Interaction of Tau with Kinesin-1: Effect of Kinesin-1 Heavy Chain Elimination on Autophagy-Mediated Mutant Tau Degradation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines Reagents, Recombinant Proteins, Plasmids, and Antibodies
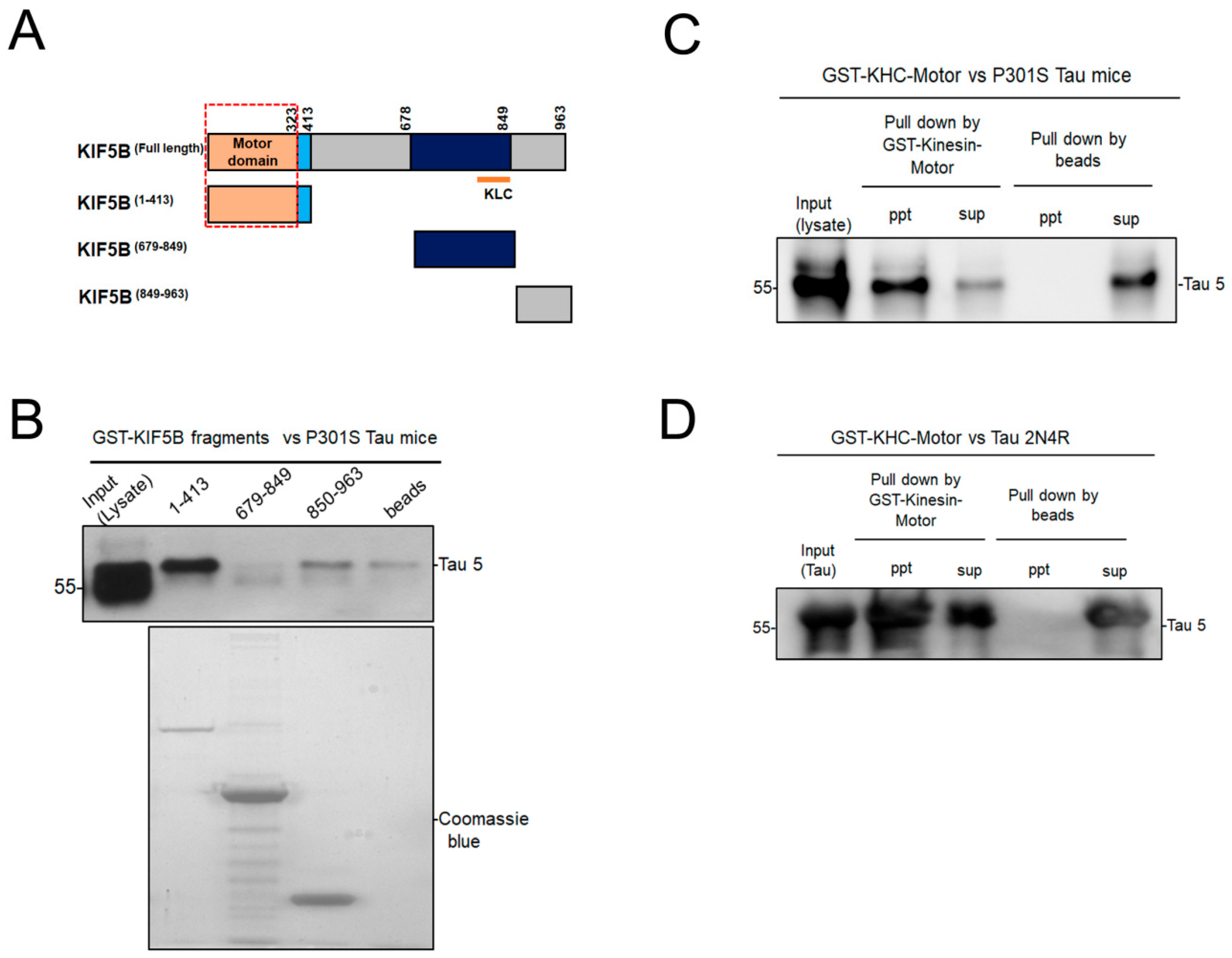
2.2. Affinity Isolation and Immobilization of GST-KIF5B Protein Fragments
2.3. Pull-Down Assay Using Tau Mouse Brain Lysates
2.4. Pull-Down of Recombinant Tau 2N-4R by Kinesin Motor Domain
2.5. ATPase Assay
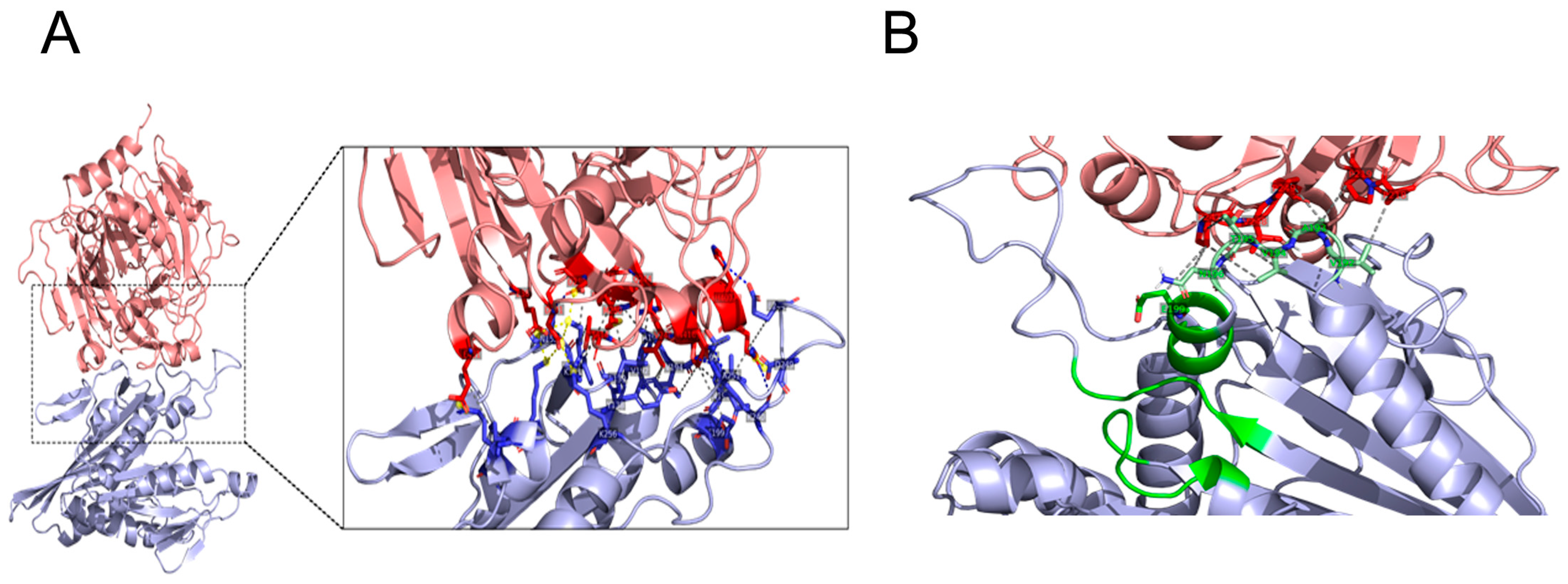
2.6. Docking of KIF5B Motor Domain with Tau
2.7. Cell Culture
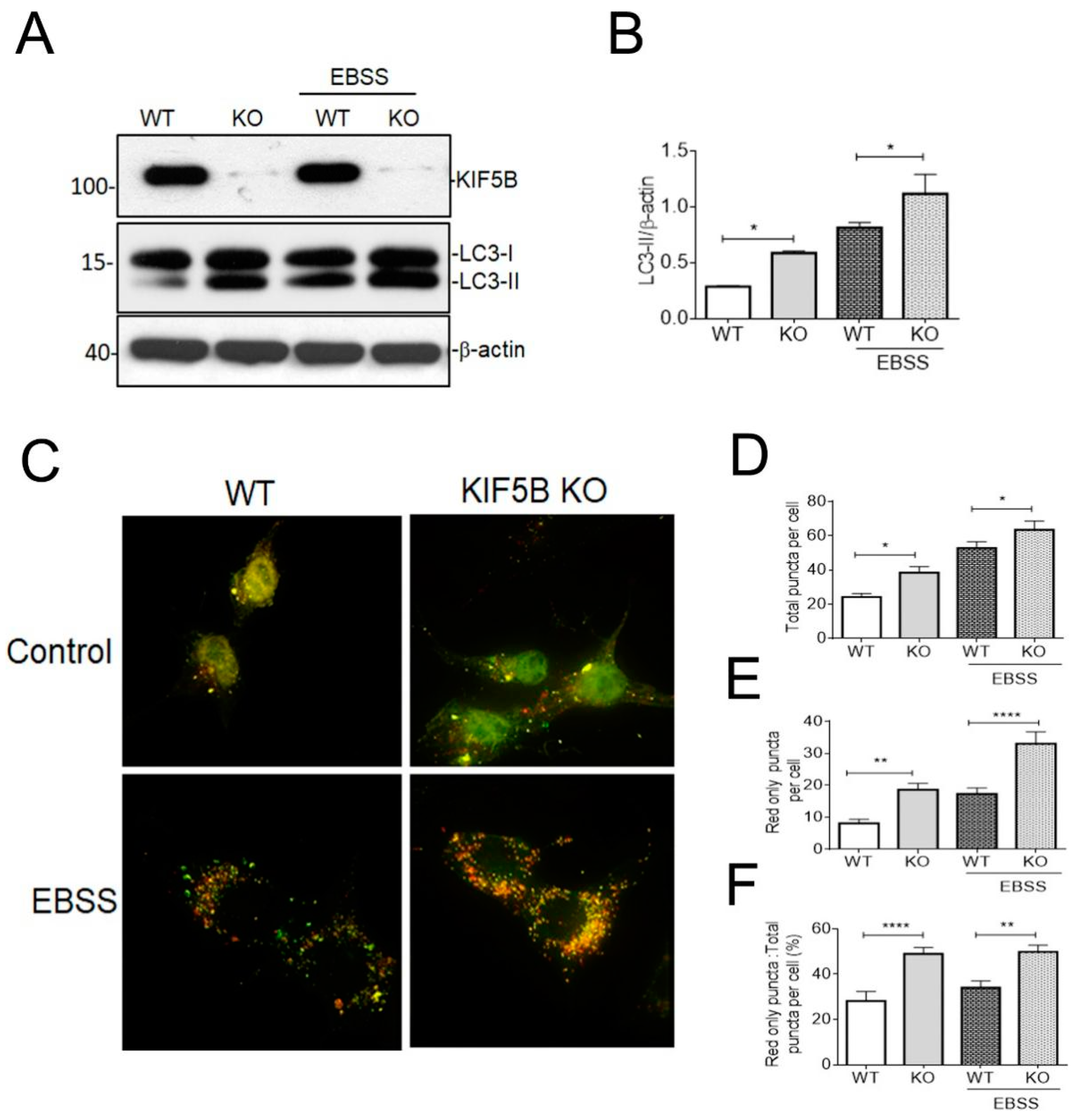
2.8. Cell Imaging and Puncta Counting
2.9. Analysis of Protein Lysates and Pull-Down Samples by Western Blotting
2.10. Statistics
3. Results
3.1. KIF5B Motor Domain Interacts with Tau
3.2. Tau Inhibits the Kinesin Motor’s Intrinsic ATPase Activity
3.3. Docking
3.4. KIF5B KO Facilitates Autophagosome Maturation
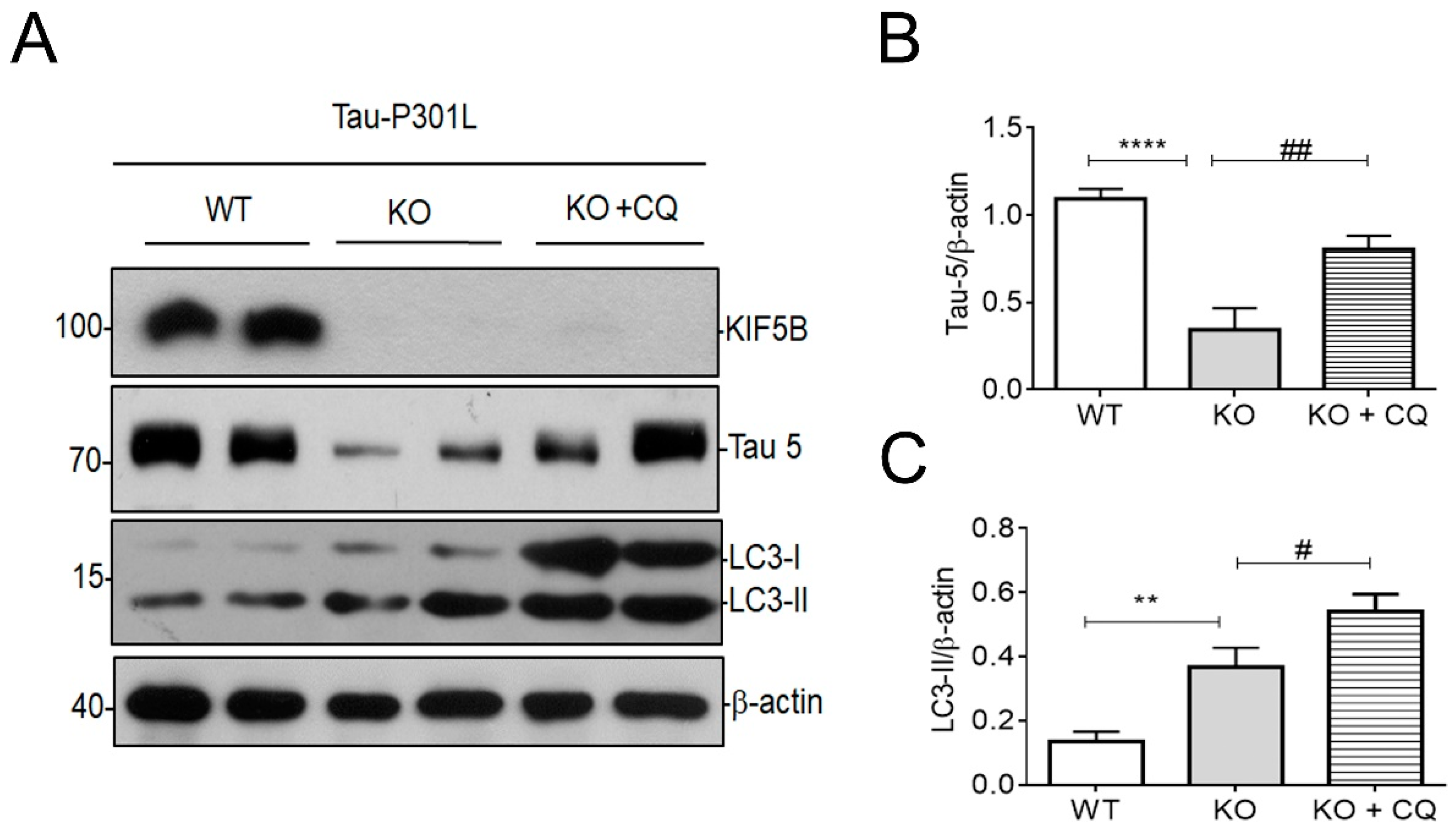
3.5. Removal of KIF5B Reduced Tau via Induction of Autophagy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hancock, W.O. Bidirectional Cargo Transport: Moving beyond Tug of War. Nat. Rev. Mol. Cell Biol. 2014, 15, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Bodakuntla, S.; Jijumon, A.S.; Villablanca, C.; Gonzalez-Billault, C.; Janke, C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends Cell Biol. 2019, 29, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Ebneth, A.; Godemann, R.; Stamer, K.; Illenberger, S.; Trinczek, B.; Mandelkow, E. Overexpression of Tau Protein Inhibits Kinesin-Dependent Trafficking of Vesicles, Mitochondria, and Endoplasmic Reticulum: Implications for Alzheimer’s Disease. J. Cell Biol. 1998, 143, 777–794. [Google Scholar] [CrossRef] [PubMed]
- Trinczek, B.; Ebneth, A.; Mandelkow, E.M.; Mandelkow, E. Tau Regulates the Attachment/Detachment but Not the Speed of Motors in Microtubule-Dependent Transport of Single Vesicles and Organelles. J. Cell Sci. 1999, 112 Pt 14, 2355–2367. [Google Scholar] [CrossRef]
