UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and siRNA
2.2. Cell Culture and Transfection
2.3. Luciferase Assay
2.4. Western Blot
2.5. Immunocytochemistry
2.6. Oxidative Stress
2.7. MTT Assay
2.8. Statistical Analysis
3. Results
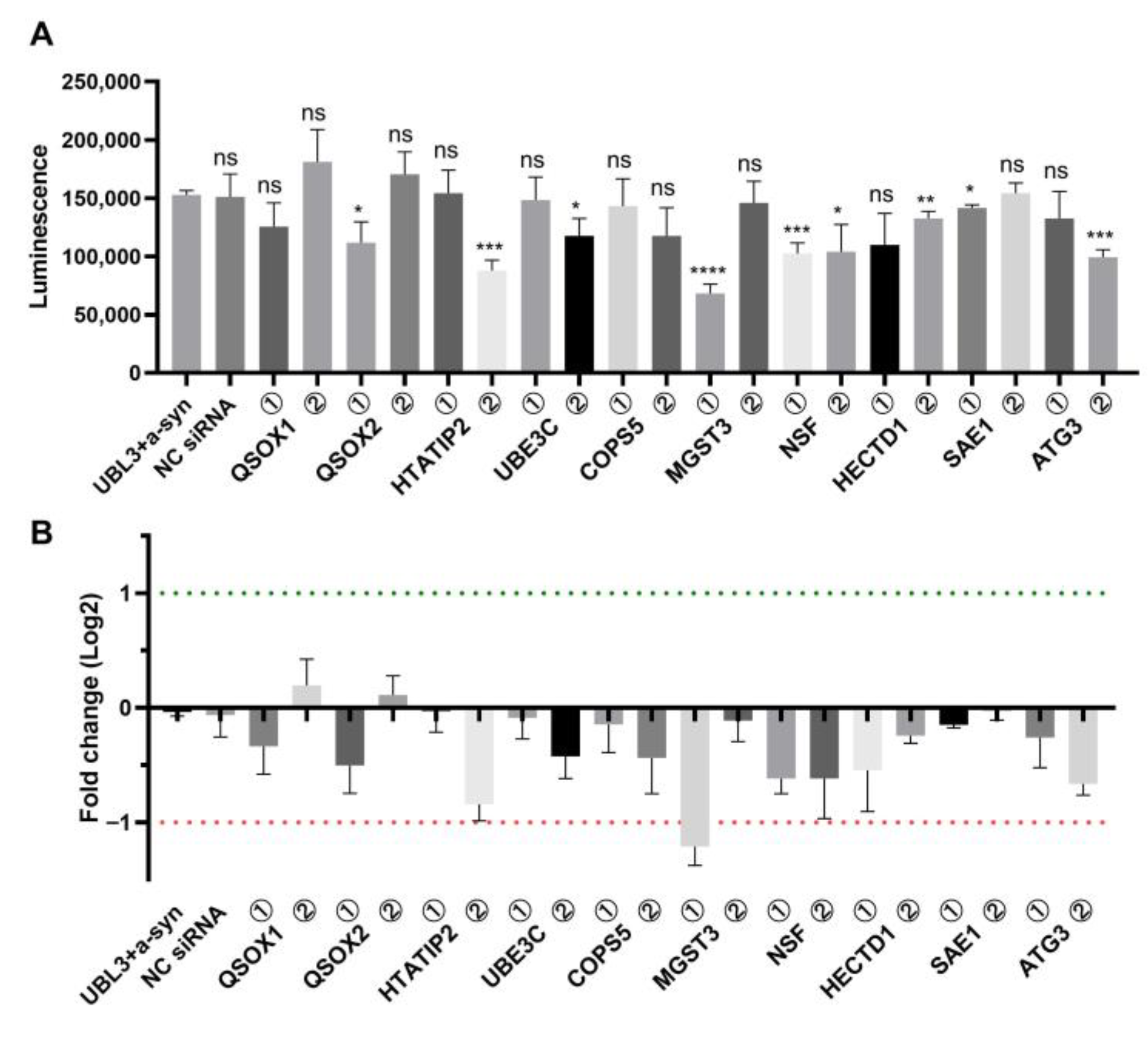
3.1. Screening of Regulators Affecting the Interaction of UBL3 with α-Syn Using Split Gaussian Luciferase Complementation Assay and RNAi Technology
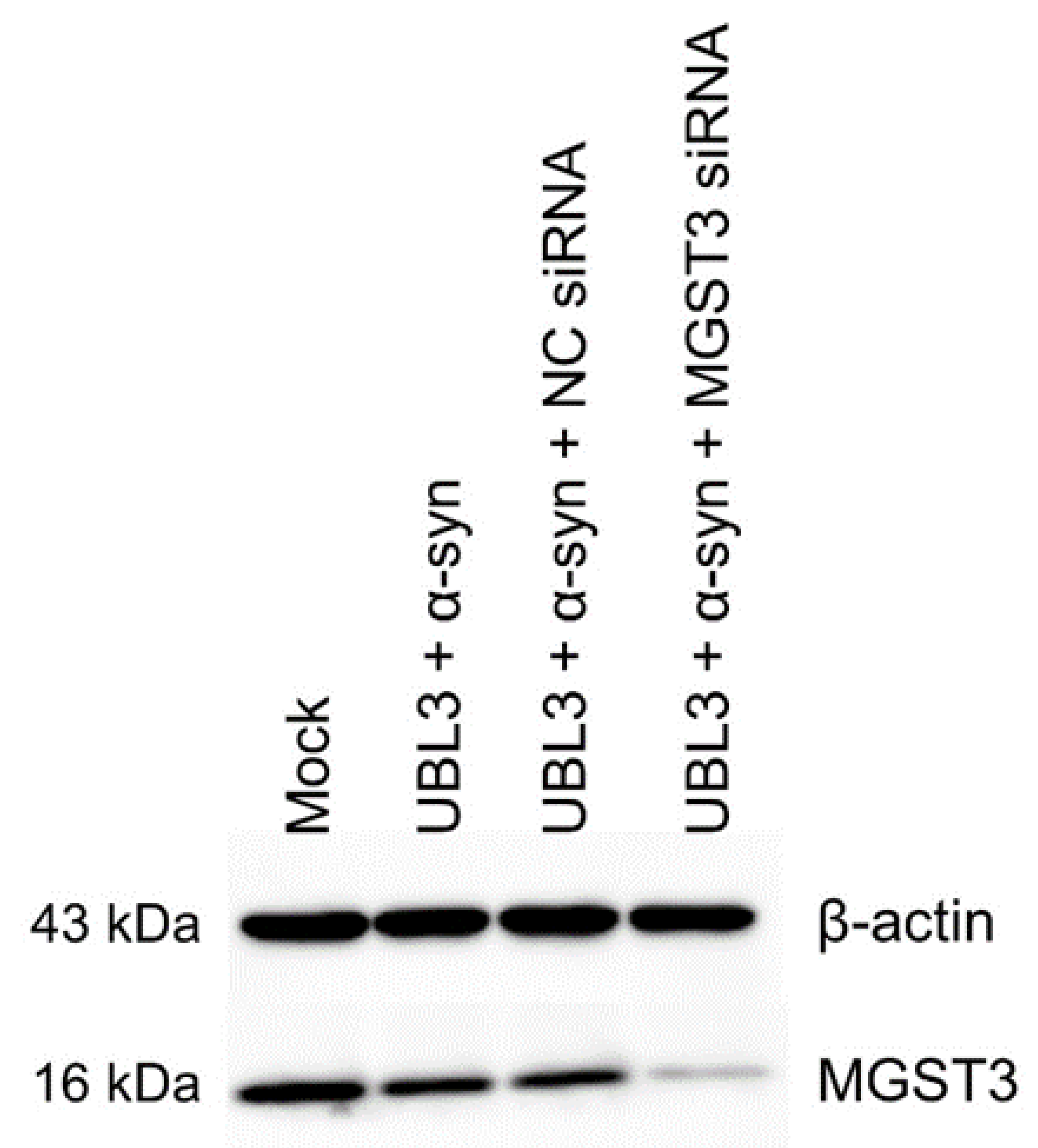
3.2. Confirmation of MGST3 Expression Silencing
3.3. Effect of Silencing MGST3 on Co-Localization of UBL3 and α-Syn
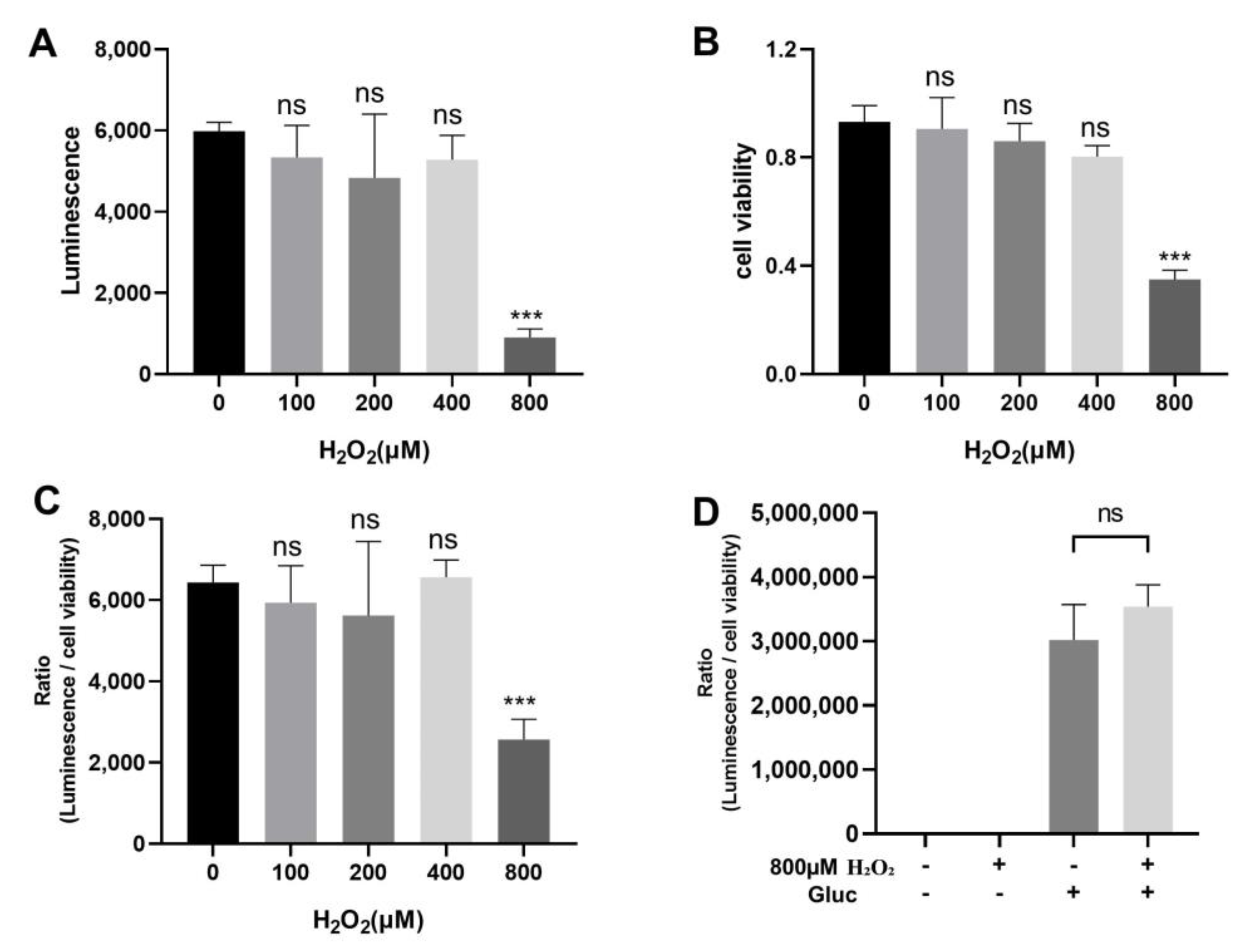
3.4. Effect of Oxidative Stress on the Interaction between UBL3 and α-Syn
3.5. Effect of Silencing MGST3 on the Interaction of UBL3 with α-Syn upon Oxidative Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Downes, B.P.; Saracco, S.A.; Lee, S.S.; Crowell, D.N.; Vierstra, R.D. MUBs, a Family of Ubiquitin-Fold Proteins That Are Plasma Membrane-Anchored by Prenylation. J. Biol. Chem. 2006, 281, 27145–27157. [Google Scholar] [CrossRef] [PubMed]
- Ageta, H.; Ageta-Ishihara, N.; Hitachi, K.; Karayel, O.; Onouchi, T.; Yamaguchi, H.; Kahyo, T.; Hatanaka, K.; Ikegami, K.; Yoshioka, Y.; et al. UBL3 Modification Influences Protein Sorting to Small Extracellular Vesicles. Nat. Commun. 2018, 9, 3936. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Wang, Y.; Zheng, J.C. Extracellular Vesicles, from the Pathogenesis to the Therapy of Neurodegenerative Diseases. Transl. Neurodegener. 2022, 11, 53. [Google Scholar] [CrossRef]
- Goedert, M. Alpha-Synuclein and Neurodegenerative Diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef]
