Abstract
The World Health Organization Classification of Tumors of the Central Nervous System recently incorporated histological features, immunophenotypes, and molecular characteristics to improve the accuracy of glioblastoma (GBM) diagnosis. FGFR3::TACC3 (F3T3) fusion has been identified as an oncogenic driver in IDH-wildtype GBMs. Recent studies have demonstrated the potential of using FGFR inhibitors in clinical trials and TACC3-targeting agents in preclinical models for GBM treatment. However, there is limited information on the clinicopathological and genetic features of IDH-wildtype GBMs with F3T3 fusion. The aim of this study was to comprehensively investigate the clinical manifestations, histological features, and mutational profiles of F3T3-positive GBMs. Between September 2017 and February 2023, 25 consecutive cases (5.0%) of F3T3-positive GBM were extracted from 504 cases of IDH-wildtype GBM. Clinicopathological information and targeted sequencing results obtained from 25 primary and 4 recurrent F3T3-positive GBMs were evaluated and compared with those from F3T3-negative GBMs. The provisional grades determined by histology only were distributed as follows: 4 (26/29; 89.7%), 3 (2/29; 6.9%), and 2 (1/29; 3.4%). Grade 2–3 tumors were ultimately diagnosed as grade 4 GBMs based on the identification of the TERT promoter mutation and the combined gain of chromosome 7 and loss of chromosome 10 (7+/10−). F3T3-positive GBMs predominantly affected women (2.6 females per male). The mean age of patients with an F3T3-positive GBM at initial diagnosis was 62 years. F3T3-positive GBMs occurred more frequently in the cortical locations compared to F3T3-negative GBMs. Imaging studies revealed that more than one-third (12/29; 41.4%) of F3T3-positive GBMs displayed a circumscribed tumor border. Seven of the seventeen patients (41.2%) whose follow-up periods exceeded 20 months died of the disease. Histologically, F3T3-positive GBMs more frequently showed curvilinear capillary proliferation, palisading nuclei, and calcification compared to F3T3-negative GBMs. Molecularly, the most common alterations observed in F3T3-positive GBMs were TERT promoter mutations and 7+/10−, whereas amplifications of EGFR, PDGFRA, and KIT were not detected at all. Other genetic alterations included CDKN2A/B deletion, PTEN mutation, TP53 mutation, CDK4 amplification, and MDM2 amplification. Our observations suggest that F3T3-positive GBM is a distinct molecular subgroup of the IDH-wildtype GBM. Both clinicians and pathologists should consider this rare entity in the differential diagnosis of diffuse astrocytic glioma to make an accurate diagnosis and to ensure appropriate therapeutic management.
1. Introduction
The 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS) incorporated histological features and immunophenotypes to improve the accuracy of glioblastoma (GBM) diagnosis [1]. With the integration of the next-generation sequencing (NGS) technique into the diagnostic process, novel molecular characteristics of GBMs have been discovered. In the 2021 WHO Classification of Tumors of the CNS, the adoption of the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) further improved the classification of gliomas, especially aiding in prognosis prediction [2].
Isocitrate dehydrogenase (IDH)-wildtype glioblastoma (GBM) is a highly heterogeneous high-grade glioma [3]. The molecular diagnosis of IDH-wildtype GBM can be established based on the following three molecular parameters: the combined gain of chromosome 7 and loss of chromosome 10 (7+/10−), epidermal growth factor receptor (EGFR) amplification, and telomerase reverse transcriptase (TERT) promoter mutation [2]. Even though research on oncogenic driver mutations applicable to specific targeted therapies for GBMs is actively ongoing, their rarity has limited their use in treatment.
Gene fusion refers to the hybridization of coding or regulatory DNA sequences between genes resulting from genomic rearrangements such as translocations, deletions, duplications, and inversions [4]. The fusion between fibroblast growth factor receptor 3 (FGFR3) and transforming acidic coiled-coil-containing protein 3 (TACC3; F3T3 fusion) has been recently identified as an oncogenic driver in 3% of IDH-wildtype GBMs [5]. This fusion gene encodes a protein located at the poles of the mitotic spindle and exhibits hyperphosphorylation and constitutive activation of the kinase domain [6]. IDH-wildtype GBMs with F3T3 fusion (F3T3-positive GBMs) have been shown to exhibit distinct histological and molecular features [6,7,8], including the palisading of monotonous, ovoid nuclei; the curvilinear proliferation of thin capillaries; calcification; desmoplastic vessels; and amplifications of cyclin-dependent kinase 4 (CDK4) and mouse double minute 2 homolog (MDM2) [9,10,11]. It has been also reported that patients with an F3T3-positive GBM show better prognoses than those with an F3T3-negative GBM [5,9,11]. Some studies have highlighted differences in methylation profiles, tumor mutational burdens (TMBs), and copy number alterations between F3T3-positive and -negative GBMs [5,12].
Some previous studies have been conducted on F3T3-positive GBMs from multi-institutional cohorts [8,13], but their clinicopathological features remain to be clarified. In this study, we aimed to comprehensively analyze the clinical manifestations, imaging findings, histological features, and molecular characteristics of 25 consecutive cases of F3T3-positive GBM diagnosed at a single institution.
2. Materials and Methods
2.1. Patient Selection
Between September 2017 and February 2023, 25 cases of diffuse astrocytic gliomas with F3T3 fusion were extracted from a cohort of 504 IDH-wildtype GBM patients (5.0%) using next-generation sequencing (NGS) analysis. Twenty-five and four tumors were diagnosed as primary and recurrent F3T3-positive GBMs, respectively. Among the 479 patients with IDH-wildtype GBM without F3T3 fusion, 40 were randomly selected to establish an age-matched control group using the simultaneously conducted NGS analysis. Clinicopathological and demographic information was collected from the electronic medical records.
2.2. Histological and Neuroimaging Analysis
Two board-certified neuropathologists (H.B. and Y.-L.S.) independently reviewed all available hematoxylin and eosin-stained slides to ensure consistency and accuracy in the assessment of histological features. The following histological features were assessed: curvilinear capillary proliferation, nuclear palisading, calcification, palisading necrosis, desmoplastic vessels, tumor giant cells, and perivascular rosette formation. They also reviewed all available brain magnetic resonance images (MRIs) and the corresponding radiological interpretations.