- Stamer, K.; Vogel, R.; Thies, E.; Mandelkow, E.; Mandelkow, E.-M. Tau Blocks Traffic of Organelles, Neurofilaments, and APP Vesicles in Neurons and Enhances Oxidative Stress. J. Cell Biol. 2002, 156, 1051–1063. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, K.-M.; Yang, L.; Dong, Q.; Yu, J.-T. Tauopathies: New Perspectives and Challenges. Mol. Neurodegener. 2022, 17, 28. [Google Scholar] [CrossRef]
- Hendricks, A.G.; Perlson, E.; Ross, J.L.; Schroeder, H.W.; Tokito, M.; Holzbaur, E.L.F. Motor Coordination via a Tug-of-War Mechanism Drives Bidirectional Vesicle Transport. Curr. Biol. 2010, 20, 697–702. [Google Scholar] [CrossRef]
- Maday, S.; Wallace, K.E.; Holzbaur, E.L.F. Autophagosomes Initiate Distally and Mature during Transport toward the Cell Soma in Primary Neurons. J. Cell Biol. 2012, 196, 407–417. [Google Scholar] [CrossRef]
- Hirokawa, N.; Niwa, S.; Tanaka, Y. Molecular Motors in Neurons: Transport Mechanisms and Roles in Brain Function, Development, and Disease. Neuron 2010, 68, 610–638. [Google Scholar] [CrossRef]
- Kreft, K.L.; van Meurs, M.; Wierenga-Wolf, A.F.; Melief, M.-J.; van Strien, M.E.; Hol, E.M.; Oostra, B.A.; Laman, J.D.; Hintzen, R.Q. Abundant Kif21b Is Associated with Accelerated Progression in Neurodegenerative Diseases. Acta Neuropathol. Commun. 2014, 2, 144. [Google Scholar] [CrossRef]
- Hares, K.; Miners, J.S.; Cook, A.J.; Rice, C.; Scolding, N.; Love, S.; Wilkins, A. Overexpression of Kinesin Superfamily Motor Proteins in Alzheimer’s Disease. J. Alzheimers Dis. 2017, 60, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Selvarasu, K.; Singh, A.K.; Iyaswamy, A.; Gopalkrishnashetty Sreenivasmurthy, S.; Krishnamoorthi, S.; Bera, A.K.; Huang, J.-D.; Durairajan, S.S.K. Reduction of Kinesin I Heavy Chain Decreases Tau Hyperphosphorylation, Aggregation, and Memory Impairment in Alzheimer’s Disease and Tauopathy Models. Front. Mol. Biosci. 2022, 9, 1050768. [Google Scholar] [CrossRef] [PubMed]
