Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential
Abstract
:1. Introduction
2. The Fundamentals of Long Non-Coding RNAs
3. Interplay of lncRNAs and Consensus Molecular Subtypes (CMSs) in CRC
4. The Role of Immunity and Inflammation in CRC Tumor Stroma
5. The Role of LncRNAs in Tumor–Stroma Immune Interplay via Extracellular Vesicles and Exosomes
6. Unraveling the Complexity: The Interplay of lncRNAs and Other ncRNAs in Cell-to-Cell Communication within the CRC Microenvironment
7. Exploring Strategies for Manipulating lncRNAs to Enhance Anti-Tumor Immunity in CRC Patients
7.1. Potential Methodologies for Manipulating Extracellularly Released lncRNAs
7.2. Immune Processes That We Can Regulate via lncRNAs
8. Conclusions: Future Perspectives on Long Non-Coding RNAs in CRC
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Patel, S.G.; Karlitz, J.J.; Yen, T.; Lieu, C.H.; Boland, C.R. The rising tide of early-onset colorectal cancer: A comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol. Hepatol. 2022, 7, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Maaser, C.; Sturm, A.; Vavricka, S.R.; Kucharzik, T.; Fiorino, G.; Annese, V.; Calabrese, E.; Baumgart, D.C.; Bettenworth, D.; Borralho Nunes, P.; et al. European Crohn’s and Colitis Organisation [ECCO] and the European Society of Gastrointestinal and Abdominal Radiology [ESGAR]. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J. Crohns Colitis 2019, 13, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Nardone, O.M.; Zammarchi, I.; Santacroce, G.; Ghosh, S.; Iacucci, M. Inflammation-Driven Colorectal Cancer Associated with Colitis: From Pathogenesis to Changing Therapy. Cancers 2023, 15, 2389. [Google Scholar] [CrossRef]
- Newman, P.; Muscat, J. Potential Role of Non-Steroidal Anti-Inflammatory Drugs in Colorectal Cancer Chemoprevention for Inflammatory Bowel Disease: An Umbrella Review. Cancers 2023, 15, 1102. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Gieniec, K.A.; Lannagan, T.R.M.; Wang, T.; Asai, N.; Mizutani, Y.; Iida, T.; Ando, R.; Thomas, E.M.; Sakai, A.; et al. The Origin and Contribution of Cancer-Associated Fibroblasts in Colorectal Carcinogenesis. Gastroenterology 2022, 162, 890–906. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Greten, F.R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Ye, Y.; Chen, Y.; Zhu, J.; Xu, L.; Cheng, W.; Lu, X.; Yan, F. Identification and validation of an inflammation-related lncRNAs signature for improving outcomes of patients in colorectal cancer. Front. Genet. 2022, 13, 955240. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Benelli, R.; Zocchi, M.R.; Poggi, A. Immune Checkpoint Receptor/Ligand Expression and Chemotherapy in Colorectal Cancer. Cancers 2023, 15, 914. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, C.; Feng, J. The role of LncRNAs in tumor immunotherapy. Cancer Cell Int. 2023, 23, 30. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.A.G.; Adachi, S.; Nong, Q.D.; Adhitama, N.; Matsuura, T.; Natsume, T.; Wada, T.; Kato, Y.; Watanabe, H. Sense-overlapping lncRNA as a decoy of translational repressor protein for dimorphic gene expression. PLoS Genet. 2021, 17, e1009683. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xiang, W.; Liu, J.; Tang, J.; Wang, J.; Liu, B.; Long, Z.; Wang, L.; Yin, G.; Liu, J. The regulatory role of antisense lncRNAs in cancer. Cancer Cell Int. 2021, 21, 459. [Google Scholar] [CrossRef]
- Tahira, A.C.; Kubrusly, M.S.; Faria, M.F.; Dazzani, B.; Fonseca, R.S.; Maracaja-Coutinho, V.; Verjovski-Almeida, S.; Machado, M.C.; Reis, E.M. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol. Cancer 2011, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.C.; Ponting, C.P. Intergenic lncRNAs and the Evolution of Gene Expression. Curr. Opin. Genet. Dev. 2014, 27, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.; Tanimoto, Y.; Takahashi, S.; Furukawa, T.; Koshiba-Takeuchi, K.; Takeuchi, J.K. Important Cardiac Transcription Factor Genes Are Accompanied by Bidirectional Long Non-Coding RNAs. BMC Genom. 2018, 19, 967. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhang, R.; Sun, X. Enhancer LncRNAs Influence Chromatin Interactions in Different Ways. Front. Genet. 2019, 10, 936. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef]
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022, 41, 100. [Google Scholar] [CrossRef]
- Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Non-Coding RNA 2021, 7, 21. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Makałowska, I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS ONE 2016, 11, e0150353. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Qu, L.; Gao, F.; Lin, J.; Liu, J.; Lin, A. LncRNAs: Architectural Scaffolds or More Potential Roles in Phase Separation. Front. Genet. 2021, 12, 626234. [Google Scholar] [CrossRef] [PubMed]
- Blythe, A.J.; Fox, A.H.; Bond, C.S. The ins and outs of lncRNA structure: How, why and what comes next? Biochim. Biophys. Acta 2016, 1859, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Guh, C.Y.; Hsieh, Y.H.; Chu, H.P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 44. [Google Scholar] [CrossRef]
- Hu, X.Y.; Hou, P.F.; Li, T.T.; Quan, H.Y.; Li, M.L.; Lin, T.; Liu, J.J.; Bai, J.; Zheng, J.N. The roles of Wnt/β-catenin signaling pathway related lncRNAs in cancer. Int. J. Biol. Sci. 2018, 14, 2003–2011. [Google Scholar] [CrossRef]
- Fu, P.F.; Zheng, X.; Fan, X.; Lin, A.F. Role of cytoplasmic lncRNAs in regulating cancer signaling pathways. J. Zhejiang Univ. Sci. B 2019, 20, 1–8. [Google Scholar] [CrossRef]
- Gong, Z.; Zhang, Y.; Yang, Y.; Yang, Y.; Zhang, J.; Wang, Y.; Zhao, L.; Yu, N.; Wu, Z.; Guo, W. LncRNA LINC01094 Promotes Cells Proliferation and Metastasis through the PTEN/AKT Pathway by Targeting AZGP1 in Gastric Cancer. Cancers 2023, 15, 1261. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, X.; Gu, X.; Li, X.; Shang, L. Progress in understanding themolecular mechanisms of the antitumour effects of the long non-coding RNA MEG3. Cancers 2023, 15, 1230. [Google Scholar] [CrossRef]
- Lingadahalli, S.; Jadhao, S.; Sung, Y.Y.; Chen, M.; Hu, L.; Chen, X.; Cheung, E. Novel lncRNA LINC00844 Regulates Prostate Cancer Cell Migration and Invasion through AR Signaling. Mol. Cancer Res. 2018, 16, 1865–1878. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, Y.; Peng, J.; Thakur, A.; Bai, N.; Yang, K.; Xu, Z. Decoding Roles of Exosomal lncRNAs in Tumor-Immune Regulation and Therapeutic Potential. Cancers 2022, 15, 286. [Google Scholar] [CrossRef]