- Kawahata, I.; Finkelstein, D.I.; Fukunaga, K. Pathogenic Impact of α-Synuclein Phosphorylation and Its Kinases in α-Synucleinopathies. Int. J. Mol. Sci. 2022, 23, 6216. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Spillantini, M.G. The Synucleinopathies: Twenty Years On. J. Parkinsons Dis. 2017, 7, S51–S69. [Google Scholar] [CrossRef]
- Masliah, E.; Rockenstein, E.; Veinbergs, I.; Sagara, Y.; Mallory, M.; Hashimoto, M.; Mucke, L. β-Amyloid Peptides Enhance α-Synuclein Accumulation and Neuronal Deficits in a Transgenic Mouse Model Linking Alzheimer’s Disease and Parkinson’s Disease. Proc. Natl. Acad. Sci. USA 2001, 98, 12245–12250. [Google Scholar] [CrossRef]
- Klucken, J.; Shin, Y.; Masliah, E.; Hyman, B.T.; McLean, P.J. Hsp70 Reduces Alpha-Synuclein Aggregation and Toxicity. J. Biol. Chem. 2004, 279, 25497–25502. [Google Scholar] [CrossRef]
- Chen, B.; Hasan, M.M.; Zhang, H.; Zhai, Q.; Waliullah, A.S.M.; Ping, Y.; Zhang, C.; Oyama, S.; Mimi, M.A.; Tomochika, Y.; et al. UBL3 Interacts with Alpha-Synuclein in Cells and the Interaction Is Downregulated by the EGFR Pathway Inhibitor Osimertinib. Biomedicines 2023, 11, 1685. [Google Scholar] [CrossRef]
- Jakobsson, P.-J.; Mancini, J.A.; Riendeau, D.; Ford-Hutchinson, A.W. Identification and Characterization of a Novel Microsomal Enzyme with Glutathione-Dependent Transferase and Peroxidase Activities *. J. Biol. Chem. 1997, 272, 22934–22939. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xiao, S.; Deng, Y.; Du, X.; Yu, Z. Cloning of a Novel Glutathione S-Transferase 3 (GST3) Gene and Expressionanalysis in Pearl Oyster, Pinctada Martensii. Fish. Shellfish Immunol. 2011, 31, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Pandey, A.K.; Houseal, M.T.; Mulligan, M.K. The Genetic Architecture of Murine Glutathione Transferases. PLoS ONE 2016, 11, e0148230. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Sakai, N.; Enokido, Y.; Uchiyama, Y.; Hatanaka, H. Free Radical-Independent Protection by Nerve Growth Factor and Bcl-2 of PC12 Cells from Hydrogen Peroxide-Triggered Apoptosis. J. Biochem. 1996, 120, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, P.J.; Morgenstern, R.; Mancini, J.; Ford-Hutchinson, A.; Persson, B. Membrane-Associated Proteins in Eicosanoid and Glutathione Metabolism (MAPEG). A Widespread Protein Superfamily. Am. J. Respir. Crit. Care Med. 2000, 161, S20–S24. [Google Scholar] [CrossRef] [PubMed]
- Bracalente, C.; Ibañez, I.L.; Berenstein, A.; Notcovich, C.; Cerda, M.B.; Klamt, F.; Chernomoretz, A.; Durán, H. Reprogramming Human A375 Amelanotic Melanoma Cells by Catalase Overexpression: Upregulation of Antioxidant Genes Correlates with Regression of Melanoma Malignancy and with Malignant Progression When Downregulated. Oncotarget 2016, 7, 41154–41171. [Google Scholar] [CrossRef]
- Ayemele, A.G.; Tilahun, M.; Lingling, S.; Elsaadawy, S.A.; Guo, Z.; Zhao, G.; Xu, J.; Bu, D. Oxidative Stress in Dairy Cows: Insights into the Mechanistic Mode of Actions and Mitigating Strategies. Antioxidants 2021, 10, 1918. [Google Scholar] [CrossRef]
- Ashbrook, D.G.; Williams, R.W.; Lu, L.; Stein, J.L.; Hibar, D.P.; Nichols, T.E.; Medland, S.E.; Thompson, P.M.; Hager, R. Joint Genetic Analysis of Hippocampal Size in Mouse and Human Identifies a Novel Gene Linked to Neurodegenerative Disease. BMC Genom. 2014, 15, 850. [Google Scholar] [CrossRef]