2.3. NGS
Formalin-fixed, paraffin-embedded tissues were microdissected manually on unstained sections under a microscope. Genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). The DNA samples were fragmented using the Covaris S2 instrument (Covaris, Woburn, MA, USA), and a library was prepared using the SureSelect XT Automation Reagent Kit (Agilent Technologies, Santa Clara, CA, USA). To ensure the appropriate fragment size, the median size of the DNA fragments was determined using the Agilent 4200 Tape Station (Agilent, Santa Clara, CA, USA), with the desired range set between 300 and 400 bp. The concentration of the DNA was measured using the Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on the NextSeq 500/550 platform using the TG NextSeq 500/550 High Output Kit v2.0 and the TG NextSeq 500/550 Mid Output Kit v2.0 (NextSeq 550 Dx, Illumina, San Diego, CA, USA). Data analysis and interpretation were conducted using BrainTumorSCAN v2.0 (Geninus, Seoul, Republic of Korea). The BrainTumorSCAN (Geninus) pipeline consisted of a sequencing panel targeting 233 glioma-related genes. The obtained sequence data were analyzed to identify clinically relevant genomic alterations, including single-nucleotide variants (SNVs), small insertions/deletions (indels), copy number alterations (CNAs), and rearrangements/fusions. SNVs and indels with a variant allele frequency (VAF) below 1% were excluded from the analysis. Copy number gains were defined as having an average copy number greater than four, while losses were defined as having a copy number less than one. Translocations were identified based on supporting reads ranging from four to twelve, depending on the sample quality. The NGS results were manually reviewed by four board-certified pathologists (H.B., B.L., S.H., and Y.-L.S.) and one bioinformatician (D.G.K.). Reported variants were cross-referenced with ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 1 December 2023) to exclude variants that were reported as benign or likely benign. The genomic data analysis visualization was performed using the cBioPortal Oncoprinter (https://www.cbioportal.org/oncoprinter, accessed on 1 December 2023).
2.4. Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics for Windows, v27.0 (IBM Corporation, Armonk, NY, USA). Categorical variables, including clinical features, histological features, and imaging findings, were compared between F3T3-positive and -negative GBMs using Fisher’s exact test or the chi-square test. Overall survival was defined as the time from initial diagnosis to death, with there being censored observations for patients who were still alive at the last follow-up. Survival curves were generated using the Kaplan–Meier method, and differences between the curves were assessed using the log-rank test. Statistical significance was defined as a p < 0.05.
3. Results
3.1. Demographics
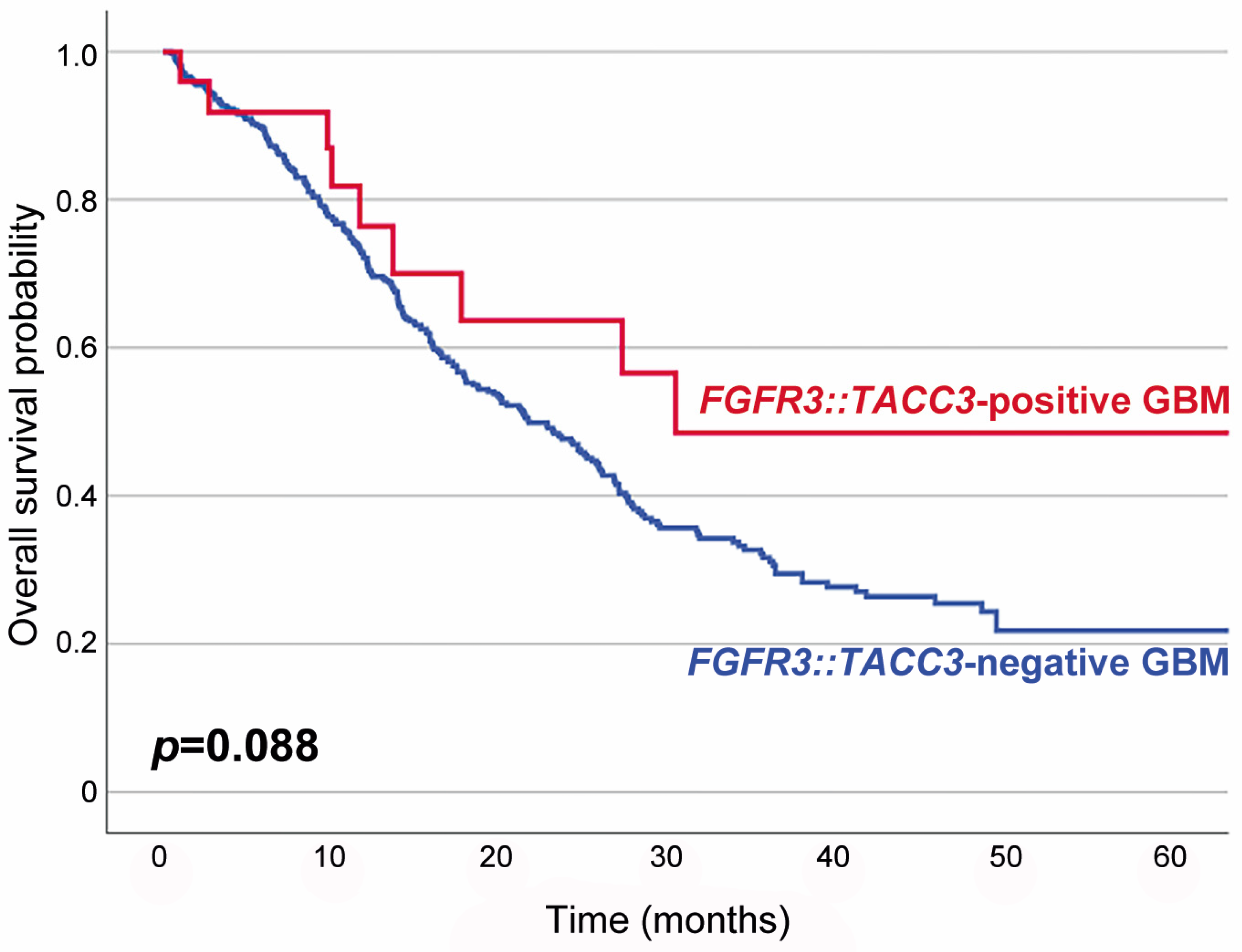
Table 1 summarizes the association between F3T3 fusion and the clinicopathological characteristics of IDH-wildtype GBMs. Twenty-five primary and four recurrent tumor specimens were obtained from 25 patients. F3T3-positive GBMs predominantly affected women (2.6 females per male; p = 0.003). The mean age of patients with an F3T3-positive GBM at initial diagnosis was 62 years (median age, 60 years; range, 40–84 years), which was similar to that of the 479 patients with an F3T3-negative GBM (60 years; p = 0.424; range, 19–87 years). Seventeen of the twenty-nine tumors (58.6%) presented as a single lesion and were totally excised. The remaining 12 tumors (41.4%) showed multiple lesions and were diagnosed with navigation-guided stereotactic biopsies. Nineteen patients (76.0%) received concurrent chemoradiation therapy with temozolomide, while two (8.0%) received radiation therapy only. The mean follow-up period for the 25 patients with an F3T3-positive GBM was 19 months (median, 12 months; range, 3–69 months). Fifteen patients (60.0%) developed tumor recurrence or progression during the follow-up period. Six of the seventeen patients whose follow-up period exceeded 20 months died of disease. Eight patients with a follow-up period shorter than 20 months were alive at the last follow-up. Survival analysis revealed that patients with an F3T3-positive GBM had a longer overall survival than those with an F3T3-negative GBM, but the difference in survival rate between the two groups was not statistically significant (p = 0.088; Figure 1).