- Kuo, F.C.; Neville, M.J.; Sabaratnam, R.; Wesolowska-Andersen, A.; Phillips, D.; Wittemans, L.B.L.; van Dam, A.D.; Loh, N.Y.; Todorčević, M.; Denton, N.; et al. HOTAIR interacts with PRC2 complex regulating the regional preadipocyte transcriptome and human fat distribution. Cell Rep. 2022, 40, 111136. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Pan, J.; Geng, Q.; Wang, G. LncRNA MALAT1 Increases the Stemness of Gastric Cancer Cells via Enhancing SOX2 mRNA Stability. FEBS Open Bio 2019, 9, 1212–1222. [Google Scholar] [CrossRef]
- Ji, Q.; Cai, G.; Liu, X.; Zhang, Y.; Wang, Y.; Zhou, L.; Sui, H.; Li, Q. MALAT1 Regulates the Transcriptional and Translational Levels of Proto-Oncogene RUNX2 in Colorectal Cancer Metastasis. Cell Death Dis. 2019, 10, 378. [Google Scholar] [CrossRef]
- Gordon, M.A.; Babbs, B.; Cochrane, D.R.; Bitler, B.G.; Richer, J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol. Carcinog. 2019, 58, 196–205. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, D.; Todoerti, K.; Nobili, L.; Tassone, P.; Amodio, N.; Neri, A. LncRNA NEAT1 in Paraspeckles: A Structural Scaffold for Cellular DNA Damage Response Systems? Noncoding RNA 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Pardini, B.; Calin, G.A. MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers 2019, 11, 378. [Google Scholar] [CrossRef]
- Abramowicz, A.; Story, M.D. The Long and Short of It: The Emerging Roles of Non-Coding RNA in Small Extracellular Vesicles. Cancers 2020, 12, 1445. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An Updated Database of Long Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An Updated Resource for Experimentally Supported lncRNA/circRNA Cancer Associations and Web Tools Based on RNA-Seq and scRNA-Seq Data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Cui, T.; Zheng, B.; Wang, N.; Luo, J.; Yang, B.; Du, M.; Cheng, J.; Dou, Y.; Wang, D. MNDR v3.0: Mammal ncRNA-Disease Repository with Increased Coverage and Annotation. Nucleic Acids Res. 2021, 49, D160–D164. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Liu, L.; Li, Z.; Liu, C.; Zou, D.; Li, Q.; Feng, C.; Jing, W.; Luo, S.; Zhang, Z.; Ma, L. LncRNAWiki 2.0: A Knowledgebase of Human Long Non-Coding RNAs with Enhanced Curation Model and Database System. Nucleic Acids Res. 2022, 50, D190–D195. [Google Scholar] [CrossRef] [PubMed]
- Bhartiya, D.; Pal, K.; Ghosh, S.; Kapoor, S.; Jalali, S.; Panwar, B.; Jain, S.; Sati, S.; Sengupta, S.; Sachidanandan, C.; et al. lncRNome: A Comprehensive Knowledgebase of Human Long Noncoding RNAs. Database 2013, 2013, bat034. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yin, X.; Xu, H.; Liu, K.; Liu, W.; Wang, L.; Zhang, C.; Bo, L.; Lan, X.; Lin, S.; et al. LncTarD 2.0: An Updated Comprehensive Database for Experimentally-Supported Functional lncRNA-Target Regulations in Human Diseases. Nucleic Acids Res. 2023, 51, D199–D207. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Ten Hoorn, S.; de Back, T.R.; Sommeijer, D.W.; Vermeulen, L. Clinical Value of Consensus Molecular Subtypes in Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2022, 114, 503–516. [Google Scholar] [CrossRef]
- Chowdhury, S.; Hofree, M.; Lin, K.; Maru, D.; Kopetz, S.; Shen, J.P. Implications of Intratumor Heterogeneity on Consensus Molecular Subtype (CMS) in Colorectal Cancer. Cancers 2021, 13, 4923. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, J.; Tang, W. The Molecular Mechanism of HOTAIR in Tumorigenesis, Metastasis, and Drug Resistance. Acta Biochim. Biophys. Sin. 2014, 46, 1011–1015. [Google Scholar] [CrossRef]
- Yu, X.; Li, Z. Long Non-Coding RNA HOTAIR: A Novel Oncogene. Mol. Med. Rep. 2015, 12, 5611–5618. [Google Scholar] [CrossRef]
- Qiu, W.; Yang, J.; Wang, B.; Yang, M.; Tian, G.; Wang, P.; Yang, J. Evaluating the Microsatellite Instability of Colorectal Cancer Based on Multimodal Deep Learning Integrating Histopathological and Molecular Data. Front. Oncol. 2022, 12, 925079. [Google Scholar] [CrossRef]
- Pashapour, S.; Shanehbandi, D.; Bornehdeli, S.; Zafari, V.; Khani, M.R.; Hashemzadeh, S.; Asvadi Kermani, T. Overexpression of HOTAIR in Tumor Tissues of Patients with Colon Cancer Correlates with Tumor Metastasis and Differentiation. Middle East. J. Cancer 2020, 11, 410–414. [Google Scholar] [CrossRef]
- Hu, Q.; Ye, Y.; Chan, L.C.; Li, Y.; Liang, K.; Lin, A.; Egranov, S.D.; Zhang, Y.; Xia, W.; Gong, J.; et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 2019, 20, 835–851. [Google Scholar] [CrossRef]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A LncRNA Interacts with PtdIns(3,4,5)P3 to Hyperactivate AKT and Confer Resistance to AKT Inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef]
- Zhao, C.; Gan, C.; Xiao, Y.; Liu, R.; Zhang, L.; Lan, T.; Ye, Y.; Tong, H.; Huang, Z.; Tang, C.; et al. High expression of long non-coding RNA Linc-A associates with poor survival in patients with colorectal cancer. Mol. Biol. Rep. 2020, 47, 7497–7504. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef]
- Javed, Z.; Khan, K.; Sadia, H.; Raza, S.; Salehi, B.; Sharifi-Rad, J.; Cho, W.C. LncRNA & Wnt signaling in colorectal cancer. Cancer Cell Int. 2020, 20, 326. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, Q.; Hann, S.S. The functions and oncogenic roles of CCAT1 in human cancer. Biomed. Pharmacother. 2019, 115, 108943. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sha, H.; Sun, X.; Zhang, Y.; Wu, Y.; Zhang, J.; Zhang, H.; Wu, J.; Feng, J. CRNDE: An Oncogenic Long Non-Coding RNA in Cancers. Cancer Cell Int. 2020, 20, 162. [Google Scholar] [CrossRef]
- Han, P.; Li, J.W.; Zhang, B.M.; Lv, J.C.; Li, Y.M.; Gu, X.Y.; Yu, Z.W.; Jia, Y.H.; Bai, X.F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef]
- Neve, B.; Jonckheere, N.; Vincent, A.; Van Seuningen, I. Single-Cell Analysis May Shed New Lights on the Role of LncRNAs in Chemoresistance in Gastrointestinal Cancers. RNA Technol. 2020, 11, 229–253. [Google Scholar] [CrossRef]
- Tang, F.; Xu, Y.; Wang, H.; Bian, E.; Zhao, B. LncRNA-ATB in Cancers: What Do We Know So Far? Mol. Biol. Rep. 2020, 47, 4077–4086. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C. Long Non-Coding RNA ATB Is Associated with Metastases and Promotes Cell Invasion in Colorectal Cancer via Sponging miR-141-3p. Exp. Ther. Med. 2020, 20, 261, Erratum in Exp. Ther. Med. 2022, 23, 238. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.-H.; Yang, F.; Wang, F.; Ma, J.-Z.; Guo, Y.-J.; Tao, Q.-F.; Liu, F.; Pan, W.; Wang, T.-T.; Zhou, C.-C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Shi, D.; Zheng, H.; Zhuo, C.; Peng, J.; Li, D.; Xu, Y.; Li, X.; Cai, G.; Cai, S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med. Oncol. 2014, 31, 31. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, G.; Liu, H.; Shan, Y.; Zhang, X. RP11-462C24.1 suppresses proliferation and invasion of colorectal carcinoma cells by regulating HSP70 through PI3K/AKT signaling pathway. Hum. Cell. 2021, 34, 132–151. [Google Scholar] [CrossRef]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. LncRNA H19: A Novel Oncogene in Multiple Cancers. Int. J. Biol. Sci. 2021, 17, 3188–3208. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Hu, Y.; Sun, D.; Liu, C.; Li, Z.; Zhu, J. Targeting non-coding RNA H19: A potential therapeutic approach in pulmonary diseases. Front. Pharmacol. 2022, 13, 978151. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Shen, J.; Chen, Z.; Yang, J.; Xie, B.; Jia, Y.; Jayasinghe, U.; Wang, J.; Zhao, W.; Xie, S.; et al. H19/let 7/Lin28 ceRNA network mediates autophagy inhibiting epithelial mesenchymal transition in breast cancer. Int. J. Oncol. 2020, 56, 794–806. [Google Scholar] [CrossRef]
