostic prediction of ASD. NeuroImage 2011, 57, 918–927. [Google Scholar] [CrossRef]
- Li, H.; Xue, Z.; Ellmore, T.M.; Frye, R.E.; Wong, S.T. Identification of faulty DTI-based sub-networks in autism using network regularized SVM. In Proceedings of the 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 2–5 May 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 550–553. [Google Scholar]
- Jin, Y.; Wee, C.Y.; Shi, F.; Thung, K.H.; Ni, D.; Yap, P.T.; Shen, D. Identification of infants at high-risk for autism spectrum disorder using multiparameter multiscale white matter connectivity networks. Hum. Brain Mapp. 2015, 36, 4880–4896. [Google Scholar] [CrossRef]
- Zhang, F.; Savadjiev, P.; Cai, W.; Song, Y.; Rathi, Y.; Tunç, B.; Parker, D.; Kapur, T.; Schultz, R.T.; Makris, N.; et al. Whole brain white matter connectivity analysis using machine learning: An application to autism. NeuroImage 2018, 172, 826–837. [Google Scholar] [CrossRef]
- Qin, B.; Wang, L.; Zhang, Y.; Cai, J.; Chen, J.; Li, T. Enhanced topological network efficiency in preschool autism spectrum disorder: A diffusion tensor imaging study. Front. Psychiatry 2018, 9, 278. [Google Scholar] [CrossRef]
- Saad, M.; Islam, S.M.R. Brain Connectivity Network Analysis and Classifications from Diffusion Tensor Imaging. In Proceedings of the 2019 International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 10–12 January 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 422–427. [Google Scholar]
- ElNakieb, Y.; Soliman, A.; Mahmoud, A.; Dekhil, O.; Shalaby, A.; Ghazal, M.; Khalil, A.; Switala, A.; Keynton, R.S.; Barnes, G.N.; et al. Autism spectrum disorder diagnosis framework using diffusion tensor imaging. In Proceedings of the 2019 IEEE International Conference on Imaging Systems and Techniques (IST), Abu Dhabi, United Arab Emirates, 9–10 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Elnakieb, Y.A.; Ali, M.T.; Soliman, A.; Mahmoud, A.H.; Shalaby, A.M.; Alghamdi, N.S.; Ghazal, M.; Khalil, A.; Switala, A.; Keynton, R.S.; et al. Computer aided autism diagnosis using diffusion tensor imaging. IEEE Access 2020, 8, 191298–191308. [Google Scholar] [CrossRef]
- ElNakieb, Y.; Ali, M.T.; Elnakib, A.; Shalaby, A.; Soliman, A.; Mahmoud, A.; Ghazal, M.; Barnes, G.N.; El-Baz, A. The Role of Diffusion Tensor MR Imaging (DTI) of the Brain in Diagnosing Autism Spectrum Disorder: Promising Results. Sensors 2021, 21, 8171. [Google Scholar] [CrossRef] [PubMed]
- An, M.; Ho, H.P.; Staib, L.; Pelphrey, K.; Duncan, J. Multimodal MRI analysis of brain subnetworks in autism using multi-view EM. In Proceedings of the 2010 Conference Record of the Forty Fourth Asilomar Conference on Signals, Systems and Computers, Pacific Grove, CA, USA, 7–10 November 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 786–789. [Google Scholar]
- Deshpande, G.; Libero, L.E.; Sreenivasan, K.R.; Deshpande, H.D.; Kana, R.K. Identification of neural connectivity signatures of autism using machine learning. Front. Hum. Neurosci. 2013, 7, 670. [Google Scholar] [CrossRef] [PubMed]
- Crimi, A.; Dodero, L.; Murino, V.; Sona, D. Case-control discrimination through effective brain connectivity. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 970–973. [Google Scholar]
- Dsouza, N.S.; Nebel, M.B.; Crocetti, D.; Robinson, J.; Mostofsky, S.; Venkataraman, A. M-gcn: A multimodal graph convolutional network to integrate functional and structural connectomics data to predict multidimensional phenotypic characterizations. In Proceedings of the Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021; Microtome Publishing: Brookline, MA, USA, 2021; pp. 119–130. [Google Scholar]