Table 1.
Association between F3T3 fusion and the clinicopathological characteristics of IDH-wildtype GBMs.
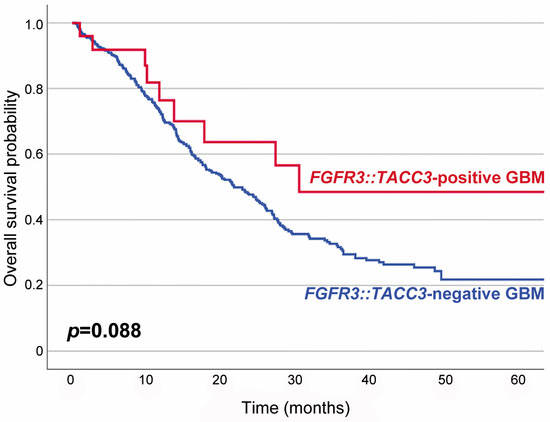
Figure 1.
Kaplan–Meier plots showing the overall survival probabilities of patients with F3T3-positive GBM (blue line) and those with F3T3-negative GBM (red line). The overall survival rate of F3T3-positive GBM patients was higher than that of F3T3-negative GBM patients, but the difference was not statistically significant (p = 0.088).
3.2. Histological Analysis
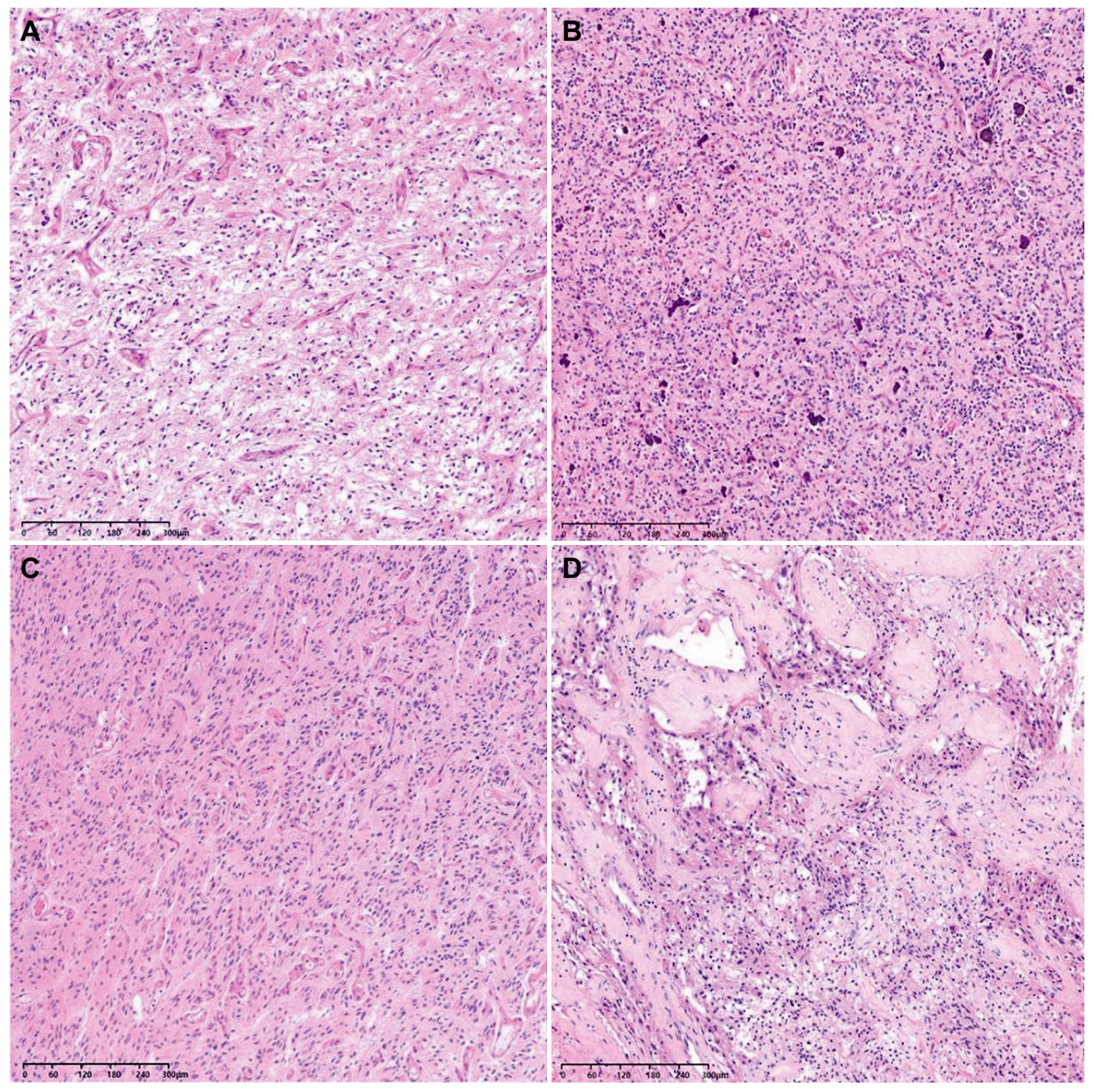
The histological grades of 29 tumor tissues were distributed as follows: 26 were grade 4 (89.7%), 2 were grade 3 (6.9%), and 1 was grade 2 (3.4%). Since the lower-grade tumors displayed neither tumor cell necrosis nor microvascular proliferation, their provisional grade was grade 2 or 3. However, they were finally confirmed as WHO grade 4 GBMs based on their NGS results showing either a TERT promoter mutation (1/3), 7+/10− (1/3), or both (1/3; Table 2). Figure 2 depicts representative photomicrographs showing the histological features of an F3T3-positive GBM.

Table 2.
Histological and molecular grades of F3T3-positive GBMs.
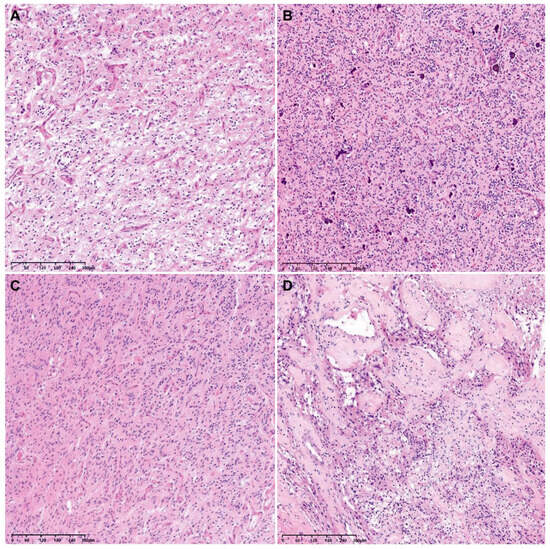
Figure 2.
Representative photomicrographs showing the histological features of an F3T3-positive GBM. The tumor tissue exhibits curvilinear capillary proliferation (A), scattered calcification within the tumor (B), nuclear palisading (C), and desmoplastic vessels (D).