- Castillo-Rangel, C.; Marin, G.; Hernández-Contreras, K.A.; Vichi-Ramírez, M.M.; Zarate-Calderon, C.; Torres-Pineda, O.; Diaz-Chiguer, D.L.; De la Mora González, D.; Gómez Apo, E.; Teco-Cortes, J.A.; et al. Neuroinflammation in Parkinson’s Disease: From Gene to Clinic: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 5792. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The Many Faces of α-Synuclein: From Structure and Toxicity to Therapeutic Target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef]
- Lu, M.; Sun, W.-L.; Shen, J.; Wei, M.; Chen, B.; Qi, Y.-J.; Xu, C.-S. LncRNA-UCA1 Promotes PD Development by Upregulating SNCA. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7908–7915. [Google Scholar] [CrossRef] [PubMed]
- Hassan, W.; Noreen, H.; Rehman, S.; Kamal, M.A.; Teixeira da Rocha, J.B. Association of Oxidative Stress with Neurological Disorders. Curr. Neuropharmacol. 2022, 20, 1046–1072. [Google Scholar] [CrossRef] [PubMed]
- Barmaki, H.; Morovati, A.; Eydivandi, Z.; Jafari Naleshkenani, F.; Saedi, S.; Musavi, H.; Abbasi, M.; Hemmati-Dinarvand, M. The Association between Serum Oxidative Stress Indexes and Pathogenesis of Parkinson’s Disease in the Northwest of Iran. Iran. J. Public Health 2021, 50, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Sian, J.; Dexter, D.T.; Lees, A.J.; Daniel, S.; Agid, Y.; Javoy-Agid, F.; Jenner, P.; Marsden, C.D. Alterations in Glutathione Levels in Parkinson’s Disease and Other Neurodegenerative Disorders Affecting Basal Ganglia. Ann. Neurol. 1994, 36, 348–355. [Google Scholar] [CrossRef]
- Bellinger, F.P.; Bellinger, M.T.; Seale, L.A.; Takemoto, A.S.; Raman, A.V.; Miki, T.; Manning-Boğ, A.B.; Berry, M.J.; White, L.R.; Ross, G.W. Glutathione Peroxidase 4 Is Associated with Neuromelanin in Substantia Nigra and Dystrophic Axons in Putamen of Parkinson’s Brain. Mol. Neurodegener. 2011, 6, 8. [Google Scholar] [CrossRef]