- Kou, N.; Liu, S.; Li, X.; Li, W.; Zhong, W.; Gui, L.; Chai, S.; Ren, X.; Na, R.; Zeng, T.; et al. H19 Facilitates Tongue Squamous Cell Carcinoma Migration and Invasion via Sponging miR-let-7. Oncol. Res. 2019, 27, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Huang, Z.; Wang, J.; Liu, H. Long non-coding RNA DANCR accelerates colorectal cancer progression via regulating the miR-185-5p/HMGA2 axis. J. Biochem. 2022, 171, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The Emerging Role of LncRNAs in Cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Huang, T.; Wang, M.; Huang, B.; Chang, A.; Liu, F.; Zhang, Y.; Jiang, B. Long Noncoding RNAs in the mTOR Signaling Network: Biomarkers and Therapeutic Targets. Apoptosis 2018, 23, 255–264. [Google Scholar] [CrossRef]
- Tu, X.; Zhang, Y.; Zheng, X.; Deng, J.; Li, H.; Kang, Z.; Cao, Z.; Huang, Z.; Ding, Z.; Dong, L.; et al. TGF-β-induced hepatocyte lincRNA-p21 contributes to liver fibrosis in mice. Sci. Rep. 2017, 7, 2957. [Google Scholar] [CrossRef]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Dong, M.; Dai, C.; Wu, S. Inflammation and Inflammatory Cytokine Contribute to the Initiation and Development of Ulcerative Colitis and Its Associated Cancer. Inflamm. Bowel Dis. 2019, 25, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.J.; Wang, Y.H.; Li, Z.G.; Wang, Y.; Li, B.Y.; Kang, H.Y.; Wu, X.Y. Immunosuppressive Effect of Exosomes from Mesenchymal Stromal Cells in Defined Medium on Experimental Colitis. Int. J. Stem Cells 2019, 12, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- Kos, K.; Salvagno, C.; Wellenstein, M.D.; Aslam, M.A.; Meijer, D.A.; Hau, C.-S.; Vrijland, K.; Kaldenbach, D.; Raeven, E.A.; Schmittnaegel, M.; et al. Tumor-associated macrophages promote intratumoral conversion of conventional CD4+ T cells into regulatory T cells via PD-1 signalling. Oncoimmunology 2022, 11, 2063225. [Google Scholar] [CrossRef]
- Mirlekar, B. Tumor promoting roles of IL-10, TGF-β, IL-4, and IL-35: Its implications in cancer immunotherapy. SAGE Open Med. 2022, 10, 20503121211069012. [Google Scholar] [CrossRef] [PubMed]
- Horzum, U.; Yanik, H.; Taskiran, E.Z.; Esendagli, G. Effector Th1 cells under PD-1 and CTLA-4 checkpoint blockade abrogate the upregulation of multiple inhibitory receptors and by-pass exhaustion. Immunology 2022, 167, 640–650. [Google Scholar] [CrossRef]
- Iglesias-Escudero, M.; Arias-González, N.; Martínez-Cáceres, E. Regulatory cells and the effect of cancer immunotherapy. Mol. Cancer 2023, 22, 26. [Google Scholar] [CrossRef]
- Tay, C.; Tanaka, A.; Sakaguchi, S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell 2023, 41, 450–465. [Google Scholar] [CrossRef]
- Ando, M.; Ito, M.; Srirat, T.; Kondo, T.; Yoshimura, A. Memory T cell, exhaustion, and tumor immunity. Immunol. Med. 2020, 43, 1–9. [Google Scholar] [CrossRef]
- Vanhaver, C.; van der Bruggen, P.; Bruger, A.M. MDSC in Mice and Men: Mechanisms of Immunosuppression in Cancer. J. Clin. Med. 2021, 10, 2872. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Sosnowska, A.; Matryba, P.; Rydzynska, Z.; Jasinski, M.; Nowis, D.; Golab, J. Myeloid Cell-Derived Arginase in Cancer Immune Response. Front. Immunol. 2020, 11, 938. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J. Roles for macrophage-polarizing interleukins in cancer immunity and immunotherapy. Cell Oncol. 2022, 45, 333–353. [Google Scholar] [CrossRef] [PubMed]
- Masoudi-Khoram, N.; Soheilifar, M.H.; Ghorbanifar, S.; Nobari, S.; Hakimi, M.; Hassani, M. Exosomes derived from cancer-associated fibroblasts mediate responseto cancer therapy. Crit. Rev. Oncol. Hematol. 2023, 185, 103967. [Google Scholar] [CrossRef]
- Tan, S.; Yang, Y.; Yang, W.; Han, Y.; Huang, L.; Yang, R.; Hu, Z.; Tao, Y.; Liu, L.; Li, Y.; et al. Exosomal cargos-mediated metabolic reprogramming in tumor microenvironment. J. Exp. Clin. Cancer Res. 2023, 42, 59. [Google Scholar] [CrossRef]
- O’Grady, T.; Njock, M.S.; Lion, M.; Bruyr, J.; Mariavelle, E.; Galvan, B.; Boeckx, A.; Struman, I.; Dequiedt, F. Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 2022, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Barreca, M.M.; Zichittella, C.; Alessandro, R.; Conigliaro, A. Molecular Mediators of RNA Loading into Extracellular Vesicles. Cells 2021, 10, 3355. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 123. [Google Scholar] [CrossRef]
- Yang, P.; Ding, J.; Bian, Y.; Ma, Z.; Wang, K.; Li, J. Long non-coding RNAs and cancer mechanisms: Immune cells and inflammatory cytokines in the tumor microenvironment. Med. Oncol. 2022, 39, 108. [Google Scholar] [CrossRef]
- Han, W.; Sulidankazha, Q.; Nie, X.; Yilidan, R.; Len, K. Pancreatic cancer cells-derived exosomal long non-coding RNA CCAT1/microRNA-138-5p/HMGA1 axis promotes tumor angiogenesis. Life Sci. 2021, 278, 119495. [Google Scholar] [CrossRef] [PubMed]
- Zong, D.; Liu, X.; Li, J.; Long, Y.; Ouyang, R.; Chen, Y. LncRNA-CCAT1/miR-152-3p is involved in CSE-induced inflammation in HBE cells via regulating ERK signaling pathway. Int. Immunopharmacol. 2022, 109, 108818. [Google Scholar] [CrossRef]
- Wang, D.; Li, Z.; Yin, H. Long Non-Coding RNA CCAT2 Activates RAB14 and Acts as an Oncogene in Colorectal Cancer. Front. Oncol. 2021, 11, 751903. [Google Scholar] [CrossRef]
- Moradi, F.; Mohajerani, F.; Sadeghizadeh, M. CCAT2 knockdown inhibits cell growth, and migration and promotes apoptosis through regulating the hsa-mir-145-5p/AKT3/mTOR axis in tamoxifen-resistant MCF7 cells. Life Sci. 2022, 311 Pt B, 121183. [Google Scholar] [CrossRef]
- Sasidharan Nair, V.; Toor, S.M.; Taha, R.Z.; Ahmed, A.A.; Kurer, M.A.; Murshed, K.; Soofi, M.E.; Ouararhni, K.; Alajez, N.M.; Abu Nada, M.; et al. Transcriptomic profiling of tumor-infiltrating CD4+TIM-3+ T cells reveals their suppressive, exhausted, and metastatic characteristics in colorectal cancer patients. Vaccines 2020, 8, 71. [Google Scholar] [CrossRef]