- Ackmann, M.; Wiech, H.; Mandelkow, E. Nonsaturable Binding Indicates Clustering of Tau on the Microtubule Surface in a Paired Helical Filament-like Conformation. J. Biol. Chem. 2000, 275, 30335–30343. [Google Scholar] [CrossRef] [PubMed]
- Monroy, B.Y.; Sawyer, D.L.; Ackermann, B.E.; Borden, M.M.; Tan, T.C.; Ori-McKenney, K.M. Competition between Microtubule-Associated Proteins Directs Motor Transport. Nat. Commun. 2018, 9, 1487. [Google Scholar] [CrossRef]
- Chen, J.; Kanai, Y.; Cowan, N.J.; Hirokawa, N. Projection Domains of MAP2 and Tau Determine Spacings between Microtubules in Dendrites and Axons. Nature 1992, 360, 674–677. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Ross, J.L.; Goldman, Y.E.; Holzbaur, E.L.F. Differential Regulation of Dynein and Kinesin Motor Proteins by Tau. Science 2008, 319, 1086–1089. [Google Scholar] [CrossRef] [PubMed]
- Vershinin, M.; Carter, B.C.; Razafsky, D.S.; King, S.J.; Gross, S.P. Multiple-Motor Based Transport and Its Regulation by Tau. Proc. Natl. Acad. Sci. USA 2007, 104, 87–92. [Google Scholar] [CrossRef]
- Stern, J.L.; Lessard, D.V.; Hoeprich, G.J.; Morfini, G.A.; Berger, C.L. Phosphoregulation of Tau Modulates Inhibition of Kinesin-1 Motility. Mol. Biol. Cell 2017, 28, 1079–1087. [Google Scholar] [CrossRef]
- Vershinin, M.; Xu, J.; Razafsky, D.S.; King, S.J.; Gross, S.P. Tuning Microtubule-Based Transport through Filamentous MAPs: The Problem of Dynein. Traffic 2008, 9, 882–892. [Google Scholar] [CrossRef]
- Maday, S.; Holzbaur, E.L.F. Autophagosome Biogenesis in Primary Neurons Follows an Ordered and Spatially Regulated Pathway. Dev. Cell 2014, 30, 71–85. [Google Scholar] [CrossRef]
- Soppina, V.; Rai, A.K.; Ramaiya, A.J.; Barak, P.; Mallik, R. Tug-of-War between Dissimilar Teams of Microtubule Motors Regulates Transport and Fission of Endosomes. Proc. Natl. Acad. Sci. USA 2009, 106, 19381–19386. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Noda, T.; Yoshimori, T. Dynein-Dependent Movement of Autophagosomes Mediates Efficient Encounters with Lysosomes. Cell Struct. Funct. 