- Irimia, A.; Lei, X.; Torgerson, C.M.; Jacokes, Z.J.; Abe, S.; Van Horn, J.D. Support vector machines, multidimensional scaling and magnetic resonance imaging reveal structural brain abnormalities associated with the interaction between autism spectrum disorder and sex. Front. Comput. Neurosci. 2018, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Eill, A.; Jahedi, A.; Gao, Y.; Kohli, J.S.; Fong, C.H.; Solders, S.; Carper, R.A.; Valafar, F.; Bailey, B.A.; Müller, R.A. Functional connectivities are more informative than anatomical variables in diagnostic classification of autism. Brain Connect. 2019, 9, 604–612. [Google Scholar] [CrossRef]
- Abraham, A.; Milham, M.P.; Di Martino, A.; Craddock, R.C.; Samaras, D.; Thirion, B.; Varoquaux, G. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. NeuroImage 2017, 147, 736–745. [Google Scholar] [CrossRef]
- Emerson, R.W.; Adams, C.; Nishino, T.; Hazlett, H.C.; Wolff, J.J.; Zwaigenbaum, L.; Constantino, J.N.; Shen, M.D.; Swanson, M.R.; Elison, J.T.; et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci. Transl. Med. 2017, 9, eaag2882. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Dominick, K.C.; Minai, A.A.; Li, H.; Erickson, C.A.; Lu, L.J. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front. Neurosci. 2017, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Jahedi, A.; Nasamran, C.A.; Faires, B.; Fan, J.; Müller, R.A. Distributed intrinsic functional connectivity patterns predict diagnostic status in large autism cohort. Brain Connect. 2017, 7, 515–525. [Google Scholar] [CrossRef]
- Kam, T.E.; Suk, H.I.; Lee, S.W. Multiple functional networks modeling for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2017, 38, 5804–5821. [Google Scholar] [CrossRef]
- Sadeghi, M.; Khosrowabadi, R.; Bakouie, F.; Mahdavi, H.; Eslahchi, C.; Pouretemad, H. Screening of autism based on task-free fmri using graph theoretical approach. Psychiatry Res. Neuroimaging 2017, 263, 48–56. [Google Scholar] [CrossRef]
- Subbaraju, V.; Suresh, M.B.; Sundaram, S.; Narasimhan, S. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Med. Image Anal. 2017, 35, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Tejwani, R.; Liska, A.; You, H.; Reinen, J.; Das, P. Autism classification using brain functional connectivity dynamics and machine learning. arXiv 2017, arXiv:1712.08041. [Google Scholar]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage Clin. 2018, 17, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.A.; Wang, Y.; Shu, Q.; Sun, Q.; Xu, Q. Classification of autism spectrum disorder using random support vector machine cluster. Front. Genet. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Fredo, A.; Jahedi, A.; Reiter, M.; Müller, R.A. Diagnostic classification of autism using resting-state fMRI data and conditional random forest. Age 2018, 12, 6–41. [Google Scholar]
- Li, X.; Dvornek, N.C.; Zhuang, J.; Ventola, P.; Duncan, J.S. Brain biomarker interpretation in ASD using deep learning and fMRI. In Proceedings of the International Conference on Medical image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018; pp. 206–214. [Google Scholar]
- Bernas, A.; Aldenkamp, A.P.; Zinger, S. Wavelet coherence-based classifier: A resting-state functional MRI study on neurodynamics in adolescents with high-functioning autism. Comput. Methods Programs Biomed. 2018, 154, 143–151. [Google Scholar] [CrossRef]