- Saadh, J.; Abedi Kiasari, B.; Shahrtash, S.A.; Arias-Gonzáles, J.L.; Chaitanya, M.; Cotrina-Aliaga, J.C.; Kadham, M.J.; Sârbu, I.; Akhavan-Sigari, R. Exosomal non-coding RNAs’ role in immune regulation and potential therapeutic applications. Pathol. Res. Pract. 2023, 247, 154522. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Wang, L.; Zhang, Z.; Li, Z.; Jia, Q. LncRNA H19 aggravates TNF-α-induced inflammatory injury via TAK1 pathway in MH7A cells. Biofactors 2020, 46, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Hirano, K.; Shichi, Y.; Gomi, F.; Yoshimura, H.; Matsushita, A.; Toyoda, M.; Ishiwata, T. Gp130-Mediated STAT3 Activation Contributes to the Aggressiveness of Pancreatic Cancer through H19 Long Non-Coding RNA Expression. Cancers 2022, 14, 2055. [Google Scholar] [CrossRef]
- Yin, L.; Yan, J.; Chen, W. Mechanism of lncRNA-H19 in Intestinal Injury of Mice with Ulcerative Colitis. Int. Arch. Allergy Immunol. 2022, 183, 985–996. [Google Scholar] [CrossRef]
- Raju, G.S.R.; Pavitra, E.; Bandaru, S.S.; Varaprasad, G.L.; Nagaraju, G.P.; Malla, R.R.; Huh, Y.S.; Han, Y.K. HOTAIR: A potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. Cancer 2023, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Obaid, M.; Udden, S.M.N.; Deb, P.; Shihabeddin, N.; Zaki, M.H.; Mandal, S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018, 8, 15670. [Google Scholar] [CrossRef]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef]
- Dong, Y.; Wei, M.H.; Lu, J.G.; Bi, C.Y. Long non-coding RNA HULC interacts with miR-613 to regulate colon cancer growth and metastasis through targeting RTKN. Biomed. Pharmacother. 2019, 109, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Shi, M.; You, Q.; Zhang, Y.; Hu, Z.; Xu, J.; Cai, Q.; Zhu, Z. Tumor- and metastasis-promoting roles of miR-488 inhibition via HULC enhancement and EZH2-mediated p53 repression in gastric cancer. Cell Biol. Toxicol. 2022, 39, 1341–1358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhong, C.; Shen, J.; Chen, S.; Jia, Y.; Duan, S. Emerging role of LINC00461 in cancer. Biomed. Pharmacother. 2022, 152, 113239. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.K.; Liu, C.; Li, X.X.; Ji, W.; Xin, C.D.; Hu, Z.Q.; Zhou, L. PHLPP2 is regulated by competing endogenous RNA network in pathogenesis of colon cancer. Aging 2020, 12, 12812–12840. [Google Scholar] [CrossRef]
- Abedini, P.; Fattahi, A.; Agah, S.; Talebi, A.; Beygi, A.H.; Amini, S.M.; Mirzaei, A.; Akbari, A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J. Cell Physiol. 2019, 234, 22028–22033. [Google Scholar] [CrossRef] [PubMed]
- Anbiyaiee, A.; Ramazii, M.; Bajestani, S.S.; Meybodi, S.M.; Keivan, M.; Khoshnam, S.E.; Farzaneh, M. The function of LncRNA-ATB in cancer. Clin. Transl. Oncol. 2023, 25, 1–9. [Google Scholar] [CrossRef]
- Eptaminitaki, G.C.; Wolff, N.; Stellas, D.; Sifakis, K.; Baritaki, S. Long Non-Coding RNAs (lncRNAs) in Response and Resistance to Cancer Immunosurveillance and Immunotherapy. Cells 2021, 10, 3313. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Attar, R.; Yulaevna, I.M.; Berardi, R. Interaction of long non-coding RNAs and circular RNAs with microRNAs for the regulation of immunological responses in human cancers. Semin. Cell Dev. Biol. 2022, 124, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Peltier, D.C.; Roberts, A.; Reddy, P. LNCing RNA to immunity. Trends Immunol. 2022, 43, 478–495. [Google Scholar] [CrossRef]
- Mekky, R.Y.; Ragab, M.F.; Manie, T.; Attia, A.A.; Youness, R.A. MALAT-1: Immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023, 31, 101653. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Azizidoost, S.; Ghaedrahmati, F.; Anbiyaee, O.; Ahmad Ali, R.; Cheraghzadeh, M.; Farzaneh, M. Emerging roles for lncRNA-NEAT1 in colorectal cancer. Cancer Cell Int. 2022, 22, 209. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, Y. Extracellular vesicles derived from M2-polarized tumor-associated macrophages promote immune escape in ovarian cancer through NEAT1/miR-101-3p/ZEB1/PD-L1 axis. Cancer Immunol. Immunother. 2023, 72, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.E.; Spencer-Merris, P.; Fox, A.H.; Petersen, J.; Michael, M.Z. The Long and the Short of It: NEAT1 and Cancer Cell Metabolism. Cancers 2022, 14, 4388. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.; Fan, W.J.; Fu, L.; Wang, X.T. LncRNA PCAT-1 regulated cell proliferation, invasion, migration, and apoptosis in colorectal cancer through targeting miR-149-5p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8310–8320. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Dashti, S.; Taheri, M. PCAT1: An oncogenic lncRNA in diverse cancers and a putative therapeutic target. Exp. Mol. Pathol. 2020, 114, 104429. [Google Scholar] [CrossRef]
- Avazpour, N.; Hajjari, M.; Kazemi Nezhad, S.R.; Tahmasebi Birgani, M. SNHG1 Long Noncoding RNA is Potentially Up-Regulated in Colorectal Adenocarcinoma. Asian Pac. J. Cancer Prev. 2020, 21, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, Z.; Zhao, Q.; Wu, T.; Zhao, Q.; Cao, Y. Knockdown of SNHG1 alleviates autophagy and apoptosis by regulating miR-362-3p/Jak2/stat3 pathway in LPS-injured PC12 cells. Neurochem. Res. 2021, 46, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Dai, W.; Guo, X.; Wang, K. LncRNA-SNHG1 promotes macrophage M2-like polarization and contributes to breast cancer growth and metastasis. Aging 2021, 13, 23169–23181. [Google Scholar] [CrossRef]
- Di, W.; Weinan, X.; Xin, L.; Zhiwei, Y.; Xinyue, G.; Jinxue, T.; Mingqi, L. Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis. 2019, 10, 514. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Hushmandi, K.; Zarrin, V.; Moghadam, E.R.; Zabolian, A.; Tavakol, S.; Samarghandian, S.; Najafi, M. PD-1/PD-L1 axis regulation in cancer therapy: The role of long non-coding RNAs and microRNAs. Life Sci. 2020, 256, 117899. [Google Scholar] [CrossRef]
- Tian, Y.; Li, L.; Lin, G.; Wang, Y.; Wang, L.; Zhao, Q.; Hu, Y.; Yong, H.; Wan, Y.; Zhang, Y. lncRNA SNHG14 promotes oncogenesis and immune evasion in diffuse large-B-cell lymphoma by sequestering miR-152-3p. Leuk. Lymphoma 2021, 62, 1574–1584. [Google Scholar] [CrossRef]
- Jiang, Y.; Hei, B.; Hao, W.; Lin, S.; Wang, Y.; Liu, X.; Meng, X.; Guan, Z. Clinical value of lncRNA SOX2-OT in pulmonary arterial hypertension and its role in pulmonary artery smooth muscle cell proliferation, migration, apoptosis, and inflammatory. Heart Lung 2022, 55, 16–23. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, Y.; Gao, Y.; Chen, Y.; Wang, X.; Chen, Z. A novel lncRNA SOX2OT promotes the malignancy of human colorectal cancer by interacting with miR-194-5p/SOX5 axis. Cell Death Dis. 2021, 12, 499. [Google Scholar] [CrossRef]
