Finding the Common Single-Nucleotide Polymorphisms in Three Autoimmune Diseases and Exploring Their Bio-Function by Using a Reporter Assay
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Selection of Candidate SNPs
2.3. DNA Extraction and Sequencing
2.4. Promoter–Reporter Construction
2.5. Cell Culture and Transient Transfections
2.6. Dual-Luciferase Reporter Assay
2.7. Statistical Analysis
3. Results
3.1. Study Subjects
3.2. Hardy–Weinberg Equilibrium Test
3.3. Allele and Genotype Analysis
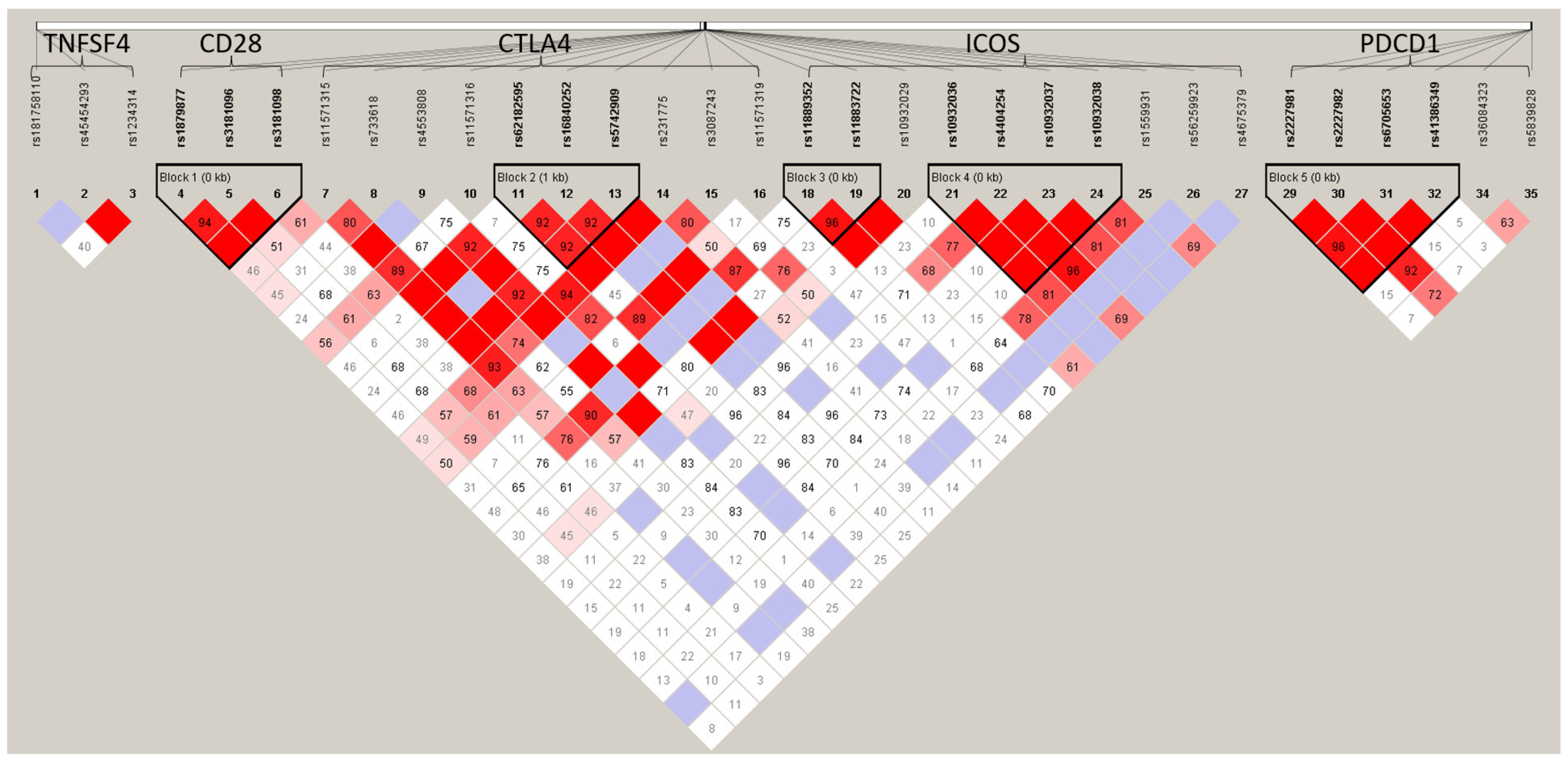
3.4. Haplotype Analysis
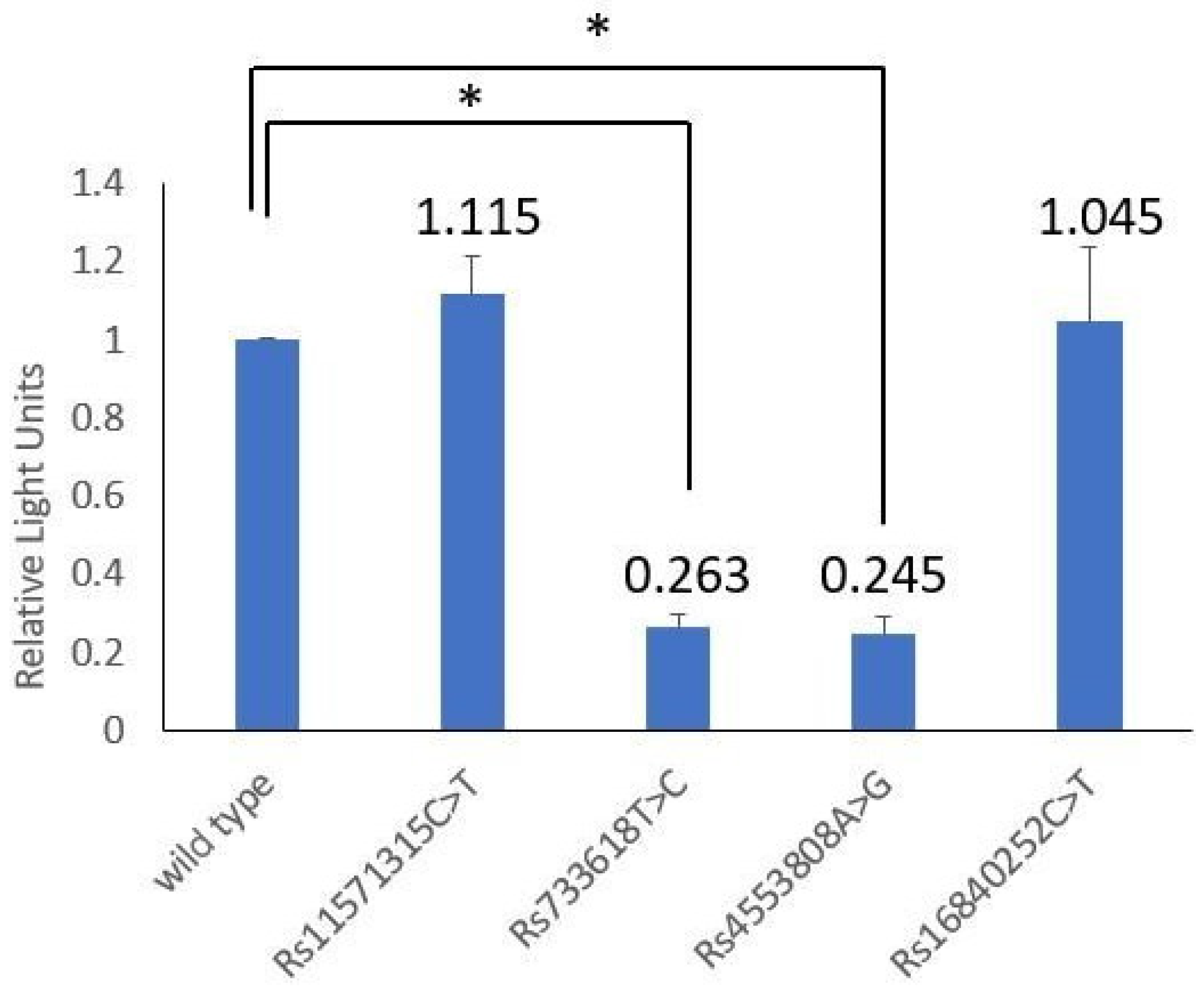
3.5. Transcriptional Activity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khoo, T.K.; Bahn, R.S. Pathogenesis of Graves’ ophthalmopathy: The role of autoantibodies. Thyroid 2007, 17, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Miao, H.B.; Lin, S.; Chen, Z. Association between rheumatoid arthritis and thyroid dysfunction: A meta-analysis and systematic review. Front. Endocrinol. 2022, 13, 1015516. [Google Scholar] [CrossRef]
- AL-Homood, I.A.; Alkhathami, R.A.; Alenazi, S.K.; Mohammed, A.A. Thyroid dysfunction among patients with systemic lupus erythematosus in Saudi Arabia. Dr. Sulaiman Al Habib Med. J. 2022, 4, 169–173. [Google Scholar] [CrossRef]
- Chan, A.T.; Al-Saffar, Z.; Bucknall, R.C. Thyroid disease in systemic lupus erythematosus and rheumatoid arthritis. Rheumatology 2001, 40, 353–354. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, W.; Doniach, D.; Roitt, I.M.; Holborow, E.J. Serological overlap between lupus erythematosus, rheumatoid arthritis, and thyroid autoimmune disease. Br. Med. J. 1961, 2, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Morand, E.F.; McCloud, P.I.; Littlejohn, G.O. Continuation of long term treatment with hydroxychloroquine in systemic lupus erythematosus and rheumatoid arthritis. Ann. Rheum. Dis. 1992, 1, 1318–1321. [Google Scholar] [CrossRef]
- Klionsky, Y.; Antonelli, M. Thyroid disease in lupus: An updated review. ACR Open Rheumatol. 2020, 2, 74–78. [Google Scholar] [CrossRef]
- Lee, C.; Chen, S.F.; Yang, Y.C.; Hsu, C.Y.; Shen, Y.C. Association between Graves’ disease and risk of incident systemic lupus erythematosus: A nationwide population-based cohort study. Int. J. Rheum. Dis. 2021, 24, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Miller, F.W.; Moore, G.F.; Weintraub, B.D.; Steinberg, A.D. Prevalence of thyroid disease and abnormal thyroid function test results in patients with systemic lupus erythematosus. Arthritis Rheum. 1987, 30, 1124–1131. [Google Scholar] [CrossRef]
- Wu, D.; Xian, W.; Hong, S.; Liu, B.; Xiao, H.; Li, Y. Graves’ disease and rheumatoid arthritis: A bidirectional mendelian randomization study. Front. Endocrinol. 2021, 12, 702482. [Google Scholar] [CrossRef]