2008, 33, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Duan, Z.; Sun, H.; Fung, M.-L.; Chen, H.; Wang, J.; Lau, C.-F.; Yang, D.; Liu, Y.; Ni, Y.; et al. Kinesin-1 Regulates Extrasynaptic Targeting of NMDARs and Neuronal Vulnerability Toward Excitotoxicity. iScience 2019, 13, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.; Ingram, E.; Takao, M.; Smith, M.J.; Jakes, R.; Virdee, K.; Yoshida, H.; Holzer, M.; Craxton, M.; Emson, P.C.; et al. Abundant Tau Filaments and Nonapoptotic Neurodegeneration in Transgenic Mice Expressing Human P301S Tau Protein. J. Neurosci. 2002, 22, 9340–9351. [Google Scholar] [CrossRef] [PubMed]
- Seitz, A.; Kojima, H.; Oiwa, K.; Mandelkow, E.-M.; Song, Y.-H.; Mandelkow, E. Single-Molecule Investigation of the Interference between Kinesin, Tau and MAP2c. EMBO J. 2002, 21, 4896–4905. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Yi, H.; Desai, R.; Hand, A.R.; Haas, A.L.; Ferreira, P.A. RANBP2 Is an Allosteric Activator of the Conventional Kinesin-1 Motor Protein, KIF5B, in a Minimal Cell-Free System. EMBO Rep. 2009, 10, 480–486. [Google Scholar] [CrossRef]
- Kull, F.J.; Sablin, E.P.; Lau, R.; Fletterick, R.J.; Vale, R.D. Crystal Structure of the Kinesin Motor Domain Reveals a Structural Similarity to Myosin. Nature 1996, 380, 550–555. [Google Scholar] [CrossRef]
- Popov, K.I.; Makepeace, K.A.T.; Petrotchenko, E.V.; Dokholyan, N.V.; Borchers, C.H. Insight into the Structure of the “Unstructured” Tau Protein. Structure 2019, 27, 1710–1715.e4. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A Web Server for Predicting the Binding Affinity of Protein-Protein Complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein-Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.-Z.; Yang, C.; Zhuang, X.-X.; Yuan, N.-N.; Wu, M.-Y.; Tan, J.-Q.; Song, J.-X.; Cheung, K.-H.; Su, H.; Wang, Y.-T.; et al. NRBF2 Is a RAB7 Effector Required for Autophagosome Maturation and Mediates the Association of APP-CTFs with Active Form of RAB7 for Degradation. Autophagy 2021, 17, 1112–1130. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N. Microtubule Organization and Dynamics Dependent on Microtubule-Associated Proteins. Curr. Opin. Cell Biol. 1994, 6, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Marx, A.; Hoenger, A.; Mandelkow, E. Structures of Kinesin Motor Proteins. Cell Motil. Cytoskelet. 2009, 66, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W.; Schulten, K. Nucleotide-Dependent Movements of the Kinesin Motor Domain Predicted by Simulated Annealing. Biophys. J. 1998, 75, 646–661. [Google Scholar] [CrossRef]
- Ohsawa, I.; Nishimaki, K.; Murakami, Y.; Suzuki, Y.; Ishikawa, M.; Ohta, S. Age-dependent neurodegeneration accompanying memory loss in transgenic mice defective in mitochondrial aldehyde dehydrogenase 2 activity. J. Neurosci. 2008, 28, 6239–6249. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef]
- Durairajan, S.S.K.; Selvarasu, K.; Bera, M.R.; Rajaram, K.; Iyaswamy, A.; Li, M. Alzheimer’s Disease and other Tauopathies: Exploring Efficacy of Medicinal Plant-derived Compounds in Alleviating Tau-mediated Neurodegeneration. Curr. Mol. Pharmacol. 2022, 15, 361–379. [Google Scholar] [CrossRef]
- Bull, N.D.; Guidi, A.; Goedert, M.; Martin, K.R.; Spillantini, M.G. Reduced axonal transport and increased excitotoxic retinal ganglion cell degeneration in mice transgenic for human mutant P301S tau. PLoS ONE 2012, 7, e34724. [Google Scholar] [CrossRef]
- Caballero, B.; Wang, Y.; Diaz, A.; Tasset, I.; Juste, Y.R.; Stiller, B.; Mandelkow, E.M.; Mandelkow, E.; Cuervo, A.M. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 2018, 17, e12692. [Google Scholar] [CrossRef]
- Hoeprich, G.J.; Thompson, A.R.; McVicker, D.P.; Hancock, W.O.; Berger, C.L. Kinesin’s Neck-Linker Determines Its Ability to Navigate Obstacles on the Microtubule Surface. Biophys. J. 2014, 106, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Siahaan, V.; Krattenmacher, J.; Hyman, A.A.; Diez, S.; Hernández-Vega, A.; Lansky, Z.; Braun, M. Kinetically