- Expression of Concern: LncRNA TUG1 promotes the progression of colorectal cancer via the miR-138-5p/ZEB2 Axis. Biosci. Rep. 2021, 41, BSR20201025. [CrossRef]
- Ren, Y.; Lyu, J.; Guo, Y.; Yao, Y.; Hu, L. Long Noncoding RNA TUG1 Inhibits Tumor Progression through Regulating Siglec-15-Related Anti-Immune Activity in Hepatocellular Carcinoma. J. Immunol. Res. 2022, 2022, 9557859. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Huang, S.; He, K.; Zhao, M.; Lin, H.; Li, D.; Qian, J.; Zhou, C.; Chen, Y.; et al. Long Non-Coding RNA Growth Arrest Specific Transcript 5 Acts as a Tumour Suppressor in Colorectal Cancer by Inhibiting Interleukin-10 and Vascular Endothelial Growth Factor Expression. Oncotarget 2017, 8, 13690–13702. [Google Scholar] [CrossRef]
- Patel, R.S.; Impreso, S.; Lui, A.; Vidyarthi, G.; Albear, P.; Patel, N.A. Long Noncoding RNA GAS5 Contained in Exosomes Derived from Human Adipose Stem Cells Promotes Repair and Modulates Inflammation in a Chronic Dermal Wound Healing Model. Biology 2022, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Stern, A.; Chiu, D.T. G6PD: A hub for metabolic reprogramming and redox signaling in cancer. Biomed. J. 2021, 44, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Valverde, A.; Naqvi, R.A.; Naqvi, A.R. Long Non-coding RNAs RN7SK and GAS5 Regulate Macrophage Polarization and Innate Immune Responses. Front. Immunol. 2020, 11, 604981. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 106. Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.J.; Wang, W.; Hann, S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019, 163, 58–72. [Google Scholar] [CrossRef]
- Poursheikhani, A.; Abbaszadegan, M.R.; Kerachian, M.A. Mechanisms of long non-coding RNA function in colorectal cancer tumorigenesis. Asia Pac. J. Clin. Oncol. 2021, 17, 7–23. [Google Scholar] [CrossRef]
- Li, J.; Han, T.; Wang, X.; Wang, Y.; Yang, Q. Identification of novel survival-related lncRNA-miRNA-mRNA competing endogenous RNA network associated with immune infiltration in colorectal cancer. Am. J. Transl. Res. 2021, 13, 5815–5834. [Google Scholar]
- Zhu, Y.; Bian, Y.; Zhang, Q.; Hu, J.; Li, L.; Yang, M.; Qian, H.; Yu, L.; Liu, B.; Qian, X. Construction and analysis of dysregulated lncRNA-associated ceRNA network in colorectal cancer. J. Cell Biochem. 2019, 120, 9250–9263. [Google Scholar] [CrossRef]
- Aydın, E.; Saus, E.; Chorostecki, U.; Gabaldón, T. A hybrid approach to assess the structural impact of long noncoding RNA mutations uncovers key NEAT1 interactions in colorectal cancer. IUBMB Life 2023, 75, 566–579. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Z.; Ji, M.; Yan, T.; Jiang, Y.; Chen, Y.; Chang, J.; Zhang, J.; Tang, D.; Zhu, D.; et al. The Establishment and Experimental Verification of an lncRNA-Derived CD8+ T Cell Infiltration ceRNA Network in Colorectal Cancer. Clin. Med. Insights Oncol. 2022, 16, 11795549221092218. [Google Scholar] [CrossRef]
- Zhou, Y.; Shao, Y.; Hu, W.; Zhang, J.; Shi, Y.; Kong, X.; Jiang, J. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ. 2023, 30, 111–124. [Google Scholar] [CrossRef]
- Guo, X.; Liang, X.; Wang, Y.; Cheng, A.; Qin, C.; Zhang, H.; Wang, Z. Construction and Comprehensive Prognostic Analysis of a lncRNA-miRNA-mRNA Regulatory Network and Tumor Immune Cell Infiltration in Colorectal Cancer. Front. Genet. 2021, 12, 652601. [Google Scholar] [CrossRef] [PubMed]
- Luan, L.; Dai, Y.; Shen, T.; Yang, C.; Chen, Z.; Liu, S.; Jia, J.; Li, Z.; Fang, S.; Qiu, H.; et al. Development of a Novel Hypoxia-Immune-Related LncRNA Risk Signature for Predicting the Prognosis and Immunotherapy Response of Colorectal Cancer. Front. Immunol. 2022, 13, 951455. [Google Scholar] [CrossRef] [PubMed]
- Mezher, M.; Abdallah, S.; Ashekyan, O.; Shoukari, A.A.; Choubassy, H.; Kurdi, A.; Temraz, S.; Nasr, R. Insights on the Biomarker Potential of Exosomal Non-Coding RNAs in Colorectal Cancer: An In Silico Characterization of Related Exosomal lncRNA/circRNA-miRNA-Target Axis. Cells 2023, 12, 1081. [Google Scholar] [CrossRef]
- Gao, Z.; Fu, P.; Yu, Z.; Zhen, F.; Gu, Y. Comprehensive Analysis of lncRNA-miRNA-mRNA Network Ascertains Prognostic Factors in Patients with Colon Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819853237. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, T.; Khalaj-Kondori, M.; Hosseinpour Feizi, M.A.; Asadi, P. lncRNA-miRNA-mRNA Interaction Network for Colorectal Cancer; An in Silico Analysis. Comput. Biol. Chem. 2020, 89, 107370. [Google Scholar] [CrossRef]
- Tan, X.; Mao, L.; Huang, C.; Yang, W.; Guo, J.; Chen, Z.; Chen, Z. Comprehensive analysis of lncRNA-miRNA-mRNA regulatory networks for microbiota-mediated colorectal cancer associated with immune cell infiltration. Bioengineered 2021, 12, 3410–3425. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, X.; Song, X.; Xie, L. Identification of Diagnostic Exosomal LncRNA-miRNA-mRNA Biomarkers in Colorectal Cancer Based on the ceRNA Network. Pathol. Oncol. Res. 2022, 28, 1610493. [Google Scholar] [CrossRef]
- Wan, X.; Du, J.; Fang, Y.; Huang, D.; Dong, H.; Cai, S. GW4869 Can Inhibit Epithelial-Mesenchymal Transition Promoted by Extracellular HSP90α in EGFR-Mutated NSCLC Cells. 2022. Available online: https://www.researchsquare.com/article/rs-1783485/v1 (accessed on 8 August 2023).
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2019, 9, 1703244. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Choi, Y.; Yim, H.Y.; Mirzaaghasi, A.; Yoo, J.K.; Choi, C. Biodistribution of Exosomes and Engineering Strategies for Targeted Delivery of Therapeutic Exosomes. Tissue Eng. Regen. Med. 2021, 18, 499–511. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, D.; Ma, X.; Wang, J.; Hou, W.; Zhang, W. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed. Pharmacother. 2020, 128, 110237. [Google Scholar] [CrossRef]
- Qi, F.; Tan, B.; Ma, F.; Zhu, B.; Zhang, L.; Liu, X.; Li, H.; Yang, J.; Cheng, B. A Synthetic Light-switchable System based on CRISPR Cas13a Regulates the Expression of LncRNA MALAT1 and Affects the Malignant Phenotype of Bladder Cancer Cells. Int. J. Biol. Sci. 2019, 15, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Bian, Y.; Ma, X.; Tang, Z.; Chen, N.; Shen, M. LncRNA H19 Knockdown in Human Amniotic Mesenchymal Stem Cells Suppresses Angiogenesis by Associating with EZH2 and Activating Vasohibin-1. Stem. Cells Dev. 2019, 28, 781–790. [Google Scholar] [CrossRef]