- Read, R.W. Clinical mini-review: Systemic lupus erythematosus and the eye. Ocul. Immunol. Inflamm. 2004, 12, 87–99. [Google Scholar] [CrossRef]
- Foster, C.S.; Vitale, A.T. Diagnosis & Treatment of Uveitis, 2nd ed.; Jaypee Brothers Medical Publishers Ltd.: New Delhi, India, 2013. [Google Scholar]
- Martin, L. The hereditary and familial aspects of exophthalmic goitre and nodular goitre. Q. J. Med. 1945, 14, 207–219. [Google Scholar] [PubMed]
- Ferrari, S.M.; Fallahi, P.; Ruffilli, I.; Elia, G.; Ragusa, F.; Benvenga, S.; Antonelli, A. The association of other autoimmune diseases in patients with Graves’ disease (with or without ophthalmopathy): Review of the literature and report of a large series. Autoimmun. Rev. 2019, 18, 287–292. [Google Scholar] [CrossRef]
- Choi, J.; Kim, S.T.; Craft, J. The pathogenesis of systemic lupus erythematosus-an update. Curr. Opin. Immunol. 2012, 24, 651–657. [Google Scholar] [CrossRef]
- Khan, M.S.; Lone, S.S.; Faiz, S.; Farooq, I.; Majid, S. Graves’ Disease: Pathophysiology, Genetics and Management; IntechOpen: London, UK, 2021; p. 67. [Google Scholar]
- Karami, J.; Aslani, S.; Jamshidi, A.; Garshasbi, M.; Mahmoudi, M. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 2019, 702, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.P.; Lin, W.T.; Yu, K.H. Investigation of the association between the genetic polymorphisms of the co-stimulatory system and systemic lupus erythematosus. Front. Immunol. 2022, 13, 946456. [Google Scholar]
- Chen, D.P.; Wen, Y.H.; Lin, W.T.; Hsu, F.P.; Yu, K.H. Exploration of the association between the single nucleotide polymorphism of co-stimulatory system and rheumatoid arthritis. Front. Immunol. 2023, 14, 1123832. [Google Scholar] [CrossRef]
- Chen, D.P.; Chu, Y.C.; Wen, Y.H.; Lin, W.T.; Hour, A.L.; Wang, W.T. Investigation of the correlation between Graves’ ophthalmopathy and CTLA4 gene polymorphism. J. Clin. Med. 2019, 8, 1842. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; Defelice, M. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
- Du, P.; Ma, X.; Wang, C. Associations of CTLA4 gene polymorphisms with Graves’ ophthalmopathy: A meta-analysis. Int. J. Genom. 2014, 2014, 537969. [Google Scholar] [CrossRef]
- Chen, P.L.; Fann, C.S.; Chang, C.C.; Wu, I.L.; Chiu, W.Y.; Lin, C.Y.; Yang, W.-S.; Chang, T.-C. Family-based association study of cytotoxic T-lymphocyte antigen-4 with susceptibility to Graves’ disease in Han population of Taiwan. Genes Immun. 2008, 9, 87–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, S.X.; Pan, C.M.; Cao, H.M.; Han, B.; Shi, J.Y.; Liang, J.; Gao, G.Q.; Peng, Y.D.; Su, Q. Association of the CTLA4 gene with Graves’ disease in the Chinese Han population. PLoS ONE 2010, 5, e9821. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Z.; Liu, M.; Li, H.; Liang, C.; Li, W.; Bao, L.; Chen, M.; Wu, G. Correlation between CTLA-4 and CD40 gene polymorphisms and their interaction in graves’ disease in a Chinese Han population. BMC Med. Genet. 2018, 19, 171. [Google Scholar] [CrossRef]