- Bhaumik, R.; Pradhan, A.; Das, S.; Bhaumik, D.K. Predicting autism spectrum disorder using domain-adaptive cross-site evaluation. Neuroinformatics 2018, 16, 197–205. [Google Scholar] [CrossRef]
- Dekhil, O.; Hajjdiab, H.; Shalaby, A.; Ali, M.T.; Ayinde, B.; Switala, A.; Elshamekh, A.; Ghazal, M.; Keynton, R.; Barnes, G.; et al. Using resting state functional mri to build a personalized autism diagnosis system. PLoS ONE 2018, 13, e0206351. [Google Scholar] [CrossRef]
- Xiao, Z.; Wang, C.; Jia, N.; Wu, J. SAE-based classification of school-aged children with autism spectrum disorders using functional magnetic resonance imaging. Multimed. Tools Appl. 2018, 77, 22809–22820. [Google Scholar] [CrossRef]
- Yang, X.; Islam, M.S.; Khaled, A.A. Functional connectivity magnetic resonance imaging classification of autism spectrum disorder using the multisite ABIDE dataset. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Wang, C.; Xiao, Z.; Wang, B.; Wu, J. Identification of autism based on SVM-RFE and stacked sparse auto-encoder. IEEE Access 2019, 7, 118030–118036. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Z.; Wu, J. Functional connectivity-based classification of autism and control using SVM-RFECV on rs-fMRI data. Phys. Medica 2019, 65, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, M.A.; Sharifi, A.; Pedram, M.M. Diagnosis of autism spectrum disorders in young children based on resting-state functional magnetic resonance imaging data using convolutional neural networks. J. Digit. Imaging 2019, 32, 899–918. [Google Scholar] [CrossRef]
- Huang, H.; Liu, X.; Jin, Y.; Lee, S.W.; Wee, C.Y.; Shen, D. Enhancing the representation of functional connectivity networks by fusing multi-view information for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2019, 40, 833–854. [Google Scholar] [CrossRef]
- Jun, E.; Kang, E.; Choi, J.; Suk, H.I. Modeling regional dynamics in low-frequency fluctuation and its application to autism spectrum disorder diagnosis. NeuroImage 2019, 184, 669–686. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front. Neuroinform. 2019, 13, 70. [Google Scholar] [CrossRef]
- Mostafa, S.; Tang, L.; Wu, F.X. Diagnosis of autism spectrum disorder based on eigenvalues of brain networks. IEEE Access 2019, 7, 128474–128486. [Google Scholar] [CrossRef]
- Song, Y.; Epalle, T.M.; Lu, H. Characterizing and predicting autism spectrum disorder by performing resting-state functional network community pattern analysis. Front. Hum. Neurosci. 2019, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Spera, G.; Retico, A.; Bosco, P.; Ferrari, E.; Palumbo, L.; Oliva, P.; Muratori, F.; Calderoni, S. Evaluation of altered functional connections in male children with autism spectrum disorders on multiple-site data optimized with machine learning. Front. Psychiatry 2019, 10, 620. [Google Scholar] [CrossRef]
- Tang, L.; Mostafa, S.; Liao, B.; Wu, F.X. A network clustering based feature selection strategy for classifying autism spectrum disorder. BMC Med. Genom. 2019, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, B.; Itahashi, T.; Fujino, J.; Ohta, H.; Nakamura, M.; Kato, N.; Mimura, M.; Hashimoto, R.i.; Aoki, Y. Machine learning approach to identify a resting-state functional connectivity pattern serving as an endophenotype of autism spectrum disorder. Brain Imaging Behav. 2019, 13, 1689–1698. [Google Scholar] [CrossRef]