Distinct Phases of Tau on Microtubules Regulate Kinesin Motors and Severing Enzymes. Nat. Cell Biol. 2019, 21, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Al-Bassam, J.; Ozer, R.S.; Safer, D.; Halpain, S.; Milligan, R.A. MAP2 and Tau Bind Longitudinally along the Outer Ridges of Microtubule Protofilaments. J. Cell Biol. 2002, 157, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Dehmelt, L.; Halpain, S. The MAP2/Tau Family of Microtubule-Associated Proteins. Genome Biol. 2005, 6, 204. [Google Scholar] [CrossRef]
- Wang, W.; Cao, L.; Wang, C.; Gigant, B.; Knossow, M. Kinesin, 30 Years Later: Recent Insights from Structural Studies. Protein Sci. 2015, 24, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhang, Y. A Mechanochemical Model of the Forward/Backward Movement of Motor Protein Kinesin-1. J. Biol. Chem. 2022, 298, 101948. [Google Scholar] [CrossRef] [PubMed]
- Schnitzer, M.J.; Block, S.M. Kinesin Hydrolyses One ATP per 8-Nm Step. Nature 1997, 388, 386–390. [Google Scholar] [CrossRef]
- Coy, D.L.; Wagenbach, M.; Howard, J. Kinesin Takes One 8-Nm Step for Each ATP That It Hydrolyzes. J. Biol. Chem. 1999, 274, 3667–3671. [Google Scholar] [CrossRef]
- Dunn, S.; Morrison, E.E.; Liverpool, T.B.; Molina-París, C.; Cross, R.A.; Alonso, M.C.; Peckham, M. Differential Trafficking of Kif5c on Tyrosinated and Detyrosinated Microtubules in Live Cells. J. Cell Sci. 2008, 121 Pt 7, 1085–1095. [Google Scholar] [CrossRef]
- Brady, S.T. A Novel Brain ATPase with Properties Expected for the Fast Axonal Transport Motor. Nature 1985, 317, 73–75. [Google Scholar] [CrossRef]
- Hammond, J.W.; Huang, C.-F.; Kaech, S.; Jacobson, C.; Banker, G.; Verhey, K.J. Posttranslational Modifications of Tubulin and the Polarized Transport of Kinesin-1 in Neurons. Mol. Biol. Cell 2010, 21, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Nirschl, J.J.; Holzbaur, E.L.F. LC3 Binding to the Scaffolding Protein JIP1 Regulates Processive Dynein-Driven Transport of Autophagosomes. Dev. Cell 2014, 29, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sato, Y.; Nixon, R.A. Lysosomal Proteolysis Inhibition Selectively Disrupts Axonal Transport of Degradative Organelles and Causes an Alzheimer’s-like Axonal Dystrophy. J. Neurosci. 