As shown in Table 3, the most common histological feature observed in the F3T3-positive group was curvilinear capillary proliferation (25/29; 86.2%), followed by desmoplastic vessels (18/29; 62.1%), palisading nuclei (16/29; 55.2%), palisading necrosis (14/29; 48.3%), and calcification within the tumor (13/29; 44.8%). Tumor giant cells and a perivascular rosette pattern were observed in 10 (34.5%) and 8 (27.6%) cases of F3T3-positive GBM, respectively. Compared to the F3T3-negative GBMs, F3T3-positive tumors showed curvilinear capillary proliferation (p < 0.001), nuclear palisading (p = 0.001), and calcification (p = 0.006) more frequently. The frequencies of desmoplastic vessels (p = 0.429), palisading necrosis (p = 0.429), tumor giant cells (p = 0.271), and perivascular rosette formation (p = 0.114) were not significantly different between F3T3-positive and -negative GBMs.

Table 3.
Association between F3T3 and the histological features of IDH-wildtype GBMs.
3.3. Neuroimaging Analysis
As shown in Table 4, F3T3-positive GBMs were most frequently found in the right cerebral hemisphere (11/29; 37.9%) and the frontal lobe (16/29; 55.2%). These tumors were not found in the deep nuclei or cerebellum. Ten tumors (34.5%) were found in the bilateral cerebral hemispheres. In 12 cases (41.4%), the tumors had multiple anatomical locations. In contrast, F3T3-negative GBMs occurred most commonly in the left cerebral hemisphere (19/40; 47.5%) and the frontal lobe (20/40; 50.0%). While the tumor laterality and location were not significantly different according to the status of F3T3 fusion, cortical localization of tumors was significantly more frequent in F3T3-positive GBMs than in tumors without F3T3 (p = 0.006). Twelve cases of F3T3-positive GBM (41.4%) showed relatively well-circumscribed tumor borders, but there was no statistical significance in tumor border configuration between the two groups (p = 0.093).

Table 4.
Association between F3T3 fusion and the locational information of IDH-wildtype GBMs.
3.4. Molecular Characteristics
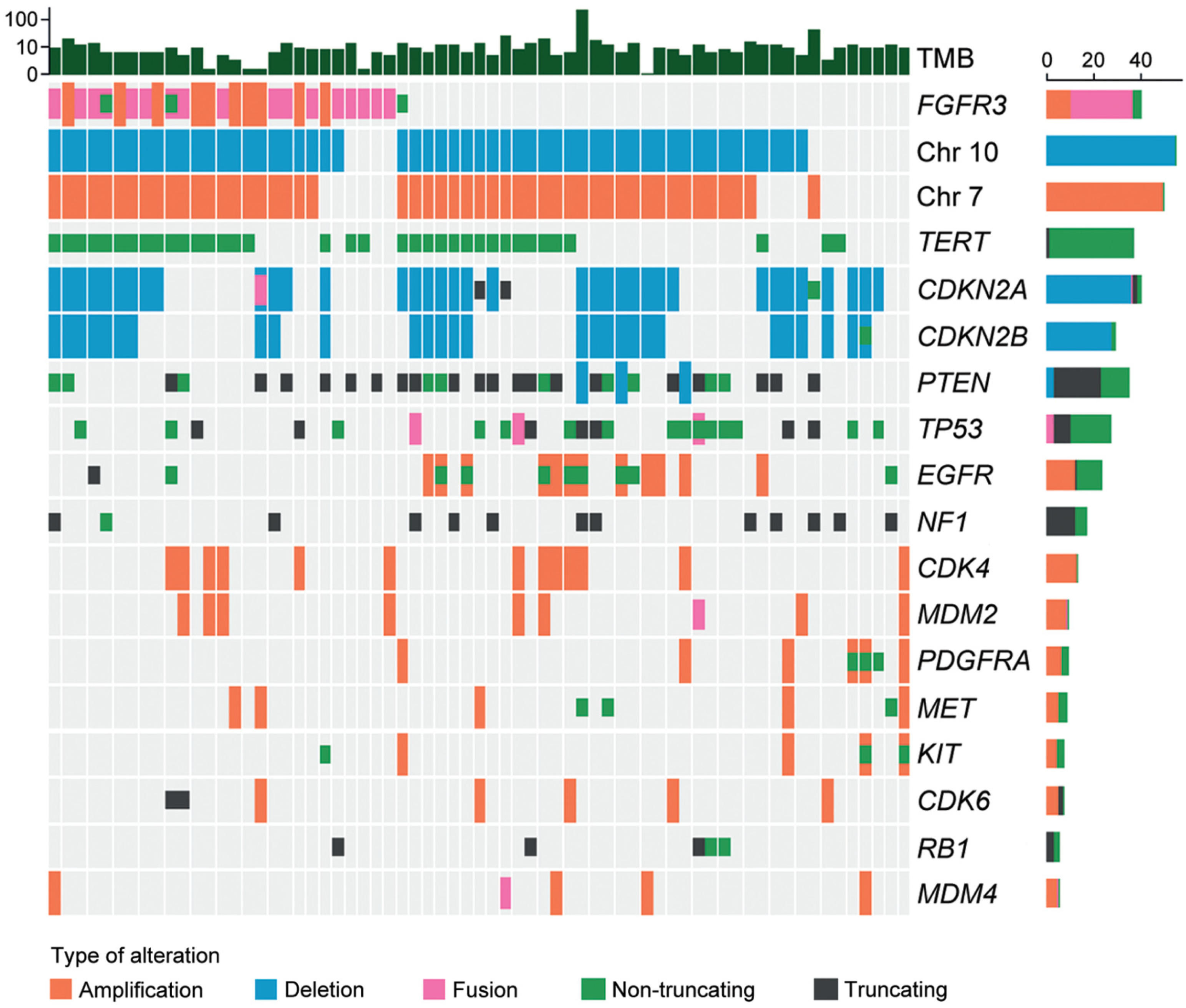
Twenty-seven tumor tissue samples were available for NGS. As shown in Figure 3, the most common fusion type was identified between FGFR3 exon 17 and TACC3 exon 11 (7/27; 25.9%), followed by the fusion between FGFR3 exon 18 and TACC3 exon 11 (6/27; 22.2%). The most common exon was exon 18 in FGFR3 (15/27; 55.6%) and exon 11 in TACC3 (13/27; 48.1%). All fusions were in-frame fusions, encoding a structural protein with consistent tyrosine kinase activity. The mean TMB of F3T3-positive GBMs was 7.08 per megabase (range of 1.51–19.67 per megabase). The most common genetic alterations detected in F3T3-positive GBMs were 7+/10− (22/27; 81.5%), followed by the TERT promoter mutation (19/27; 70.4%). None of the F3T3-positive GBMs demonstrated amplifications in EGFR, platelet-derived growth factor receptor A (PDGFRA), or KIT. Other alterations included CDK inhibitor 2A (CDKN2A) deletion (13/27; 48.1%), CDK inhibitor 2B (CDKN2B) deletion (10/27; 37.0%), phosphatase and tensin homolog deletion in chromosome 10 (PTEN) mutation (9/27; 33.3%), CDK4 amplification (6/27; 22.2%), TP53 mutation (5/27; 18.5%), and murine double minute 2 (MDM2) amplification (4/27; 14.8%). The information on O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation was available in 25 F3T3-positive GBMs. Thirteen F3T3-positive tumors (52.0%) had a methylated MGMT promoter.