- Chaitra, N.; Vijaya, P.; Deshpande, G. Diagnostic prediction of autism spectrum disorder using complex network measures in a machine learning framework. Biomed. Signal Process. Control 2020, 62, 102099. [Google Scholar] [CrossRef]
- Fan, G.; Chen, Y.; Chen, Y.; Yang, M.; Wang, J.; Li, C.; Li, Y.; Liu, T. Abnormal brain regions in two-group cross-location dynamics model of autism. IEEE Access 2020, 8, 94526–94534. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Li, J.; Yu, J.; Yu, X. Attentional connectivity-based prediction of autism using heterogeneous rs-fMRI data from CC200 atlas. Exp. Neurobiol. 2020, 29, 27. [Google Scholar] [CrossRef]
- Hu, J.; Cao, L.; Li, T.; Liao, B.; Dong, S.; Li, P. Interpretable learning approaches in resting-state functional connectivity analysis: The case of autism spectrum disorder. Comput. Math. Methods Med. 2020, 2020, 1394830. [Google Scholar] [CrossRef]
- Sherkatghanad, Z.; Akhondzadeh, M.; Salari, S.; Zomorodi-Moghadam, M.; Abdar, M.; Acharya, U.R.; Khosrowabadi, R.; Salari, V. Automated detection of autism spectrum disorder using a convolutional neural network. Front. Neurosci. 2020, 13, 1325. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.M.; Gallo, S.; Cerliani, L.; Zhutovsky, P.; El-Gazzar, A.; Van Wingen, G. Classifying autism spectrum disorder using the temporal statistics of resting-state functional MRI data with 3D convolutional neural networks. Front. Psychiatry 2020, 11, 440. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Li, H.; Fan, Y. Improving diagnosis of autism spectrum disorder and disentangling its heterogeneous functional connectivity patterns using capsule networks. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1331–1334. [Google Scholar]
- Liu, W.; Liu, M.; Yang, D.; Wang, M.; Tao, T. Automatic diagnosis of autism based on functional magnetic resonance imaging and elastic net. In Proceedings of the 2020 IEEE 5th Information Technology and Mechatronics Engineering Conference (ITOEC), Chongqing, China, 12–14 June 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 104–108. [Google Scholar]
- Liu, J.; Sheng, Y.; Lan, W.; Guo, R.; Wang, Y.; Wang, J. Improved ASD classification using dynamic functional connectivity and multi-task feature selection. Pattern Recognit. Lett. 2020, 138, 82–87. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.H.; Li, L. Diagnosing autism spectrum disorder using brain entropy: A fast entropy method. Comput. Methods Programs Biomed. 2020, 190, 105240. [Google Scholar] [CrossRef]
- Ronicko, J.F.A.; Thomas, J.; Thangavel, P.; Koneru, V.; Langs, G.; Dauwels, J. Diagnostic classification of autism using resting-state fMRI data improves with full correlation functional brain connectivity compared to partial correlation. J. Neurosci. Methods 2020, 345, 108884. [Google Scholar] [CrossRef]
- Khan, N.A.; Waheeb, S.A.; Riaz, A.; Shang, X. A three-stage teacher, student neural networks and sequential feed forward selection-based feature selection approach for the classification of autism spectrum disorder. Brain Sci. 2020, 10, 754. [Google Scholar] [CrossRef]
- Reiter, M.A.; Jahedi, A.; Fredo, A.; Fishman, I.; Bailey, B.; Müller, R.A. Performance of machine learning classification models of autism using resting-state fMRI is contingent on sample heterogeneity. Neural Comput. Appl. 2021, 33, 3299–3310. [Google Scholar] [CrossRef]