- Kaykhaei, M.; Moghadam, H.; Dabiri, S.; Salimi, S.; Jahantigh, D.; Tamandani, D.M.K.; Rasouli, A. Association of CTLA4 (rs4553808) and PTPN22 (rs2476601) gene polymorphisms with Hashimoto’s thyroiditis disease: A case-control study and an In-silico analysis. Meta Gene 2020, 24, 100693. [Google Scholar] [CrossRef]
- Zou, C.; Qiu, H.; Tang, W.; Wang, Y.; Lan, B.; Chen, Y. CTLA4 tagging polymorphisms and risk of colorectal cancer: A case-control study involving 2,306 subjects. Onco Targets Ther. 2018, 11, 4609–4619. [Google Scholar] [CrossRef]
- Huang, X.; Liu, X.; Ye, Y.; Zhang, T.; Mei, S.; Zhu, T.; Peng, S.; Cai, J.; Yan, Z.; Zeng, K.; et al. Polymorphisms and circulating plasma protein levels of immune checkpoints (CTLA-4 and PD-1) are associated with Posner-Schlossman syndrome in Southern Chinese. Front. Immunol. 2021, 12, 607966. [Google Scholar] [CrossRef] [PubMed]
- Bouwhuis, M.G.; Gast, A.; Figl, A.; Eggermont, A.M.; Hemminki, K.; Schadendorf, D.; Kumar, R. Polymorphisms in the CD28/CTLA4/ICOS genes: Role in malignant melanoma susceptibility and prognosis? Cancer Immunol. Immunother. 2010, 59, 303–312. [Google Scholar] [CrossRef]
- Haimila, K. Genetics of T Cell Co-Stimulatory Receptors: CD28, CTLA4, ICOS and PDCD1 in Immunity and Transplantation; Academic Dissertations from the Finnish Red Cross Blood Transfusion Service, no. 54; Finnish Red Cross Blood Service: Helsinki, Finland, 2009. [Google Scholar]
- Kawabata, M.; Inoue, N.; Watanabe, M.; Kobayashi, A.; Hidaka, Y.; Miyauchi, A.; Iwatani, Y. PD-1 gene polymorphisms and thyroid expression of PD-1 ligands differ between Graves’ and Hashimoto’s diseases. Autoimmunity 2021, 54, 450–459. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 pathways: Similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.P.; Wen, Y.H.; Wang, W.T.; Lin, W.T. Exploring the bio-functional effect of single nucleotide polymorphisms in the promoter region of the TNFSF4, CD28, and PDCD1 genes. J. Clin. Med. 2023, 12, 2157. [Google Scholar] [CrossRef]
- Ligers, A.; Teleshova, N.; Masterman, T.; Huang, W.X.; Hillert, J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001, 2, 145–152. [Google Scholar] [CrossRef]
- Wang, X.B.; Zhao, X.; Giscombe, R.; Lefvert, A.K. A CTLA-4 gene polymorphism at position -318 in the promoter region affects the expression of protein. Genes Immun. 2002, 3, 233–234. [Google Scholar] [CrossRef]