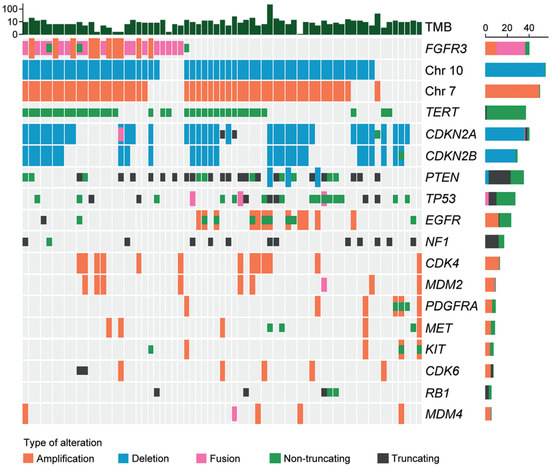
Figure 3.
Molecular characteristics of F3T3-positive GBMs.
Table 5 summarizes the difference in the frequencies of genetic alterations between F3T3-positive and -negative GBMs. In total, 28 and 17 of the 40 F3T3-negative GBMs had 7+/10− and the TERT promoter mutation, respectively. There was no significant difference in the alteration frequencies according to the presence of F3T3 fusion (p = 0.842 and p = 0.773). The EGFR and PDGFRA amplifications were detected in 12 (p = 0.002) and 6 (p = 0.035) F3T3-negative GBMs, respectively. There were no significant differences in the frequencies of other mutations and the degree of MGMT promoter methylation between the two groups.

Table 5.
Differences in the frequencies of genetic alterations between F3T3-positive and -negative GBMs.
3.5. Brief Presentation of a Case of F3T3-Positive GBM Showing Unusual Histology
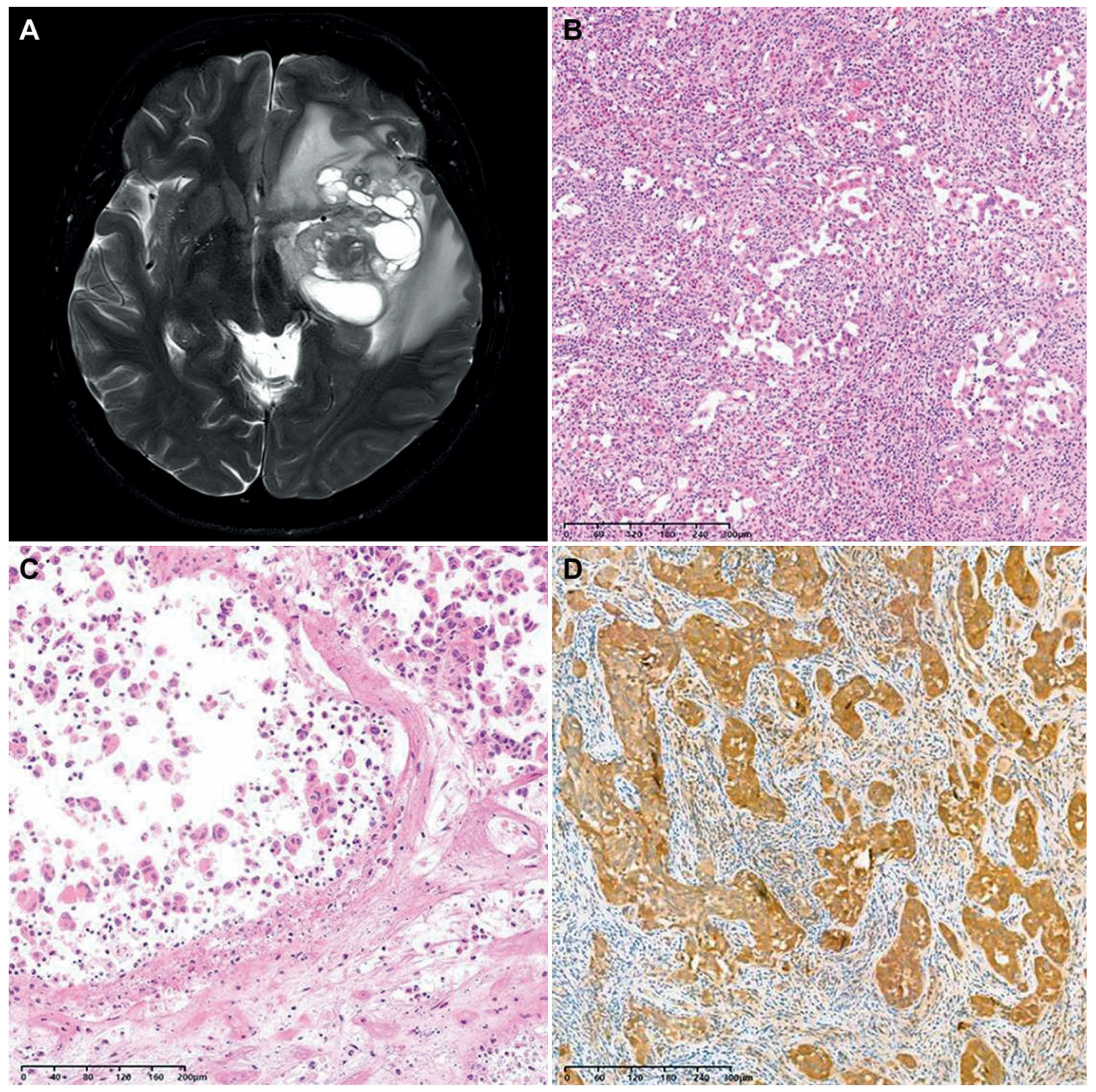
We encountered a case of an F3T3-positive GBM displaying unusual histology. A 40-year-old woman presented with a 6.7 cm sized multiloculated mass in the left temporal lobe (Figure 4A). She underwent gross total resection. The tumor exhibited a relatively well-circumscribed border. Histologically, the tumor consisted of round-to-oval cells arranged in an alveolar or micropapillary pattern (Figure 4B). The tumor cells possessed pleomorphism nuclei with vesicular chromatin, conspicuous nucleoli, and an eosinophilic cytoplasm. The peritumoral area showed significant inflammatory infiltrates with eosinophils. The tumor lacked the characteristic histological features typically seen in F3T3-positive GBMs. Instead, tumor cell necrosis, frequent mitotic figures, and a cell-in-cell phenomenon were observed (Figure 4C). Immunostaining revealed that the tumor cells were negative for glial fibrillary acidic protein and oligodendrocyte transcription factor 2, but they were diffusely positive for CD56 and S100 protein (Figure 4D). NGS analysis revealed the presence of 7+/10−, F3T3 fusion, and amplifications of CDK4 and MDM2, compatible with the diagnosis of IDH-wildtype GBM with F3T3 fusion.