- Devika, K.; Oruganti, V.R.M. A Machine Learning Approach for Diagnosing Neurological Disorders using Longitudinal Resting-State fMRI. In Proceedings of the 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 28–29 January 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 494–499. [Google Scholar]
- Ahammed, M.S.; Niu, S.; Ahmed, M.R.; Dong, J.; Gao, X.; Chen, Y. Bag-of-features model for asd fmri classification using svm. In Proceedings of the 2021 Asia-Pacific Conference on Communications Technology and Computer Science (ACCTCS), Shenyang, China, 22–24 January 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 52–57. [Google Scholar]
- Ahammed, M.S.; Niu, S.; Ahmed, M.R.; Dong, J.; Gao, X.; Chen, Y. Darkasdnet: Classification of asd on functional mri using deep neural network. Front. Neuroinform. 2021, 15, 635657. [Google Scholar] [CrossRef] [PubMed]
- Graña, M.; Silva, M. Impact of machine learning pipeline choices in autism prediction from functional connectivity data. Int. J. Neural Syst. 2021, 31, 2150009. [Google Scholar] [CrossRef]
- Al-Hiyali, M.I.; Yahya, N.; Faye, I.; Khan, Z.; Alsaih, K. Classification of BOLD FMRI signals using wavelet transform and transfer learning for detection of autism spectrum disorder. In Proceedings of the 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Langkawi Island, Malaysia, 1–3 March 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 94–98. [Google Scholar]
- Pominova, M.; Kondrateva, E.; Sharaev, M.; Bernstein, A.; Burnaev, E. Fader networks for domain adaptation on fMRI: ABIDE-II study. In Thirteenth International Conference on Machine Vision; SPIE: Bellingham, WA, USA, 2021; Volume 11605, pp. 570–577. [Google Scholar]
- Yin, W.; Mostafa, S.; Wu, F.X. Diagnosis of autism spectrum disorder based on functional brain networks with deep learning. J. Comput. Biol. 2021, 28, 146–165. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Wang, G.; Cao, L.; Qiao, L.; Liu, M. Multi-Scale Graph Representation Learning for Autism Identification with Functional MRI. Front. Neuroinform. 2022, 15, 802305. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, N.; Schrader, P. A study of brain networks for autism spectrum disorder classification using resting-state functional connectivity. Mach. Learn. Appl. 2022, 8, 100290. [Google Scholar] [CrossRef]
| Article | Dataset | ASD | HC | Age | Sex (Male%) | Feature Selection | ML Classifier | Goal/Findings | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| An et al., 2010 DTI+ fMRI [65] | Clinical | n.s. | n.s. | Child-control dataset | n.s. |
|
|
| Classification error: 8.55% |
| Ingalhalikar et al., 2011 [56] | Clinical | 45 | 30 | 10.5 ± 2.5 | 74.6% | FA, MD | SVM |
| 80% |
| Li et al., 2012 [57] | Clinical and simulated | 10 | 10 | 7–14 | n.s. | Brain connectivity network | SVM-recursive feature elimination (RFE) |
| 100% |
| Deshpande et al., 2013 DTI + fMRI [66] | Clinical | 15 | 15 | 21.1 ± 0.9 | n.s. |
|
|
| 95.9% |
| Jin et al., 2015 [58] | NDAR | 40 High-risk infants | 40 Low-risk infants | 6-month-old infants | 70% | FA, MD, and fiber length | Multikernel SVM |
| 76% |
| Crimi et al., 2017 sMRI+ DTI [67] | ABIDE-II | 31 | 23 | n.s. | n.s. |
| SVM |
|
|
| Zhang et al., 2018 [59] | Clinical | 70 | 79 | 11.0 ± 2.6 | 100% |
| SVM |
| 78.3% |
| Irimia et al., 2018 sMRI+ DTI [69] | Clinical | 110 | 83 | 12.74 | 50% |
| SVM |
| 94.82% |
| Qin et al., 2018 [60] | Clinical | 39 | 19 | 2.89 ± 0.97 | 82% | Graph-theory-based features | Edges and nodes |
| — |