- Anjos, S.M.; Tessier, M.C.; Polychronakos, C. Association of the cytotoxic T lymphocyte-associated antigen 4 gene with type 1 diabetes: Evidence for independent effects of two polymorphisms on the same haplotype block. J. Clin. Endocrinol. Metab. 2004, 89, 6257–6265. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, L.; Yang, X.H.; Kang, Y.N.; Wen, W.; Zhang, F.Q.; Yue, W.H.; Zhang, Q.; Chen, F.; Cao, W.; Yue, J.; et al. Association of the soluble CTLA4 with schizophrenia: An observational study. J. Bio-X Res. 2020, 3, 116–122. [Google Scholar] [CrossRef]
- GTEx consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Yao, L.; Liu, B.; Jiang, L.; Zhou, L.; Liu, X. Association of cytotoxic T-lymphocyte antigen 4 gene with immune thrombocytopenia in Chinese Han children. Hematology 2019, 24, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, J.; Chen, Y.; Tang, W.; Liu, C.; Sun, Y.; Chen, J. Association of CTLA-4 tagging polymorphisms and haplotypes with hepatocellular carcinoma risk: A case-control study. Medicine 2019, 98, e16266. [Google Scholar] [CrossRef]
- Chen, D.P.; Chang, S.W.; Wang, P.N.; Lin, W.T.; Hsu, F.P.; Wang, W.T.; Tseng, C.P. The association between single-nucleotide polymorphisms of co-stimulatory genes within non-HLA region and the prognosis of leukemia patients with hematopoietic stem cell transplantation. Front. Immunol. 2021, 12, 730507. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, Y.; Chen, S.; Lin, J.; Chen, B.; Yu, S.; Kang, M. Investigation of cytotoxic T-lymphocyte antigen 4 polymorphisms in gastric cardia adenocarcinoma. Scand. J. Immunol. 2016, 83, 212–218. [Google Scholar] [CrossRef]
- Kumar, V.; Abbas, A.K.; Fausto, N.; Mitchell, R.N. Robbins Basic Pathology, 8th ed.; Saunders Elsevier: Philadelphia, PA, USA, 2007. [Google Scholar]
- Xue, K.; Niu, W.Q.; Cui, Y. Association of HLA-DR3 and HLA-DR15 Polymorphisms with Risk of Systemic Lupus Erythematosus. Chin. Med. J. 2018, 131, 2844–2851. [Google Scholar]
- Chowdhary, V.R.; Dai, C.; Tilahun, A.Y.; Hanson, J.A.; Smart, M.K.; Grande, J.P.; Rajagopalan, G. A central role for hla-dr3 in anti-smith antibody responses and glomerulonephritis in a transgenic mouse model of spontaneous lupus. J. Immunol. 2015, 195, 4660–4667. [Google Scholar] [CrossRef] [PubMed]
- Rykova, E.; Ershov, N.; Damarov, I.; Merkulova, T. SNPs in 3’UTR miRNA target sequences associated with individual drug susceptibility. Int. J. Mol. Sci. 2022, 23, 13725. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sequence | Position | Size |
|---|---|---|---|
| KpnI–CTLA4F | 5′-ACAT GGTACC CTTGCTGCTAAGAGCATC-3′ | 203865939- 203867940 | 2001 bp |
| EcoRV–CTLA4R | 5′-AGTA GATATCGGGCTTTATGGGAGCGGT-3′ | ||