2011, 31, 7817–7830. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.M.P.; Groth-Pedersen, L.; Høyer-Hansen, M.; Kirkegaard, T.; Corcelle, E.; Andersen, J.S.; Jäättelä, M.; Nylandsted, J. Depletion of Kinesin 5B Affects Lysosomal Distribution and Stability and Induces Peri-Nuclear Accumulation of Autophagosomes in Cancer Cells. PLoS ONE 2009, 4, e4424. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Su, Q.P.; Chen, Y.; Zhu, Y.; Jiang, D.; Rong, Y.; Zhang, S.; Zhang, Y.; Ren, H.; Zhang, C.; et al. Kinesin 1 Drives Autolysosome Tubulation. Dev. Cell 2016, 37, 326–336. [Google Scholar] [CrossRef]
- Rong, Y.; Liu, M.; Ma, L.; Du, W.; Zhang, H.; Tian, Y.; Cao, Z.; Li, Y.; Ren, H.; Zhang, C.; et al. Clathrin and Phosphatidylinositol-4,5-Bisphosphate Regulate Autophagic Lysosome Reformation. Nat. Cell Biol. 2012, 14, 924–934. [Google Scholar] [CrossRef]
- Trimmer, P.A.; Borland, M.K. Differentiated Alzheimer’s Disease Transmitochondrial Cybrid Cell Lines Exhibit Reduced Organelle Movement. Antioxid. Redox Signal 2005, 7, 1101–1109. [Google Scholar] [CrossRef]
- Kar, S.; Fan, J.; Smith, M.J.; Goedert, M.; Amos, L.A. Repeat Motifs of Tau Bind to the Insides of Microtubules in the Absence of Taxol. EMBO J. 2003, 22, 70–77. [Google Scholar] [CrossRef]
- McVicker, D.P.; Chrin, L.R.; Berger, C.L. The Nucleotide-Binding State of Microtubules Modulates Kinesin Processivity and the Ability of Tau to Inhibit Kinesin-Mediated Transport. J. Biol. Chem. 2011, 286, 42873–42880. [Google Scholar] [CrossRef]
- Xu, J.; King, S.J.; Lapierre-Landry, M.; Nemec, B. Interplay between Velocity and Travel Distance of Kinesin-Based Transport in the Presence of Tau. Biophys. J. 2013, 105, L23–L25. [Google Scholar] [CrossRef][Green Version]