| Payabvash 2019 [32] | Clinical | 14 | 33 | 8.9 ± 2.7 | 100% |
|
|
| 75.3% in EDI using RF |
| Saad et al., 2019 [61] | USC Multimodal Connectivity Database | 51 | 41 | n.s. | n.s. | Graph-theory-based features | SVM |
| 75% |
| Eill et al., 2019 aMRI+ DTI+ fMRI [70] | Clinical | 46 | 47 | 13.63 ± 2.81 | 84.8% |
| Conditional random forest (CRF) |
|
|
| ElNakieb et al., 2019 [62] | NADR | 122 | 141 | 8–17.9 | 50% | Global and local extraction of FA, MD, AD, RD, and skewness features | SVM |
| 71% |
| ElNakieb et al., 2020 [63] | NADR | 124 | 139 | 8–17.9 | 50% | Global and local extraction of FA, MD, AD, RD, and skewness features | SVM |
| 73% |
| ElNakieb et al., 2021 [64] | ABIDE-II | 125 | 100 | 5.1–46.6 | n.s. | Global and local extraction of FA, MD, AD, RD, and skewness features | Linear and non-linear classifiers |
| 99% |
| D’Souza et al., 2021 DTI + rs-fMRI [68] | Clinical | 57 | 275 | n.s. | n.s. | Phenotypic measures of rs-MRI connectivity. DTI tractography | Multimodal graph convolutional network (M-GCN) |
| — |
| Article | Dataset | ASD | HC | Age | Sex (Male%) | Feature Selection | ML Classifier | Goal/Findings | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| Abraham et al., 2017 rs-fMRI [71] | ABIDE | 403 | 468 | n.s. | 83.5% |
|
|
| 67% |
| Emerson et al., 2017 [72] fcMRI | Clinical | 11 high-risk infants | 48 | 6–24 months high-risk | 69.5% |
|
|
| 96.6% |
| Guo et al., 2017 rs-fMRI [73] | ABIDE I | 55 | 55 | 12.7 ± 2.4 | 76.4% |
|
|
| 86.36% |
| Jahedi A 2017 fcMRI [74] | ABIDE | 126 | 126 | 17.3 ± 6.0 | 80.6% |
|
|
| 92.7% |
| Kam et al., 2017 rs-fMRI [75] | ABIDE | 119 | 144 | <20 | n.s. |
|
|
|
|
| Sadeghi et al., 2017 rs-fMRI [76] | ABIDE | 31 | 29 | 20.49 ± 6.16 | 100% |
|
|
| 92% |
| Subbaraju et al., 2017 rs-fMRI [77] | ABIDE, PCP | 505 | 530 | 6.5-58 | 84.8% |
|
|
| 78.6–95% |
| Tejwani et al., 2017 rs-fMRI [78] | ABIDE | 147 | 146 | n.s. | n.s. |
|
|
| 65% |
| Heinsfeld et al., 2018 rs-fMRI [79] | ABIDE I | 505 | 530 | Site-specific | Site-specific |
|
|
| 70% |
| Bi et al., 2018 rs-fMRI [80] | ABIDE | 45 | 39 | 13.4 ± 2.4 | 88% |
|
|
| 96.15% |
| Fredo et al., 2018 rs-fMR [81] | ABIDE I, II | 160 | 160 | 12.16 ± 2.76 | 100% |
|
|
| 65% |
| Li et al., 2018 T-fMRI+rs-fMRI [82] |
|
|
| n.s. | n.s. | — |
|
|
|
| Bernas et al., 2018 rs-fMRI [83] | ABIDE | 24 | 39 | 15.5 ± 1.0 | 87% |
|
|
| 86.7% |
| Bhaumik et al., 2018 rs-fMRI [84] | ABIDE | 167 | 205 | 13.4 ± 5.1 | 81.7% |
|
|
| 62% |
| Dekhil et al., 2018 rs-fMRI [85] | NDAR | 123 | 160 | 12.9 ± 3 | 53.3% |
|
|
| 91% |
| Xiao et al., 2018 fMRI [86] | ABIDE | 42 | 42 | 9.78 ± 1.5 | 82.1% |
|
|
| 87.2% |
| Yang et al., 2019 rs-fMRI [87] | ABIDE | 505 | 530 | 6–6.4 | 84.8% |
|
|
| 71.98% with Ridge |
| Wang et al., 2019 (a) rs-fMRI [88] | ABIDE I, II | 255 | 276 | Site-specific | Site-specific |
|
|
| 90.6% |
| Wang et al., 2019 (b) rs-fMRI [89] | ABIDE | 501 | 533 | n.s. | n.s. |
|
|
| 93.59% |
| Aghdam et al. 2019 rs-fMRI [90] | ABIDE I, II | 210 | 249 | 5–10 | 72.1% |
|
|
| 70.5% |
| Huang et al., 2019 rs-fMRI [91] | ABIDE | 45 | 47 | 11.1 ± 2.3 | 80.4% |
|
|
| 79.4% |
| Jun et al., 2019 rs-fMRI [92] | ABIDE | 121 | 171 | 14.4 ± 5.8 | 78.4% |
|
|
| 84.6% |
| Eslami et al., 2019 rs-fMRI [93] | ABIDE I | 505 | 530 | Site-specific | Site-specific |
|
|
| 70.3% |
| Mostafa et al., 2019 rs-fMRI [94] | ABIDE I | 403 | 468 | Site-specific | Site-specific |
|
|
| 77.7% |
| Song et al., 2019 rs-fMRI [95] | ABIDE | 119 | 116 | Site-specific | Site-specific |