| Rs11571315TF | 5′-GCT CCT CTA CAT AAT ACT TCA ATT CCA GCA TTG-3′ | 203866178 | |
| Rs11571315TR | 5′-CAA TGC TGG AAT TGA AGT ATT ATG TAG AGG AGC-3′ | ||
| Rs733618CF | 5′-TCA TGG GTT TAG CTG CCT GTC CCT GCC ACT-3′ | 203866221 | |
| Rs733618CR | 5′-AGT GGC AGG GAC AGG CAG CTA AAC CCA TGA-3′ | ||
| Rs4553808GF | 5′-CAC TTT TTG AAA AAC CTC TGT TGC CCA GTC TGG C-3′ | 203866282 | |
| Rs4553808GR | 5′-GCC AGA CTG GGC AAC AGA GGT TTT TCA AAA AGT G-3′ | ||
| Rs16840252TF | 5′-AAT GGG AAA CCA TGG ATG GAC TGG AGT AGG CA-3′ | 203866796 | |
| Rs16840252TR | 5′-TGC CTA CTC CAG TCC ATC CAT GGT TTC CCA TT-3′ |
| SNP | Position | Allele | Minor Allele Frequency | HWE p-Value | Odds Ratio | pa Value | |
|---|---|---|---|---|---|---|---|
| Patient | Control | (95% CI) | |||||
| CTLA4 | |||||||
| rs11571315 | 203866178 | C/T | 0.163 | 0.4 | 0.925 | 0.291 (0.138–0.612) | 0.001 * |
| rs733618 | 203866221 | T/C | 0.412 | 0.65 | 0.998 | 0.378 (0.199–0.717) | 0.003 * |
| rs4553808 | 203866282 | A/G | 0.038 | 0.138 | 0.602 | 0.244 (0.065–0.912) | 0.025 * |
| rs11571316 | 203866366 | A/G | 0.113 | 0.263 | 0.980 | 0.356 (0.152–0.836) | 0.015 * |
| rs62182595 | 203866465 | A/G | 0.038 | 0.138 | 0.949 | 0.244 (0.065–0.912) | 0.025 * |
| rs16840252 | 203866796 | C/T | 0.038 | 0.138 | 0.602 | 0.244 (0.065–0.912) | 0.025 * |
| rs5742909 | 203867624 | C/T | 0.05 | 0.125 | 0.665 | 0.368 (0.111–1.228) | 0.093 |
| rs231775 | 203867991 | A/G | 0.213 | 0.355 | 0.990 | 0.530 (0.261–1.076) | 0.077 |
| rs3087243 | 203874196 | G/A | 0.163 | 0.263 | 0.980 | 0.545 (0.251–1.183) | 0.122 |
| rs11571319 | 203874215 | G/A | 0.075 | 0.313 | 0.102 | 0.178 (0.069–0.464) | <0.001 * |
| CD28 | |||||||
| rs1879877 | 203705277 | G/T | 0.321 | 0.449 | 0.759 | 0.580 (0.302–1.112) | 0.100 |
| rs3181096 | 203705369 | C/T | 0.175 | 0.269 | 0.036 * | 0.576 (0.268–1.235) | 0.154 |
| rs3181097 | 203705416 | G/A | 0.325 | 0.615 | 0.988 | 0.301 (0.157–0.578) | <0.001 * |
| rs3181098 | 203705655 | G/A | 0.163 | 0.244 | 0.066 | 0.563 (0.257–1.230) | 0.147 |
| PDCD1 | |||||||
| rs5839828 | 241859601 | G/GG | 0.419 | 0.311 | 0.948 | 1.599 (0.814–3.140) | 0.172 |
| rs36084323 | 241859444 | C/T | 0.541 | 0.365 | 0.999 | 2.048 (1.061–3.955) | 0.032 * |
| rs41386349 | 241851697 | G/A | 0.175 | 0.200 | 0.384 | 0.848 (0.383–1.880) | 0.685 |
| rs6705653 | 241851407 | T/C | 0.25 | 0.218 | 0.559 | 1.196 (0.572–2.503) | 0.634 |
| rs2227982 | 241851281 | G/A | 0.513 | 0.397 | 0.994 | 1.594 (0.848–2.995) | 0.147 |
| rs2227981 | 241851121 | A/G | 0.25 | 0.218 | 0.559 | 1.196 (0.572–2.503) | 0.634 |
| rs10204525 | 241850169 | C/T | 0.359 | 0.188 | 0.827 | 2.427 (1.172–5.023) | 0.015 * |
| ICOS | |||||||
| rs11571305 | 203935403 | G/A | 0.359 | 0.313 | 0.016 * | 1.232 (0.636–2.387) | 0.536 |