- Sandeep; Sahu, M.R.; Rani, L.; Kharat, A.S.; Mondal, A.C. Could Vitamins Have a Positive Impact on the Treatment of Parkinson’s Disease? Brain Sci. 2023, 13, 272. [Google Scholar] [CrossRef]
- Bosco, D.A.; Fowler, D.M.; Zhang, Q.; Nieva, J.; Powers, E.T.; Wentworth, P.; Lerner, R.A.; Kelly, J.W. Elevated Levels of Oxidized Cholesterol Metabolites in Lewy Body Disease Brains Accelerate α-Synuclein Fibrilization. Nat. Chem. Biol. 2006, 2, 249–253. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Zhang, H.; Tomochika, Y.; Chen, B.; Ping, Y.; Islam, M.S.; Aramaki, S.; Sato, T.; Nagashima, Y.; Nakamura, T.; et al. UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3. Biomedicines 2023, 11, 2491. https://doi.org/10.3390/biomedicines11092491
Yan J, Zhang H, Tomochika Y, Chen B, Ping Y, Islam MS, Aramaki S, Sato T, Nagashima Y, Nakamura T, et al. UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3. Biomedicines. 2023; 11(9):2491. https://doi.org/10.3390/biomedicines11092491
Chicago/Turabian StyleYan, Jing, Hengsen Zhang, Yuna Tomochika, Bin Chen, Yashuang Ping, Md. Shoriful Islam, Shuhei Aramaki, Tomohito Sato, Yu Nagashima, Tomohiko Nakamura, and et al. 2023. "UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3" Biomedicines 11, no. 9: 2491. https://doi.org/10.3390/biomedicines11092491
APA StyleYan, J., Zhang, H., Tomochika, Y., Chen, B., Ping, Y., Islam, M. S., Aramaki, S., Sato, T., Nagashima, Y., Nakamura, T., Kahyo, T., Kaneda, D., Ogawa, K., Yoshida, M., & Setou, M. (2023). UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3. Biomedicines, 11(9), 2491. https://doi.org/10.3390/biomedicines11092491