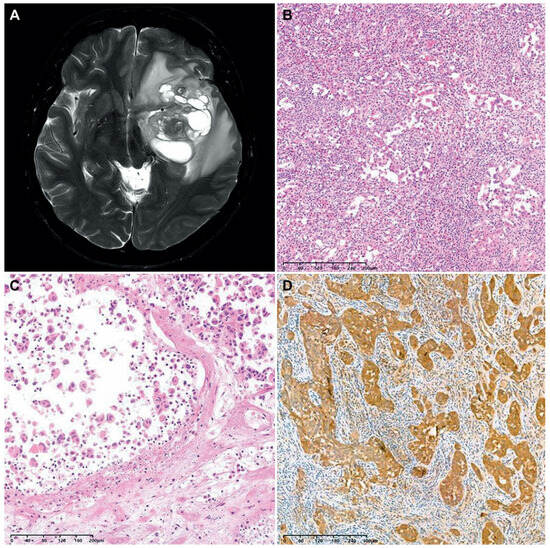
Figure 4.
A case of F3T3-positive GBM showing unusual histology. (A) The brain MRI revealed a relatively well-circumscribed, multiloculated cystic mass in the left temporal lobe. (B) The tumor cells were arranged in an alveolar or micropapillary pattern. The peritumoral area exhibited mixed inflammatory infiltrates, including eosinophils. (C) In some microscopic foci, individually dispersed tumor cells displayed a cell-in-cell phenomenon. (D) Immunostaining revealed that the tumor cells diffusely and strongly expressed the S100 protein.
4. Discussion
Diffuse gliomas are categorized into three groups: adult-type diffuse gliomas, pediatric-type diffuse low-grade gliomas, and pediatric-type diffuse high-grade gliomas [8]. Extensive molecular research has been conducted in the field of brain tumors to achieve an integrated diagnosis, leading to a comprehensive understanding of the molecular landscape [14,15,16]. Even though there is a pressing need for novel drug research specific to brain tumors, patients with a GBM still present dismal prognoses and often develop resistance to conventional therapy [5]. Clinical trials targeting several oncogenic mutations and cellular signaling pathways frequently observed in GBMs, such as EGFR mutation or amplification, the phosphoinositide 3-kinase/Akt/mammalian target of the rapamycin pathway, the p53 pathway, and the retinoblastoma (RB) pathway, have failed to improve outcomes due to challenges related to blood–brain barrier penetration, drug stability, and safety concerns [17]. While FGFR alterations are frequently observed in GBMs, their clinical significance may be limited since therapeutically meaningful alterations, such as F3T3 fusion, are rare [18].
FGFR proteins modulate various signaling pathways through molecular alterations and have significant importance in cancer biology. Multiple FGFR inhibitors currently in clinical development demonstrate therapeutic relevance and scalability [19]. The TACC3 gene has been identified as an important partner for FGFR fusions and is associated with the pathogenesis of several solid tumors [19]. The TACC3 protein possesses a coiled-coil domain at its C terminus, which promotes stability and organization of the mitotic spindle [20]. Since F3T3 fusion was first reported in GBMs and urothelial carcinomas of the urinary bladder [21,22], it has now been observed in pulmonary adenocarcinoma and squamous cell carcinoma, nasopharyngeal carcinoma, and uterine cervical squamous cell carcinoma [23,24,25,26,27]. The mechanism of action of this fusion protein involves delaying mitotic progression and inducing aneuploidy by increasing downstream signaling through the mitogen-activated protein kinase pathway, activating mitochondrial biogenesis and metabolism, and recruiting endogenous TACC3 from the mitotic spindle. These processes are believed to enhance malignant transformation [19,28,29]. The discovery of oncogenic gene fusions has significant clinical implications, suggesting the possibility of targeted therapy. A personalized approach utilizing FGFR fusion-targeted inhibitors may improve the prognosis of patients with gliomas [30]. Tabernero et al. [31] reported partial responses among patients with F3T3-positive tumors who were treated with the FGFR inhibitor erdafitinib. Several targeted therapies have been evaluated in clinical trials for patients with gliomas with FGFR fusion, including erdafitinib, ponatinib, and infigratinib [1]. These studies indicated that F3T3 fusion is a potential therapeutic target. Particularly, tumors with F3T3 fusion appear to be more sensitive to FGFR-targeted therapy compared to those with other FGFR abnormalities. This finding is meaningful because treatment options for patients with aggressive tumors with F3T3 fusion, such as a GBM and urothelial carcinoma, are limited [32].
F3T3 fusion has been detected in 3–12% of adult GBM cases [5,11,12,33]. Consistent with previous data, in this study, we detected F3T3 fusion in 5.0% (25/504) of the IDH-wildtype GBMs. Notably, we observed a higher frequency of F3T3 fusion in women (72.0%) compared to men (28.0%). When compared to F3T3-negative GBMs, a significant female predilection was observed (2.6 females per male; p = 0.003). In contrast, three previous studies reported either equal rates of F3T3 fusion in men and women [9] or slightly higher rates in men (1.47 male per female and 1.2 male per female) [5,8]. Since the previous studies showing equal gender distribution were conducted in Western populations, we considered that the difference in gender distribution may reflect racial difference. It is reasonable to assume that the relative incidence of F3T3-positive GBM may vary by ethnicity. Both domestic multi-institutional and global multinational studies are necessary to confirm or disprove this assumption and to explore the underlying factors contributing to this gender-based difference in the occurrence of F3T3 fusion.
Our study provides the first evidence that GBMs with F3T3 fusion are more frequently located in the cerebral cortical area compared to F3T3-negative tumors. Twelve of the twenty-nine F3T3-positive GBMs (41.3%) were located in the cerebral cortex or subcortical area. Consistent with our results, Di Stefano et al. [30] investigated the radiological profiles of gliomas with F3T3 fusion and found they more often affected the insula and temporal lobe. In contrast, Roux et al. [34] analyzed MRI scans of 392 GBM patients with the IDH wildtype and reported 63.3% (248/392) of GBMs were located in the subcortical white matter of the subventricular zones of both hemispheres. Since they did not mention the presence of absence of F3T3 fusion in their cases, we cannot determine whether they examined F3T3-positive tumors, F3T3-negative tumors, or a mixture of both types.