- Shui, X.; Chen, S.; Lin, J.; Kong, J.; Zhou, C.; Wu, J. Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T cell differentiation through reducing the STAT3 protein level. J. Cell Physiol. 2019, 234, 22477–22484. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, Z.; Tan, X.; Jiang, H.; Xu, Z.; Fang, Y.; Han, D.; Hong, W.; Wei, W.; Tu, J. CAR-macrophage: A new immunotherapy candidate against solid tumors. Biomed. Pharmacother. 2021, 139, 111605. [Google Scholar] [CrossRef]
- Zamani, F.; Suzuki, T. Synthetic RNA Modulators in Drug Discovery. J. Med. Chem. 2021, 64, 7110–7155. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, A.; Czerniak, A.; Levy, T.; Amiur, S.; Gallula, J.; Matouk, I.; Abu-lail, R.; Sorin, V.; Birman, T.; de Groot, N.; et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Rosso, G.; Cauda, V. Biomimicking Extracellular Vesicles with Fully Artificial Ones: A Rational Design of EV-BIOMIMETICS toward Effective Theranostic Tools in Nanomedicine. ACS Biomater. Sci. Eng. 2022. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Gao, W.; Xie, N. Exosomes as Anticancer Drug Delivery Vehicles: Prospects and Challenges. Front. Biosci. 2022, 27, 293. [Google Scholar] [CrossRef]
- Hazekawa, M.; Nishinakagawa, T.; Hosokawa, M.; Ishibashi, D. Development of an Organ-Directed Exosome-Based siRNA-Carrier Derived from Autologous Serum for Lung Metastases and Testing in the B16/BL6 Spontaneous Lung Metastasis Model. Pharmaceutics 2022, 14, 815. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, L.; Zhang, Y.; Lu, R. Plant-Derived Exosomes as a Drug-Delivery Approach for the Treatment of Inflammatory Bowel Disease and Colitis-Associated Cancer. Pharmaceutics 2022, 14, 822. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, Y.; Ni, X.; Yang, Y.; Fu, Z.; Liu, R.; Zhang, C.Y.; Chen, X. In vivo self-assembled small RNA targets H19 lncRNA for the treatment of colorectal cancer. J. Control. Release 2023, 358, 142–160. [Google Scholar] [CrossRef]
- Hattab, D.; Gazzali, A.M.; Bakhtiar, A. Clinical Advances of siRNA-Based Nanotherapeutics for Cancer Treatment. Pharmaceutics 2021, 13, 1009. [Google Scholar] [CrossRef]
- Roy, R.K.; Yadav, R.; Sharma, U.; Wasson, M.K.; Sharma, A.; Tanwar, P.; Jain, A.; Prakash, H. Impact of noncoding RNAs on cancer directed immune therapies: Now then and forever. Int. J. Cancer 2022, 151, 981–992. [Google Scholar] [CrossRef]
- Franz, C.; Wuehrl, M.; Hartmann, S.; Klupp, F.; Schmidt, T.; Schneider, M. Long non-coding RNAs CCAT1 and CCAT2 in colorectal liver metastases are tumor-suppressive via MYC interaction and might predict patient outcomes. PLoS ONE 2023, 18, e0286486. [Google Scholar] [CrossRef]
- Barczak, W.; Carr, S.M.; Liu, G.; Munro, S.; Nicastri, A.; Lee, L.N.; Hutchings, C.; Ternette, N.; Klenerman, P.; Kanapin, A.; et al. Long non-coding RNA-derived peptides are immunogenic and drive a potent anti-tumour response. Nat. Commun. 2023, 14, 1078. [Google Scholar] [CrossRef]
- Munteanu, M.C.; Sethuraman, S.N.; Singh, M.P.; Malayer, J.; Ranjan, A. LncRNA FENDRR Expression Correlates with Tumor Immunogenicity. Genes 2021, 12, 897. [Google Scholar] [CrossRef]
- Li, G.; Kryczek, I.; Nam, J.; Li, X.; Li, S.; Li, J.; Wei, S.; Grove, S.; Vatan, L.; Zhou, J.; et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat. Cell Biol. 2021, 23, 526–537. [Google Scholar] [CrossRef]
- Toker, J.; Iorgulescu, J.B.; Ling, A.L.; Villa, G.R.; Gadet, J.A.M.A.; Parida, L.; Getz, G.; Wu, C.J.; Reardon, D.A.; Chiocca, E.A.; et al. Clinical Importance of the lncRNA NEAT1 in Cancer Patients Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2023, 29, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Xia, J.; Jiao, M.; Zhao, P.; Wang, Z.; Lin, S.; Xing, Y.; Li, Y.; Lu, Z.; Zhong, Z.; et al. Exosomal lncRNA HOTAIR induces PDL1+ B cells to impede anti-tumor immunity in colorectal cancer. Biochem. Biophys. Res. Commun. 2023, 644, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Chen, Y.; Yang, L.; Xu, T.; Wang, F.; Chen, L.; Liu, J.; Liu, G. LncRNA SNHG4 promotes malignant biological behaviors and immune escape of colorectal cancer cells by regulating the miR-144-3p/MET axis. Am. J. Transl. Res. 2021, 13, 11144–11161. [Google Scholar] [PubMed]
- Xian, D.; Niu, L.; Zeng, J.; Wang, L. LncRNA KCNQ1OT1 Secreted by Tumor Cell-Derived Exosomes Mediates Immune Escape in Colorectal Cancer by Regulating PD-L1 Ubiquitination via MiR-30a-5p/USP22. Front. Cell Dev. Biol. 2021, 9, 653808. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Tang, Y.; Yang, M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342-3p/ANXA2 axis. J. Transl. Med. 2021, 19, 8. [Google Scholar] [CrossRef]
- Zheng, J.; Dou, R.; Zhang, X.; Zhong, B.; Fang, C.; Xu, Q.; Di, Z.; Huang, S.; Lin, Z.; Song, J.; et al. LINC00543 promotes colorectal cancer metastasis by driving EMT and inducing the M2 polarization of tumor-associated macrophages. J. Transl. Med. 2023, 21, 153. [Google Scholar] [CrossRef]
- Chen, R.; Wei, J.M. Integrated analysis identifies oxidative stress-related lncRNAs associated with progression and prognosis in colorectal cancer. BMC Bioinform. 2023, 24, 76. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhang, D.; Wang, T.; Ji, J.; Jin, C.; Peng, C.; Tan, Y.; Zhou, J.; Wang, L.; Feng, Y.; et al. CAF-derived exosomal WEE2-AS1 facilitates colorectal cancer progression via promoting degradation of MOB1A to inhibit the Hippo pathway. Cell Death Dis. 2022, 13, 796. [Google Scholar] [CrossRef]
- de Lima, D.S.; Cardozo, L.E.; Maracaja-Coutinho, V.; Suhrbier, A.; Mane, K.; Jeffries, D.; Silveira, E.L.V.; Amaral, P.P.; Rappuoli, R.; de Silva, T.I.; et al. Long noncoding RNAs are involved in multiple immunological pathways in response to vaccination. Proc. Natl. Acad. Sci. USA 2019, 116, 17121–17126. [Google Scholar] [CrossRef] [PubMed]
- Lüscher-Dias, T.; Conceição, I.M.; Schuch, V.; Maracaja-Coutinho, V.; Amaral, P.P.; Nakaya, H.I. Long non-coding RNAs associated with infection and vaccine-induced immunity. Essays Biochem. 2021, 65, 657–669. [Google Scholar] [CrossRef]
- Ventura, M.I.; Azizian, A.; Evans, S.E.; Velasquez, S.; Arguello, J.C.; Warburton, K. Vaccine breakthrough infections with SARS-CoV-2: Why older adults need booster vaccinations. Public Health Pract. 2022, 4, 100307. [Google Scholar] [CrossRef]
- Chattopadhyay, P.; Mishra, P.; Mehta, P.; Soni, J.; Gupta, R.; Tarai, B.; Budhiraja, S.; Pandey, R. Transcriptomic study reveals lncRNA-mediated downregulation of innate immune and inflammatory response in the SARS-CoV-2 vaccination breakthrough infections. Front. Immunol. 2022, 13, 1035111. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Noncoding RNAs: Regulatory Molecules of Host-Microbiome Crosstalk. Trends Microbiol. 2021, 29, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, M.; Zhang, J.; Guo, B.; Singh, S.; Lin, X.; Xiong, H.; Ju, S.; Wang, L.; Zhou, Y.; et al. The role of lncRNA H19 in tumorigenesis and drug resistance of human Cancers. Front. Genet. 2022, 13, 1005522. [Google Scholar] [CrossRef]