- Chaudhary, A.R.; Berger, F.; Berger, C.L.; Hendricks, A.G. Tau Directs Intracellular Trafficking by Regulating the Forces Exerted by Kinesin and Dynein Teams. Traffic 2018, 19, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ali, M.Y.; Bookwalter, C.S.; Warshaw, D.M.; Trybus, K.M. Diffusive Movement of Processive Kinesin-1 on Microtubules. Traffic 2009, 10, 1429–1438. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selvarasu, K.; Singh, A.K.; Dakshinamoorthy, A.; Sreenivasmurthy, S.G.; Iyaswamy, A.; Radhakrishnan, M.; Patnaik, S.; Huang, J.-D.; Williams, L.L.; Senapati, S.; et al. Interaction of Tau with Kinesin-1: Effect of Kinesin-1 Heavy Chain Elimination on Autophagy-Mediated Mutant Tau Degradation. Biomedicines 2024, 12, 5. https://doi.org/10.3390/biomedicines12010005
Selvarasu K, Singh AK, Dakshinamoorthy A, Sreenivasmurthy SG, Iyaswamy A, Radhakrishnan M, Patnaik S, Huang J-D, Williams LL, Senapati S, et al. Interaction of Tau with Kinesin-1: Effect of Kinesin-1 Heavy Chain Elimination on Autophagy-Mediated Mutant Tau Degradation. Biomedicines. 2024; 12(1):5. https://doi.org/10.3390/biomedicines12010005
Chicago/Turabian StyleSelvarasu, Karthikeyan, Abhay Kumar Singh, Avinash Dakshinamoorthy, Sravan Gopalkrishnashetty Sreenivasmurthy, Ashok Iyaswamy, Moorthi Radhakrishnan, Supriti Patnaik, Jian-Dong Huang, Leonard L. Williams, Sanjib Senapati, and et al. 2024. "Interaction of Tau with Kinesin-1: Effect of Kinesin-1 Heavy Chain Elimination on Autophagy-Mediated Mutant Tau Degradation" Biomedicines 12, no. 1: 5. https://doi.org/10.3390/biomedicines12010005
APA StyleSelvarasu, K., Singh, A. K., Dakshinamoorthy, A., Sreenivasmurthy, S. G., Iyaswamy, A., Radhakrishnan, M., Patnaik, S., Huang, J.-D., Williams, L. L., Senapati, S., & Durairajan, S. S. K. (2024). Interaction of Tau with Kinesin-1: Effect of Kinesin-1 Heavy Chain Elimination on Autophagy-Mediated Mutant Tau Degradation. Biomedicines, 12(1), 5. https://doi.org/10.3390/biomedicines12010005