| rs11889352 | 203935948 | T/A | 0.397 | 0.225 | 0.679 | 2.272 (1.135–4.546) | 0.019 * |
| rs11883722 | 203936122 | G/A | 0.449 | 0.375 | 0.208 | 1.357 (0.718–2.561) | 0.346 |
| rs10932029 | 203937045 | T/C | 0.138 | 0.118 | 0.074 | 1.187 (0.462–3.047) | 0.722 |
| rs10932035 | 203959929 | G/A | 0.329 | 0.477 | 0.036 * | 0.537 (0.251–1.149) | 0.107 |
| rs10932036 | 203960458 | A/T | 0.077 | 0.041 | 0.968 | 1.972 (0.475–8.193) | 0.496 |
| rs4404254 | 203960563 | T/C | 0.256 | 0.263 | 0.954 | 1.138 (0.558–2.322) | 0.722 |
| rs10932037 | 203960623 | C/T | 0.077 | 0.053 | 0.943 | 1.500 (0.406–5.541) | 0.746 |
| rs10932038 | 203960861 | A/G | 0.077 | 0.042 | 0.967 | 1.917 (0.461–7.967) | 0.498 |
| rs1559931 | 203961006 | G/A | 0.256 | 0.229 | 0.987 | 1.164 (0.547–2.475) | 0.693 |
| rs4675379 | 203961372 | G/C | 0.179 | 0.286 | 0.186 | 0.547 (0.226–1.325) | 0.178 |
| TNFSF4 | |||||||
| rs1234314 | 173208253 | C/G | 0.375 | 0.35 | 0.822 | 1.114 (0.585–2.124) | 0.742 |
| rs45454293 | 173208097 | A/G | 0.163 | 0.15 | 0.394 | 1.100 (0.468–2.583) | 0.828 |
| SNP | Genotype/Allele | Patient | Control | OR (95% CI) | p-Value | Q-Value |
|---|---|---|---|---|---|---|
| N = 40 | N = 40 | |||||
| CTLA4 | ||||||
| rs11571315 | CC vs. CT vs. TT | 0.006 * | 0.0720 | |||
| TT | 28 | 15 | Ref. | 1.000 | ||
| CT | 11 | 18 | 0.327 (0.123–0.870) | 0.023 | 0.1712 | |
| CC | 1 | 7 | 0.077 (0.009–0.682) | 0.015 | 0.1675 | |
| TT vs. CT + CC | 0.257 (0.101–0.652) | 0.004 | 0.0766 | |||
| TT + CT vs. CC | 0.121 (0.014–1.034) | 0.057 | 0.2634 | |||
| rs733618 | CC vs. CT vs. TT | 0.011 * | 0.0977 | |||
| CC | 13 | 5 | Ref. | 1.000 | ||
| CT | 21 | 18 | 0.449 (0.134–1.502) | 0.189 | 0.4715 | |
| TT | 6 | 17 | 0.136 (0.034–0.545) | 0.003 | 0.0766 | |
| CC vs. CT + TT | 0.297 (0.094–0.934) | 0.032 | 0.1787 | |||
| CC + CT vs. TT | 0.239 (0.082–0.696) | 0.007 | 0.1173 | |||
| rs4553808 | AA vs. AG vs. GG | 0.019 * | 0.0977 | |||
| AA | 37 | 29 | Ref. | 1.000 | ||
| AG | 3 | 11 | 0.214 (0.055–0.838) | 0.019 * | 0.1675 | |
| GG | 0 | 0 | NA | NA | ||
| AA vs. AG + GG | 0.214 (0.055–0.838) | 0.019 * | 0.1675 | |||
| AA + AG vs. GG | NA | NA | ||||
| rs11571316 | GG vs. AG vs. AA | 0.056 | 0.1833 | |||
| GG | 32 | 22 | Ref. | 1.000 | ||
| AG | 7 | 15 | 0.321 (0.112–0.916) | 0.030 * | 0.1787 | |
| AA | 1 | 3 | 0.229 (0.022–2.349) | 0.305 | 0.6089 | |
| GG vs. AG + AA | 0.306 (0.113–0.826) | 0.017 * | 0.1675 | |||
| GG + AG vs. AA | 0.316 (0.031–3.178) | 0.615 | 0.8171 | |||
| rs16840252 | CC vs. CT vs. TT | 0.019 * | 0.0977 | |||
| CC | 37 | 29 | Ref. | 1.000 | ||
| CT | 3 | 11 | 0.214 (0.055–0.838) | 0.019 * | 0.1675 | |
| TT | 0 | 0 | NA | NA | ||
| CC vs. CT + TT | 0.214 (0.055–0.838) | 0.019 * | 0.1675 | |||
| CC + CT vs. TT | NA | NA | ||||