Bielle et al. [9] reported that F3T3-positive GBMs are histologically characterized by monomorphic, ovoid nuclei; nuclear palisading; a curvilinear capillary network; calcification; and desmoplastic vessels. We found that curvilinear capillary networks (p < 0.001), nuclear palisading (p = 0.001), and calcification (p = 0.004) were more frequently identified in F3T3-positive GBMs compared to the control group. In cases where these three recurrent characteristic histological findings are encountered in the diagnosis of IDH-wildtype GBM, pathologists should consider the possibility of an F3T3 fusion and perform a molecular test. Additionally, FGFR3 immunostaining has been recommended as a screening tool for detecting F3T3 fusion, with a sensitivity of 100% and a specificity of 92% [9]. In F3T3-positive cases, FGFR3 immunostaining consistently revealed a positive expression, although heterogeneous staining patterns were occasionally observed. Particularly, areas exhibiting the characteristic histological features exhibited a stronger staining intensity [9]. Therefore, FGFR3 immunostaining can facilitate the efficient prediction of F3T3 fusion, making the diagnostic process easier.
We found that none of the IDH-wildtype GBMs with F3T3 fusion had EGFR amplification, which is consistent with the findings of a previous study by Di Stefano et al. [11]. These observations suggest that F3T3 fusion is mutually exclusive with IDH mutation and EGFR amplification. Similarly, none of the F3T3-positive GBMs presented with KIT or PDGFRA amplifications, while 83 (17.4%) and 63 (13.2%) of the 479 F3T3-negative GBMs had PDGFRA and KIT amplifications, respectively. The absence of the amplification of these genes in our F3T3-positive GBM cases raises the possibility of mutual exclusivity between KIT/PDGFRA and F3T3 fusion, similar to that of IDH mutation and EGFR amplification. Consistent with our data, Mata et al. [5] also reported a lack of PDGFRA and KIT amplifications in their F3T3-positive GBMs, demonstrating significant differences from that of F3T3-negative GBMs. In contrast, according to the 2021 WHO Classification of Tumors of the CNS, PDGFRA alteration has been reported in 10–15% of GBM cases [14,21,35]. Nobusawa et al. [36] observed the amplifications of PDGFRA, KIT, and KDR in 33 (8.5%), 17 (4.4%), and 13 (3.3%) of 390 GBMs, respectively. Further studies are warranted to investigate whether the absence of EGFR, PDGFRA, and KIT amplifications is due to the rarity of reported GBM cases with F3T3 fusion or is due to these alterations and F3T3 fusion being mutually exclusive events. We found no significant difference in the frequency of TP53 alteration between the F3T3-positive and -negative groups. Even though we further analyzed whether the difference exists according to the type of TP53 mutation, Fisher’s exact test revealed that non-truncating mutation (p = 0.102), truncating mutation (p = 0.583), and fusion (p = 0.372) of TP53 did not show any statistical significance. Further investigations using a larger cohort are necessary to compare both the mutational profiles of TP53 and the expression patterns of p53 protein between F3T3-positive and -negative GBMs.
Unlike the previous reports by Bielle et al. [9] and Di Stefano et al. [11], this study represents a large series of 25 consecutive F3T3-positive GBM cases diagnosed in a single institution. We comprehensively analyzed a wide range of clinicopathological characteristics, including clinical features, radiological characteristics, histological findings, and molecular characteristics. Bielle et al. [9] described a multi-institutional series of 30 patients with diffuse gliomas, including low-grade (grade 2) tumors. They demonstrated recurrent morphological features and molecular characteristics. Similarly, Di Stefano et al. [11] collected 80 F3T3-positive glioma cases from multiple institutions encompassing low-grade (grades 2 and 3) gliomas. They investigated the radiological features and molecular characteristics. In this study, by combining the clinical, pathological, and imaging features, we provided strong, unique insights into understanding the implications of F3T3 fusion in IDH-wildtype GBMs.
In summary, our findings suggest that F3T3-positive GBM is a distinct molecular subgroup within the IDH-wildtype GBM group. Both clinicians and pathologists should consider this rare entity in the differential diagnosis of diffuse astrocytic glioma to make an accurate diagnosis and to ensure appropriate therapeutic management. Identifying this rare entity based on its distinctive imaging findings and histological features allows for the potential utilization of FGFR3 inhibitors or TACC3-targeting agents, opening up additional treatment options for patients. As clinical trials investigating novel therapeutic agents progress, it will be meaningful to assess their efficacy in F3T3-positive GBMs and to compare their impact on survival rates with those in the non-fusion group.
Author Contributions
Conceptualization, H.B. and Y.-L.S.; methodology, H.B. and B.L.; software, H.B.; validation, J.L. and H.-S.K.; formal analysis, H.B.; investigation, S.H.; resources, Y.-L.S.; data curation, B.L.; writing—original draft preparation, H.B. and Y.-L.S.; writing—review and editing, H.-S.K. and Y.-L.S.; visualization, H.B. and H.-S.K.; supervision, Y.-L.S.; funding acquisition, H.-S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (2023R1A2C2006223).
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Samsung Medical Center (protocol code: 2022–09-065-002; date of approval: 27 September 2022).
Informed Consent Statement
Regarding the retrospective nature of this study, the Institutional Review Board waived the requirement for the investigators to obtain signed informed consent.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to thank Deok Geun Kim for their technical support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- WHO Classification of Tumours of the Central Nervous System, 4th ed.; Louis, D.N., Ohgaki, H., Wiestler, O.D., Cavenee, W.K., Eds.; WHO: Lyon, France, 2016. [Google Scholar]
- WHO Classification of Tumors Editorial Board (Ed.) WHO Classification of Tumours: Central Nervous System Tumours, 5th ed.; WHO: Lyon, France, 2021. [Google Scholar]
- Polivka, J.; Svajdler, M.; Priban, V.; Mracek, J.; Kasik, P.; Martinek, P.; Ptakova, N.; Sharif Bagheri, M.; Shetti, D.; Pesta, M.; et al. Oncogenic fusions in gliomas: An institutional experience. Anticancer Res. 2022, 42, 1933–1939. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wang, H.; Huang, X.; Li, W.; Huang, Q.; Yan, Y.; Chen, J. Gene fusion in malignant glioma: An emerging target for next-generation personalized treatment. Transl. Oncol. 2018, 11, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Mata, D.A.; Benhamida, J.K.; Lin, A.L.; Vanderbilt, C.M.; Yang, S.R.; Villafania, L.B.; Ferguson, D.C.; Jonsson, P.; Miller, A.M.; Tabar, V.; et al. Genetic and epigenetic landscape of IDH-wildtype glioblastomas with FGFR3-TACC3 fusions. Acta Neuropathol. Commun. 2020, 8, 186. [Google Scholar] [CrossRef]