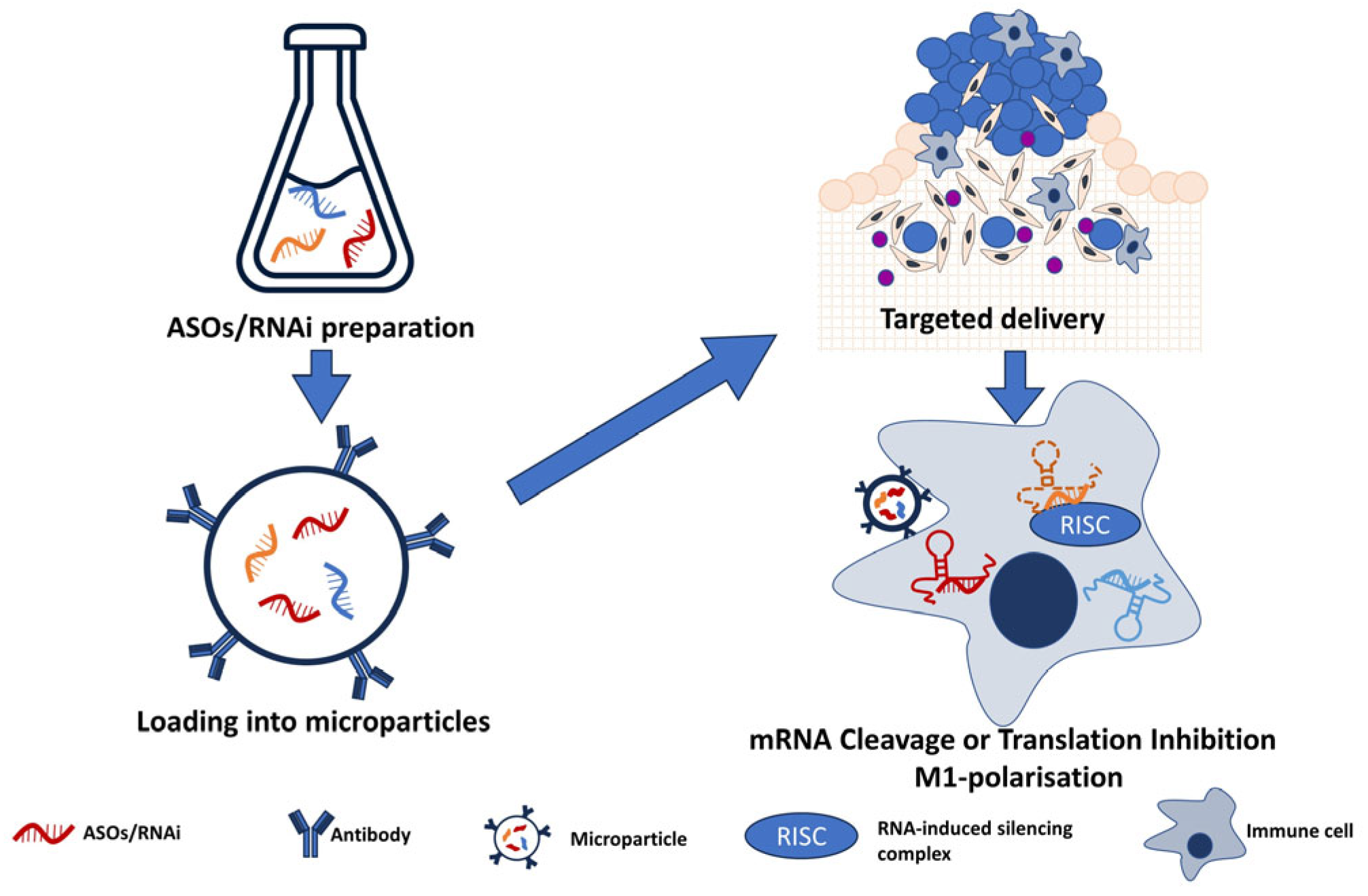
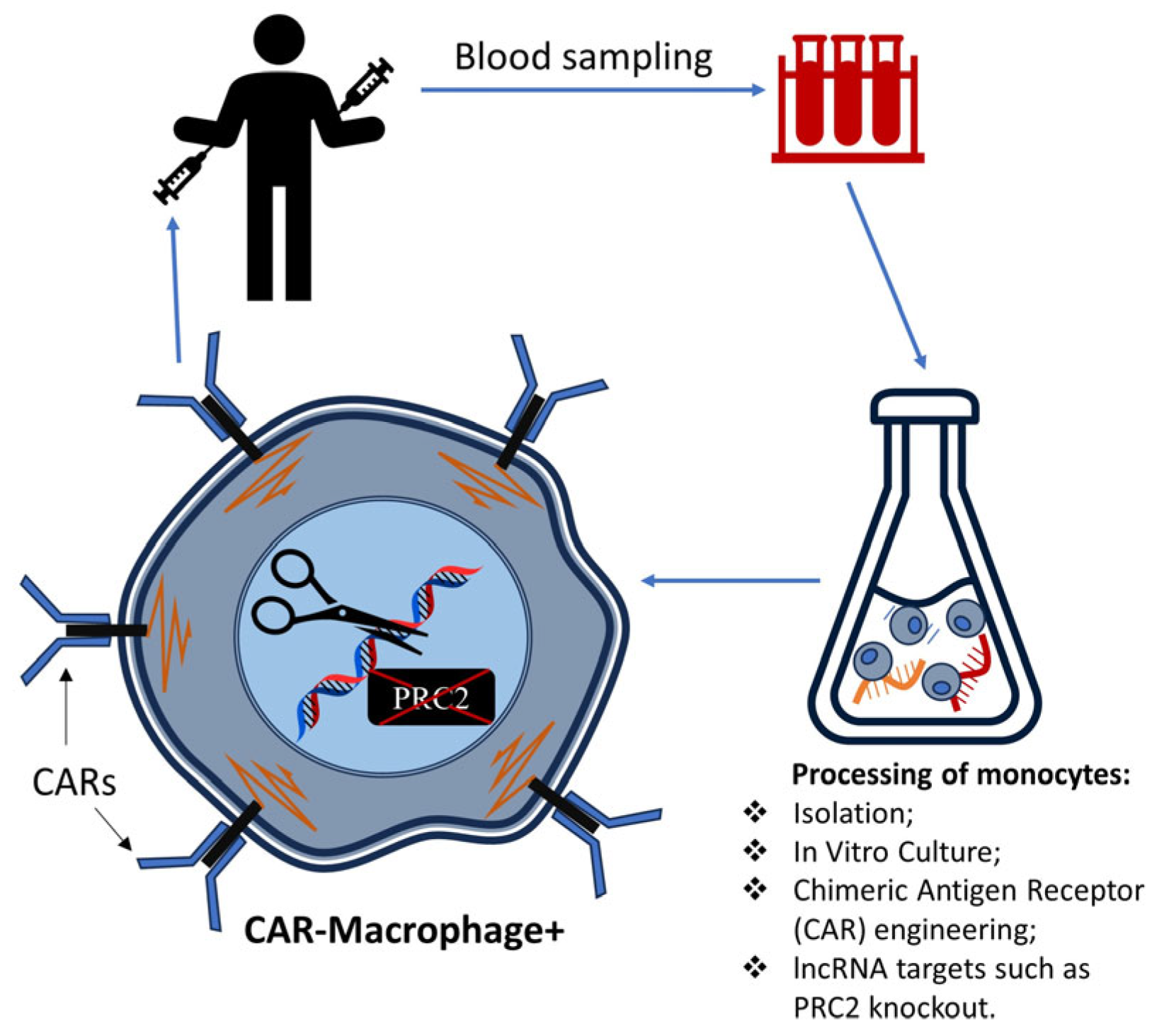
| Classification | Description |
|---|---|
| Sense lncRNAs | Transcribed from the same strand as a protein-coding gene and may overlap entirely or partially with the gene [12]. |
| Antisense lncRNAs | Transcribed from the opposite strand of a protein-coding gene and may overlap with exons or introns [13]. |
| Intronic lncRNAs | Located within the introns of a protein-coding gene but transcribed independently [14]. |
| Intergenic lncRNAs | Situated between protein-coding genes and do not overlap with them. Also known as long intergenic non-coding RNAs (lincRNAs) [15]. |
| Bidirectional lncRNAs | Transcribed in close proximity to a protein-coding gene but in the opposite direction [16]. |
| Enhancer lncRNAs (eRNAs) | Associated with enhancer regions and may regulate the activity of enhancers, influencing gene expression [17]. |
| Broad Function | Specific Mechanism | Description |
|---|---|---|
| Gene Expression Regulation | Transcriptional Control | Involves the activation/repression of transcription, enhancer activity, RNA polymerase interference, chromatin remodeling, histone modification, and DNA methylation [18,19,20]. |
| Post-transcriptional Control | Includes the regulation of splicing, mRNA stability, and translation [21,22]. | |
| RNA Interactions | miRNA Sponging | lncRNAs may sequester miRNAs away from their target mRNAs [23]. |
| RNA-RNA Interactions | Includes base pairing with other RNAs, affecting function or stability [24]. | |
| RNA–Protein Interactions | Scaffolding and Sequestration | lncRNAs can act as scaffolds for protein complexes or sequester proteins away from functional locations. May overlap with gene expression regulation and RNA interactions [25,26]. |
| Structural Roles | Nuclear Architecture | Contributes to the organization of nuclear structures. May have indirect effects on gene regulation [27]. |
| Signaling Regulation | Pathway Modulation | Involves interactions with signaling molecules or pathway components, potentially impacting various cellular processes, including gene expression, growth, and stress [28,29]. |
| CMS Subtype | Typical Molecular Genetic Alterations |
|---|---|
| CMS1 (MSI Immune) | High microsatellite instability (MSI-H), DNA mismatch repair (MMR) deficiency, hypermutated phenotype, and a high neoantigen load. |
| CMS2 (Canonical) | Chromosomal instability, a high level of somatic copy number alterations, the activation of the WNT and MYC signaling pathways, and mutations in APC and TP53. |
| CMS3 (Metabolic) | Microsatellite stable (MSS), metabolic dysregulation, KRAS mutations, and involvement in the PI3K/AKT signaling pathway and possibly others that affect metabolism, such as CDK2 signaling. |
| CMS4 (Mesenchymal) | Stromal invasion and involvement in the RAS/MAPK, Rb/E2F, CDK8/β-catenin, and Raf/ERK pathways. There is a focus on the epithelial-to-mesenchymal transition (EMT) and the regulation of pathways related to cell growth and migration. |
| lncRNA | CMS Subtype | Main Characteristics | References |
|---|---|---|---|
| HOTAIR | CMS1 (MSI Immune) | Antisense lncRNA. Gene expression regulation. Oncogenic. Interacts with PRC2 and LSD1 to modulate H3K27 methylation, affecting gene silencing. Consequences: PTEN methylation, PI3K/p-AKT/p-MDM2/p53, and PI3K/AKT/mTOR pathways in tumorigenesis; regulates ASTN1, PCDHA1, and MUC5AC in metastasis. | [49,50,53,54] |
| LINK-A (LINC01139) | CMS1 (MSI Immune) | Intergenic lncRNA. Pathway modulation. Oncogenic. Facilitates crosstalk between the PIP3 and GPCR pathways, attenuating PKA activity on TRIM71, leading to the degradation of PLC and tumor suppressors Rb and p53. Directly binds to phosphatidylcholine, AKT, and PIP3, causing AKT hyperactivation and tumorigenesis. | [55,56,57,58] |
| CCAT1 | CMS2 (Canonical) | Intergenic lncRNA. Nuclear architecture; scaffolding. Oncogenic. Mediates chromosome looping with CTCF, affecting c-Myc promoter and promoting c-Myc expression. Acts as a ceRNA; serves as a scaffold for epigenetic complexes, with chromosome looping central to interaction. | [59,60] |
| CRNDE | CMS2 (Canonical) | Intergenic lncRNA. miRNA sponging; pathway modulation; transcriptional control. Oncogenic. Molecular sponge for miRNAs; promotes cell growth. Activates/inhibits the Wnt/β-catenin, PI3K/AKT/mTOR, Ras/MAPK, and Notch1 signaling pathways. Binds to EZH2. | [61,62,63] |
| lncRNA-ATB | CMS3 (Metabolic) | Intergenic lncRNA. miRNA sponging. Oncogenic. Interacts with miR-141-3p and miR-200c, influencing the CDK2 pathway, affecting EMT process, and contributing to cancer progression. | [64,65,66,67] |
| RP11-462C24.1 (RPL34-DT) | CMS3 (Metabolic) | Intergenic lncRNA. Pathway modulation; transcriptional control. Oncosuppressive. Upregulates HSP70; inhibits the PI3K/AKT signaling pathway. | [68,69] |
| H19 | CMS4 (Mesenchymal) | Intergenic lncRNA. RNA interactions; pathway modulation. Oncogenic. Promotes CRC progression by targeting RB with miR-675, sponging miR-200a and miR-138, leading to HMGA2 upregulation. Activates the RAS/MAPK, Rb/E2F, CDK8/β-catenin, and Raf/ERK pathways. | [70,71,72,73,74] |
| lincRNA-p21 (TP53COR1) | CMS4 (Mesenchymal) | Intergenic lncRNA. RNA–RNA interactions; RNA–protein interactions. Oncosuppressive. Interacts with the JUNB and CTNNB1 mRNAs, reducing translation. Antagonism via HuR. mTOR/lincRNA-p21 involved in carcinogenesis, progression, metastasis. Part of the p53 network. | [75,76,77,78] |
| Mechanism | Key Components | Effect | Key Details |
|---|---|---|---|
| The secretion of pro-inflammatory cytokines | CAFs and TAMs | Induces inflammation | CAFs and TAMs secrete IL-6 and TNF-α, which can foster chronic inflammation, paradoxically promoting tumor progression. |
| The secretion of immunosuppressive factors | CAFs and TAMs | Immunosuppression | CAFs and TAMs secrete TGF-β, IL-10, and PD-L1, which inhibit T cells and promote Tregs. |
| T cell exhaustion | T cells | Immunosuppression | Chronic exposure to tumor antigens and inflammatory signals can lead to a state of T cell dysfunction characterized by sustained expression of inhibitory receptors (PD-1 and CTLA-4). |
| The recruitment of regulatory immune cells | MDSCs and Tregs | Immunosuppression | The tumor stroma can attract immunosuppressive cell types like MDSCs and Tregs, which suppress cytotoxic T cells and NK cells. |
| Metabolic reprogramming | Tumor cells and stromal cells | Immunosuppression | Tumor cells and stromal cells can alter the metabolic landscape of the TME, creating conditions like hypoxia and nutrient deprivation that negatively impact immune cell function. |