| rs11571319 | GG vs. AG vs. AA | <0.001 * | 0.0178 | |||
| GG | 34 | 16 | Ref. | 1.000 | ||
| AG | 6 | 23 | 0.123 (0.042–0.360) | <0.001 * | 0.0302 | |
| AA | 0 | 1 | NA | 0.333 | 0.6375 | |
| GG vs. AG + AA | 0.118 (0.040–0.344) | <0.001 * | 0.0302 | |||
| GG + AG vs. AA | NA | 1.000 | 1.0000 | |||
| CD28 | ||||||
| rs3181097 | GG vs. AG vs. AA | <0.001 * | 0.0178 | |||
| AA | 15 | 6 | Ref. | 1.000 | ||
| AG | 24 | 18 | 0.533 (0.173–1.646) | 0.271 | 0.5857 | |
| GG | 1 | 15 | 0.027 (0.003–0.249) | <0.001 * | 0.0302 | |
| GG vs. AG + AA | 0.303 (0.103–0.892) | 0.026 * | 0.1742 | |||
| GG + AG vs. AA | 0.041 (0.005–0.331) | <0.001 * | 0.0302 | |||
| rs3181098 | GG vs. AG vs. AA | 0.055 | 0.1833 | |||
| GG | 27 | 25 | Ref. | 1.000 | ||
| AG | 13 | 9 | 1.337 (0.488–3.669) | 0.572 | 0.8171 | |
| AA | 0 | 5 | NA | 0.053 | 0.2630 | |
| GG vs. AG + AA | 0.860 (0.339–2.180) | 0.750 | 0.9105 | |||
| GG + AG vs. AA | NA | 0.026 * | 0.1742 | |||
| PDCD1 | ||||||
| rs36084323 | TT vs. CT vs. CC | 0.110 | 0.2565 | |||
| TT | 8 | 15 | Ref. | 1.000 | ||
| CT | 18 | 17 | 1.985 (0.671–5.871) | 0.212 | ||
| CC | 11 | 5 | 4.125 (1.057–16.097) | 0.037 * | ||
| TT + CT vs. CC | 2.472 (0.890–6.864) | 0.079 | ||||
| TT vs. CT + CC | 2.708 (0.835–8.785) | 0.090 | ||||
| rs10204525 | TT vs. CT vs. CC | 0.075 | 0.2132 | |||
| TT | 17 | 27 | Ref. | 1.000 | ||
| CT | 16 | 11 | 2.310 (0.868–6.146) | 0.091 | 0.5073 | |
| CC | 6 | 2 | 4.765 (0.860–26.383) | 0.118 | 0.1983 | |
| TT + CT vs. CC | 2.688 (1.076–6.715) | 0.032 * | 0.2786 | |||
| TT vs. CT + CC | 3.455 (0.652–18.294) | 0.154 | 0.2974 | |||
| ICOS | ||||||
| rs11889352 | AA vs. AT vs. TT | 0.045 * | 0.1800 | |||
| AA | 15 | 23 | Ref. | 1.000 | ||
| AT | 17 | 16 | 1.629 (0.635–4.183) | 0.309 | 0.6089 | |
| TT | 7 | 1 | 10.733 (1.197–96.283) | 0.020 * | 0.1675 | |
| AA vs. AT + TT | 2.165 (0.881–5.322) | 0.090 | 0.2974 | |||
| AA + AT vs. TT | 8.531 (0.997–73.006) | 0.029 * | 0.1787 | |||
| rs4675379 | GG vs. CG vs. CC | 0.042 * | 0.1800 | |||
| GG | 27 | 9 | Ref. | 1.000 | ||
| CG | 10 | 12 | 0.278 (0.090–0.859) | 0.023 * | 0.1712 | |
| CC | 2 | 0 | NA | 1.000 | 1.0000 | |
| GG vs. CG + CC | 0.333 (0.111–1.001) | 0.047 * | 0.2422 | |||
| GG + CG vs. CC | NA | 0.537 | 0.7995 |
| Haplotypes | Freq. Cases | Freq. Controls | OR | 95% CI | p-Value |
|---|---|---|---|---|---|
| CTLA4 | |||||
| Ars62182595Trs16840252Crs5742909 | 0.075 | 0.250 | 0.243 | 0.061–0.964 | 0.034 |
| Ars62182595Trs16840252Trs5742909 | 0.075 | 0.250 | 0.243 | 0.061–0.964 | 0.034 |
| Ars62182595Crs16840252Crs5742909 | 0.075 | 0.250 | 0.243 | 0.061–0.964 | 0.034 |
| Ars62182595Crs16840252Trs5742909 | 0.075 | 0.250 | 0.243 | 0.061–0.964 | 0.034 |