- Gott, H.; Uhl, E. FGFR3-TACCs3 fusions and their clinical relevance in human glioblastoma. Int. J. Mol. Sci. 2022, 23, 8675. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Sanson, M.; Iavarone, A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017, 19, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Metais, A.; Tauziede-Espariat, A.; Garcia, J.; Appay, R.; Uro-Coste, E.; Meyronet, D.; Maurage, C.A.; Vandenbos, F.; Rigau, V.; Chiforeanu, D.C.; et al. Clinico-pathological and epigenetic heterogeneity of diffuse gliomas with FGFR3::TACC3 fusion. Acta Neuropathol. Commun. 2023, 11, 14. [Google Scholar] [CrossRef]
- Bielle, F.; Di Stefano, A.L.; Meyronet, D.; Picca, A.; Villa, C.; Bernier, M.; Schmitt, Y.; Giry, M.; Rousseau, A.; Figarella-Branger, D.; et al. Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol. 2018, 28, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Piombino, E.; Altieri, R.; Romano, C.; Certo, F.; Barbagallo, G.M.V.; Vigneri, P.; Condorelli, D.; Colarossi, L.; Colarossi, C.; et al. Glioblastoma, IDH-Wild type with FGFR3-TACC3 fusion: When morphology may reliably predict the molecular profile of a tumor. A case report and literature review. Front. Neurol. 2022, 13, 823015. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.L.; Picca, A.; Saragoussi, E.; Bielle, F.; Ducray, F.; Villa, C.; Eoli, M.; Paterra, R.; Bellu, L.; Mathon, B.; et al. Clinical, molecular, and radiomic profile of gliomas with FGFR3-TACC3 fusions. Neuro Oncol. 2020, 22, 1614–1624. [Google Scholar] [CrossRef]
- Wu, Z.; Lopes Abath Neto, O.; Bale, T.A.; Benhamida, J.; Mata, D.; Turakulov, R.; Abdullaev, Z.; Marker, D.; Ketchum, C.; Chung, H.J.; et al. DNA methylation analysis of glioblastomas harboring FGFR3-TACC3 fusions identifies a methylation subclass with better patient survival. Acta Neuropathol. 2022, 144, 155–157. [Google Scholar] [CrossRef]
- Dono, A.; El Achi, H.; Bundrant, B.E.; Goli, P.S.; Zhu, P.; Ozkizilkaya, H.I.; Esquenazi, Y.; Ballester, L.Y. Infiltrating gliomas with FGFR alterations: Histologic features, genetic alterations, and potential clinical implications. Cancer Biomark. 2023, 36, 117–131. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Trifonov, V.; Chan, J.M.; Castano, A.; Lia, M.; Abate, F.; Keir, S.T.; Ji, A.X.; Zoppoli, P.; Niola, F.; et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013, 45, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019, 80, 101896. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wu, Z.; Zhang, H.; Zhang, N.; Wu, W.; Wang, Z.; Dai, Z.; Zhang, X.; Zhang, L.; Peng, Y.; et al. Glioma targeted therapy: Insight into future of molecular approaches. Mol. Cancer 2022, 21, 39. [Google Scholar] [CrossRef]
- Costa, R.; Carneiro, B.A.; Taxter, T.; Tavora, F.A.; Kalyan, A.; Pai, S.A.; Chae, Y.K.; Giles, F.J. FGFR3-TACC3 fusion in solid tumors: Mini review. Oncotarget 2016, 7, 55924–55938. [Google Scholar] [CrossRef]
- Hood, F.E.; Royle, S.J. Pulling it together: The mitotic function of TACC3. Bioarchitecture 2011, 1, 105–109. [Google Scholar] [CrossRef]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef]
- Williams, S.V.; Hurst, C.D.; Knowles, M.A. Oncogenic FGFR3 gene fusions in bladder cancer. Hum. Mol. Genet. 2013, 22, 795–803. [Google Scholar] [CrossRef]
- Capelletti, M.; Dodge, M.E.; Ercan, D.; Hammerman, P.S.; Park, S.I.; Kim, J.; Sasaki, H.; Jablons, D.M.; Lipson, D.; Young, L.; et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin. Cancer Res. 2014, 20, 6551–6558. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; Elvin, J.A.; Kamath, S.D.; Ali, S.M.; Paintal, A.S.; Restrepo, A.; Berry, E.; Giles, F.J.; Johnson, M.L. FGFR3-TACC3: A novel gene fusion in cervical cancer. Gynecol. Oncol. Rep. 2015, 13, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kato, H.; Matsuura, S.; Inoue, Y.; Igarashi, H.; Nagura, K.; Nakamura, S.; Maruyama, K.; Tajima, M.; Funai, K.; et al. A novel somatic FGFR3 mutation in primary lung cancer. Oncol. Rep. 2014, 31, 1219–1224. [Google Scholar] [CrossRef]
- Tamura, R.; Yoshihara, K.; Saito, T.; Ishimura, R.; Martinez-Ledesma, J.E.; Xin, H.; Ishiguro, T.; Mori, Y.; Yamawaki, K.; Suda, K.; et al. Novel therapeutic strategy for cervical cancer harboring FGFR3-TACC3 fusions. Oncogenesis 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, Z.H.; Lin, Z.R.; Xu, L.H.; Zhong, Q.; Zeng, M.S. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol. Ther. 2014, 15, 1613–1621. [Google Scholar] [CrossRef]
- Frattini, V.; Pagnotta, S.M.; Tala; Fan, J.J.; Russo, M.V.; Lee, S.B.; Garofano, L.; Zhang, J.; Shi, P.; Lewis, G.; et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature 2018, 553, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Ryan, E.L.; Royle, S.J. FGFR3-TACC3 cancer gene fusions cause mitotic defects by removal of endogenous TACC3 from the mitotic spindle. Open Biol. 2017, 7, 170080. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.L.; Fucci, A.; Frattini, V.; Labussiere, M.; Mokhtari, K.; Zoppoli, P.; Marie, Y.; Bruno, A.; Boisselier, B.; Giry, M.; et al. Detection, characterization, and inhibition of FGFR-TACC Fusions in IDH wild-type glioma. Clin. Cancer Res. 2015, 21, 3307–3317. [Google Scholar] [CrossRef]
- Tabernero, J.; Bahleda, R.; Dienstmann, R.; Infante, J.R.; Mita, A.; Italiano, A.; Calvo, E.; Moreno, V.; Adamo, B.; Gazzah, A.; et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2015, 33, 3401–3408. [Google Scholar] [CrossRef]
- Parish, A.; Schwaederle, M.; Daniels, G.; Piccioni, D.; Fanta, P.; Schwab, R.; Shimabukuro, K.; Parker, B.A.; Helsten, T.; Kurzrock, R. Fibroblast growth factor family aberrations in cancers: Clinical and molecular characteristics. Cell Cycle 2015, 14, 2121–2128. [Google Scholar] [CrossRef]
- Asif, S.; Fatima, R.; Krc, R.; Bennett, J.; Raza, S. Comparative proteogenomic characterization of glioblastoma. CNS Oncol. 2019, 8, CNS37. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Roca, P.; Edjlali, M.; Sato, K.; Zanello, M.; Dezamis, E.; Gori, P.; Lion, S.; Fleury, A.; Dhermain, F.; et al. MRI atlas of IDH wild-type supratentorial glioblastoma: Probabilistic maps of phenotype, management, and outcomes. Radiology 2019, 293, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, S.; Stawski, R.; Kim, Y.H.; Nakazato, Y.; Ohgaki, H. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: A population-based study. Neuropathology 2011, 31, 583–588. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).