| The modulation of extracellular matrix (ECM) | CAFs | Creates a physical barrier | CAFs can remodel the ECM, creating a physical barrier that hinders immune cell infiltration and access to tumor cells. |
| Cell polarization | TAMs, CD4+ T cells, MDSCs, and DCs | Immunosuppression | TAMs adopt an M2 polarization state, CD4+ T cells can be polarized into Tregs, MDSCs suppress T cell function, and DCs can become tolerogenic. |
| lncRNA Name (Genome Type) | Mechanism of Action | Effect on Cancer Progression | Interaction with Tumor Microenvironment |
|---|---|---|---|
| CCAT1 (Intergenic) | Influences inflammation, angiogenesis, and immune regulation via the microRNA-138-5p–HMGA1 axis in exosomes. Promotes immune cell polarization and pro-inflammatory cytokine release. | Promoting | Mediates angiogenesis and influences immune interactions in CRC. |
| CCAT2 (Sense) | Facilitates progression via PI3K/AKT/mTOR signaling. Enhances growth and metastasis via interactions with TAF15 to stimulate RAB14 transcription, triggering AKT/GSK3β signaling. Modulates the hsa-miR-145-5p/AKT3/mTOR axis in MCF7 cells. | Promoting | Plays a role in tumor–stroma immune interplay. |
| CRNDE (Intergenic) | Influences inflammation and immune evasion by releasing immunosuppressive factors, inducing T cell exhaustion, and recruiting regulatory immune cells. Activates NF-κB and JAK/STAT signaling. Participates in Th17 differentiation via CRNDE-h isoform interactions with RORγt. | Promoting | Mediates tumor–stroma immune interplay. |
| H19 (Intergenic) | Guides inflammation, pro-inflammatory cytokine release, and extracellular matrix remodeling via the upregulation of TNF-α. Involved in the STAT3 pathway. | Promoting | Facilitates interactions with immune cells in CRC. |
| HOTAIR (Antisense) | Connected with tumor grade and prognosis. Influences B cells toward a regulatory role via PDL1, suppressing CD8+ T cell activity. May stimulate pro-inflammatory cytokines and extracellular matrix remodeling. | Promoting | Suppresses CD8+ T cell activity. |
| HULC (Antisense) | Influences immune response, enhancing EZH2-H3K27me3 enrichment, and targets miR-613 and miR-488, promoting cell proliferation and suppressing p53 expression, which may facilitate tumor growth and metastasis. | Promoting | Influences immune response, promotes tumor growth and metastasis. |
| LINC00461 (Intergenic) | Mixed effects on CRC development and immunity. Promotes tumor growth and proliferation via the miR-323b-3p/NFIB axis. Acts as a competitive endogenous RNA (ceRNA) for PHLPP2, a colon cancer tumor suppressor. | Mixed | Influences cell migration, invasion, and transition, and the epithelial–mesenchymal transition. |
| lnc-ATB (Intergenic) | Involved in cancer progression, particularly in CRC, stimulating the release of pro-inflammatory cytokines and enhancing metastasis through pathways involving CDK2 and miR-200c. | Promoting | Enhances cancer metastasis and induces the EMT. |
| lnc-EGFR or EGILA (Antisense) | Facilitates immune evasion in CRC through the EGFR signaling pathway, potentially inducing T cell exhaustion and enhancing Treg differentiation. | Promoting | Enhances tumor immune escape. |
| MALAT1 (Intergenic) | Modulates T cell function, induces pro-inflammatory cytokines, and plays roles in immune evasion and inflammation in various cancers, potentially impacting NF-κB signaling. | Promoting | Suppresses innate and adaptive immune responses. |
| NEAT1 (Intergenic) | Contributes to immunosuppression and stemness maintenance through the regulation of ALDH1 and c-Myc. NEAT1 also influences metabolic and mitochondrial homeostasis, Fosters immune evasion through its expression in M2-polarized tumor-associated macrophages. | Promoting | Promotes immune evasion. |
| PCAT-1 (Antisense) | Enhances pro-inflammatory cytokine secretion and affects cellular processes like proliferation, invasion, and apoptosis by targeting miR-149-5p. | Promoting | Fosters an inflammatory tumor microenvironment. |
| SNHG1 (Intergenic) | Promotes pro-inflammatory cytokine secretion, cell proliferation, migration, and the EMT. It is linked with the Wnt/β-catenin pathway and molecules like MYC and SLC3A2, affecting immune responses and tumor progression. | Mixed | Influences immune response and induces the EMT. |
| SNHG14 (Sense) | Promotes CRC progression by negatively regulating EPHA7 through an EZH2-dependent pathway, enhancing methylation on the EPHA7 promoter and stabilizing EZH2 mRNA by interacting with FUS and freeing it from miR-186-5p-induced silence. | Promoting | Regulator of the immune response. |
| SOX2OT (Sense) | Related to inflammation and oncogenesis. It regulates pro-inflammatory cytokines and is linked with SOX2. Its silencing suppresses CRC cell growth and alters miR-194-5p | Mixed | Affects cell behavior in the TME. |
| TUG1 (Intergenic) | Interacts with the miR-138-5p/ZEB2 axis, promoting the EMT and, on the other hand, an immunosuppressive environment. | Mixed | Affects cancer cell behavior in the TME. |
| GAS5 (Antisense) | Downregulates IL-10 and VEGF-A via the NF-κB and Erk1/2 pathways. By inhibiting these cytokines, GAS5 suppresses tumor cell proliferation and promotes the tumor-suppressive function of M1 macrophages | Inhibiting | Reduces tumor immune escape, potentially reducing angiogenesis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shakhpazyan, N.K.; Mikhaleva, L.M.; Bedzhanyan, A.L.; Sadykhov, N.K.; Midiber, K.Y.; Konyukova, A.K.; Kontorschikov, A.S.; Maslenkina, K.S.; Orekhov, A.N. Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential. Biomedicines 2023, 11, 2411. https://doi.org/10.3390/biomedicines11092411
Shakhpazyan NK, Mikhaleva LM, Bedzhanyan AL, Sadykhov NK, Midiber KY, Konyukova AK, Kontorschikov AS, Maslenkina KS, Orekhov AN. Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential. Biomedicines. 2023; 11(9):2411. https://doi.org/10.3390/biomedicines11092411
Chicago/Turabian StyleShakhpazyan, Nikolay K., Liudmila M. Mikhaleva, Arcady L. Bedzhanyan, Nikolay K. Sadykhov, Konstantin Y. Midiber, Alexandra K. Konyukova, Andrey S. Kontorschikov, Ksenia S. Maslenkina, and Alexander N. Orekhov. 2023. "Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential" Biomedicines 11, no. 9: 2411. https://doi.org/10.3390/biomedicines11092411
APA StyleShakhpazyan, N. K., Mikhaleva, L. M., Bedzhanyan, A. L., Sadykhov, N. K., Midiber, K. Y., Konyukova, A. K., Kontorschikov, A. S., Maslenkina, K. S., & Orekhov, A. N. (2023). Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential. Biomedicines, 11(9), 2411. https://doi.org/10.3390/biomedicines11092411