|
|
|
|
| Spera et al., 2019 rs-fMRI [96] | ABIDE | 102 | 88 | 6.5–13 | 100% |
|
|
| 71% |
| Tang et al., 2019 rs-fMRI [97] | ABIDE | 42 | 37 | n.s. | n.s. |
|
|
| AUC = 62.6 |
| Yamagata et al., 2019 rs-fMRI [98] | Clinical | 15 | 45 | 28.3 ± 6.1 | 100% |
|
|
| 75% |
| Chaitra et al., 2020 fMRI [99] | ABIDE | 432 | 556 | n.s. | n.s. |
|
|
| 70.1% |
| Fan et al., 2020 [100] | ABIDE | 145 | 157 | 16.4 ± 6.5 | 100% |
|
|
| 74.9% |
| Liu et al., 2020 [101] | ABIDE | 506 | 548 | 16.6 ± 8.1 | 85.3% |
|
|
| 76.8% |
| Hu et al., 2020 [102] | ABIDE | 403 | 468 | n.s. | n.s. |
|
|
| 69.8% |
| Sherkatghanad et al., 2020 [103] | ABIDE I | 505 | 530 | Site-specific | Site-specific |
|
|
| 70.2% |
| Thomas et al., 2020 [104] | ABIDE I and II | 620 | 542 | 5–64 Median = 13 | 80% |
|
|
| 64% |
| Jiao et al., 2020 [105] | ABIDE I | 505 | 530 | n.s. | n.s. |
|
|
| 71% |
| Liu et al., 2020 [106] | ABIDE | 250 | 218 | Center specific | 84.8% |
|
|
| 83.33% |
| Liu et al., 2020 [107] | ABIDE I | 403 | 468 | 17.07 ± 7.95 | 83.5% |
|
|
| 76.8% |
| Zhang et al., 2020 [108] | ABIDE I | 21 | 26 | 25.3 ± 6.3 | 100% |
|
|
| AUC = 62 |
| Ronicko et al., 2020 [109] | ABIDE I, II | 300 | 300 | 11.87 ± 2.8 | 80.5% |
|
|
| 70.3% |
| Khan et al., 2020 [110] | ABIDE | 505 | 530 | Site-specific | Site-specific |
|
|
| 82% |
| Reiter et al., 2021 [111] | ABIDE | 306 | 350 | 6–18 | n.s. |
|
|
| 73.75% |
| Devika, K., and Oruganti, V. R. M. 2021 [112] | ABIDE II | 23 | 15 | n.s. | 84.2% |
|
|
| 80.76% |
| Ahammed et al., 2021 [113] | ABIDE I | 19 | 19 | 15–35 | 78.9% |
|
|
| 81% |
| Ahammed et al., 2021 [114] | ABIDE I | 79 | 105 | 15.25 ± 6.58 | 81% |
|
|
| 94.7% |
| Graña, M., and Silva, M. 2021 [115] | ABIDE | 408 | 476 | Site-specific | Site-specific |
|
|
| Best median AUC = 0.767 |
| Al-Hiyali et al., 2021 [116] | ABIDE | 41 | 41 | n.s. | n.s. |
|
|
| 85.9% with KNN |
| Pominova et al., 2021 [117] | ABIDE II | 184 | 168 | 10.15 ± 2.98 | n.s. |
|
|
| — |
| Yin et al., 2021 [118] | ABIDE I | 403 | 468 | — | — |
|
|
| 79.2% |
| Chu et al., 2022 [119] | ABIDE | 79 | 105 | 14.51 ± 6.23 | 79.9% |
|
|
| 0.795 |
| Yang et al., 2022 [120] | ABIDE I | 403 | 468 | 6–58 | — |
|
|
| 69.43% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helmy, E.; Elnakib, A.; ElNakieb, Y.; Khudri, M.; Abdelrahim, M.; Yousaf, J.; Ghazal, M.; Contractor, S.; Barnes, G.N.; El-Baz, A. Role of Artificial Intelligence for Autism Diagnosis Using DTI and fMRI: A Survey. Biomedicines 2023, 11, 1858. https://doi.org/10.3390/biomedicines11071858
Helmy E, Elnakib A, ElNakieb Y, Khudri M, Abdelrahim M, Yousaf J, Ghazal M, Contractor S, Barnes GN, El-Baz A. Role of Artificial Intelligence for Autism Diagnosis Using DTI and fMRI: A Survey. Biomedicines. 2023; 11(7):1858. https://doi.org/10.3390/biomedicines11071858
Chicago/Turabian StyleHelmy, Eman, Ahmed Elnakib, Yaser ElNakieb, Mohamed Khudri, Mostafa Abdelrahim, Jawad Yousaf, Mohammed Ghazal, Sohail Contractor, Gregory Neal Barnes, and Ayman El-Baz. 2023. "Role of Artificial Intelligence for Autism Diagnosis Using DTI and fMRI: A Survey" Biomedicines 11, no. 7: 1858. https://doi.org/10.3390/biomedicines11071858
APA StyleHelmy, E., Elnakib, A., ElNakieb, Y., Khudri, M., Abdelrahim, M., Yousaf, J., Ghazal, M., Contractor, S., Barnes, G. N., & El-Baz, A. (2023). Role of Artificial Intelligence for Autism Diagnosis Using DTI and fMRI: A Survey. Biomedicines, 11(7), 1858. https://doi.org/10.3390/biomedicines11071858