| Grs62182595Trs16840252Crs5742909 | 0.075 | 0.250 | 0.243 | 0.061–0.964 | 0.034 |
| ICOS | |||||
| Ars11889352C11883722 | 0.750 | 0.925 | 0.243 | 0.061–0.964 | 0.034 |
| GO Case | Control | SLE Case | Control | RA Case | Control | |
|---|---|---|---|---|---|---|
| rs11571315 | ||||||
| TT | 28 | 15 | 53 | 33 | 69 | 47 |
| CT | 11 | 18 | 15 | 31 | 33 | 41 |
| CC | 1 | 7 | 3 | 11 | 17 | 12 |
| rs733618 | ||||||
| CC | 13 | 5 | 33 | 15 | 36 | 18 |
| CT | 21 | 18 | 18 | 34 | 44 | 46 |
| TT | 6 | 17 | 21 | 26 | 41 | 36 |
| rs4553808 | ||||||
| AA | 37 | 29 | 71 | 55 | 103 | 77 |
| AG | 3 | 11 | 1 | 20 | 16 | 23 |
| GG | 0 | 0 | 0 | 0 | 4 | 0 |
| rs16840252 | ||||||
| CC | 37 | 29 | 69 | 53 | 105 | 75 |
| CT | 3 | 11 | 1 | 22 | 14 | 25 |
| TT | 0 | 0 | 1 | 0 | 4 | 0 |
| rs11571319 | ||||||
| GG | 34 | 16 | 58 | 40 | 91 | 61 |
| AG | 6 | 23 | 2 | 28 | 18 | 38 |
| AA | 0 | 1 | 8 | 7 | 15 | 1 |
| rs36084323 | ||||||
| TT | 8 | 15 | 19 | 33 | 32 | 40 |
| CT | 18 | 17 | 34 | 31 | 62 | 43 |
| CC | 11 | 5 | 18 | 7 | 30 | 13 |
| Reporter Contractions | RLU | Mean | SD | p | |||||
|---|---|---|---|---|---|---|---|---|---|
| CTLA4 wild type | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | Ref. |
| rs11571315 C > T | 0.947 | 1.174 | 1.160 | 1.181 | 1.114 | - | 1.115 | 0.098 | 0.328 |
| rs733618 T > C | 0.218 | 0.293 | 0.234 | 0.291 | 0.256 | 0.286 | 0.263 | 0.032 | <0.001 * |
| rs4553808 A > G | 0.250 | 0.212 | 0.206 | 0.300 | 0.205 | 0.297 | 0.245 | 0.045 | <0.001 * |
| rs16840252 C > T | 0.819 | 1.093 | 0.799 | 1.188 | 1.141 | 1.232 | 1.045 | 0.189 | 0.929 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, Y.-C.; Yu, K.-H.; Lin, W.-T.; Wang, W.-T.; Chen, D.-P. Finding the Common Single-Nucleotide Polymorphisms in Three Autoimmune Diseases and Exploring Their Bio-Function by Using a Reporter Assay. Biomedicines 2023, 11, 2426. https://doi.org/10.3390/biomedicines11092426
Chu Y-C, Yu K-H, Lin W-T, Wang W-T, Chen D-P. Finding the Common Single-Nucleotide Polymorphisms in Three Autoimmune Diseases and Exploring Their Bio-Function by Using a Reporter Assay. Biomedicines. 2023; 11(9):2426. https://doi.org/10.3390/biomedicines11092426
Chicago/Turabian StyleChu, Yen-Chang, Kuang-Hui Yu, Wei-Tzu Lin, Wei-Ting Wang, and Ding-Ping Chen. 2023. "Finding the Common Single-Nucleotide Polymorphisms in Three Autoimmune Diseases and Exploring Their Bio-Function by Using a Reporter Assay" Biomedicines 11, no. 9: 2426. https://doi.org/10.3390/biomedicines11092426
APA StyleChu, Y.-C., Yu, K.-H., Lin, W.-T., Wang, W.-T., & Chen, D.-P. (2023). Finding the Common Single-Nucleotide Polymorphisms in Three Autoimmune Diseases and Exploring Their Bio-Function by Using a Reporter Assay. Biomedicines, 11(9), 2426. https://doi.org/10.3390/biomedicines11092426





