Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animal
2.3. Breeding of Zebrafish
2.4. Medium Used for Embryo Development
2.5. Aspartame Treatment
2.6. Determination of Methanol Level Using HPLC
2.7. Analysis of Toxicity of Aspartame on Zebrafish Embryo
2.8. Identification of Developmental Stages of Zebrafish
2.9. Determination of Survival Rate
2.10. Determination of Somite’s Development
2.11. Analysis of Heart Rate in Zebrafish
2.12. Measurement of Axial and Head Length of Zebrafish
2.13. Analysis of Yolk Sac Edema
2.14. Analysis of Locomotor Activity
2.15. Detection of Cartilage Development in Zebrafish Embryos Using Alcian Blue Staining
2.16. Analysis of Protein Levels via Western Blot and Dot Blot
2.17. Immunocytochemistry
2.18. In Vitro Cell Culture and Cell Treatment
2.19. Transmission Electron Microscopy (TEM)
2.20. Dataset
2.21. Differential Gene Expression Analysis
2.22. Analysis of Protein-Protein Interaction via STRING Database
2.23. Enrichment Analysis Using ClueGO
2.24. Single Cell Transcriptomic Analysis
2.25. Molecular Docking Analysis
2.26. Statistical Analysis
3. Results
3.1. Aspartame Exposure Reduces the Formation of Larvae and the Development of Cartilage in Zebrafish
3.2. Aspartame Delays Post-Fertilization Development by Altering the Head Length and Locomotor Behavior of Zebrafish
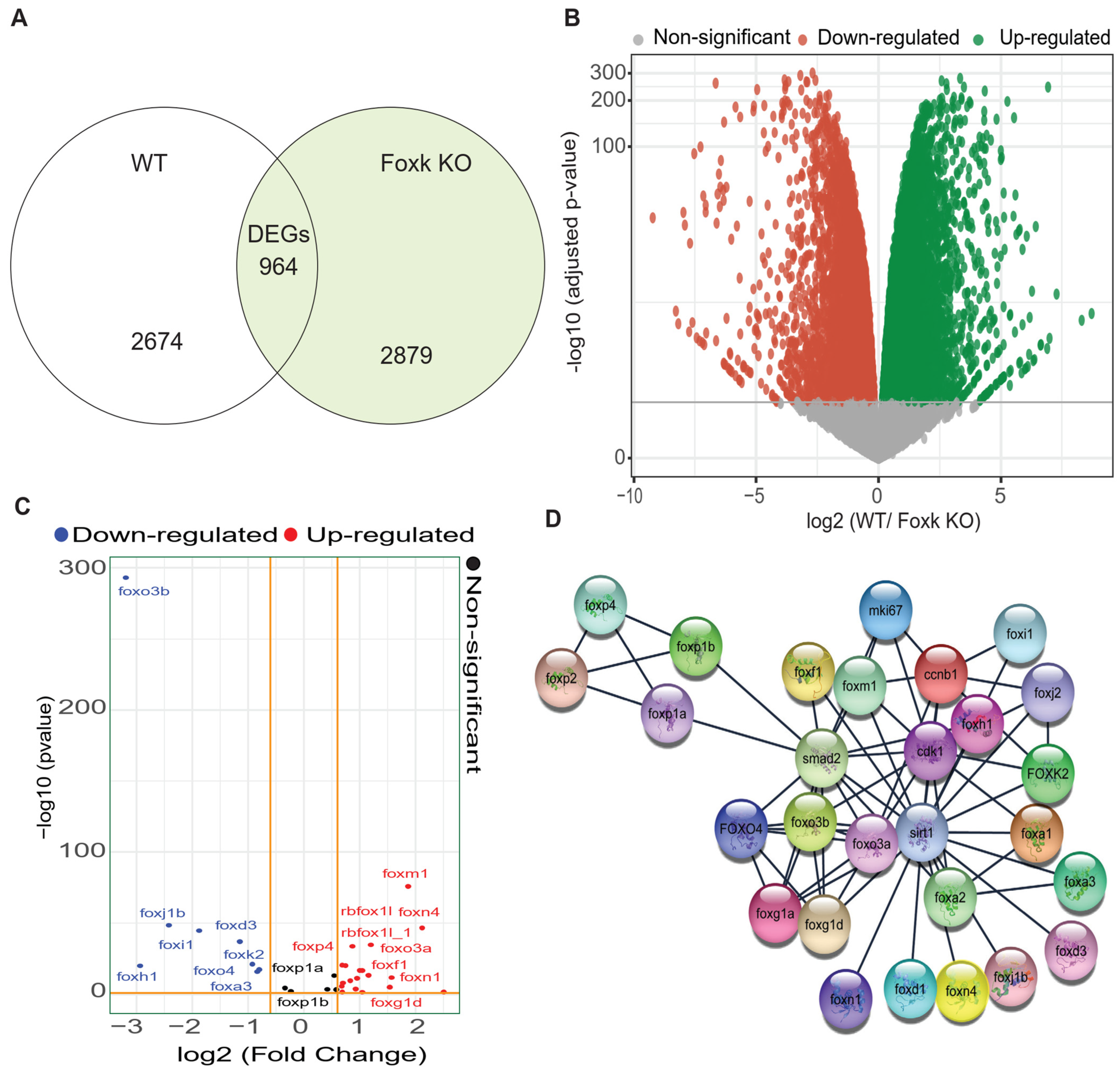
3.3. RNA-Sequencing-Based DEG Analysis Shows SIRT1 and FOXO3a Are Involved in the Neurodevelopment
3.4. Aspartame Could Target and Reduce the Expression of SIRT1 and FOXO3a Proteins in Neuron Cells: In Silico and In Vitro Evidence
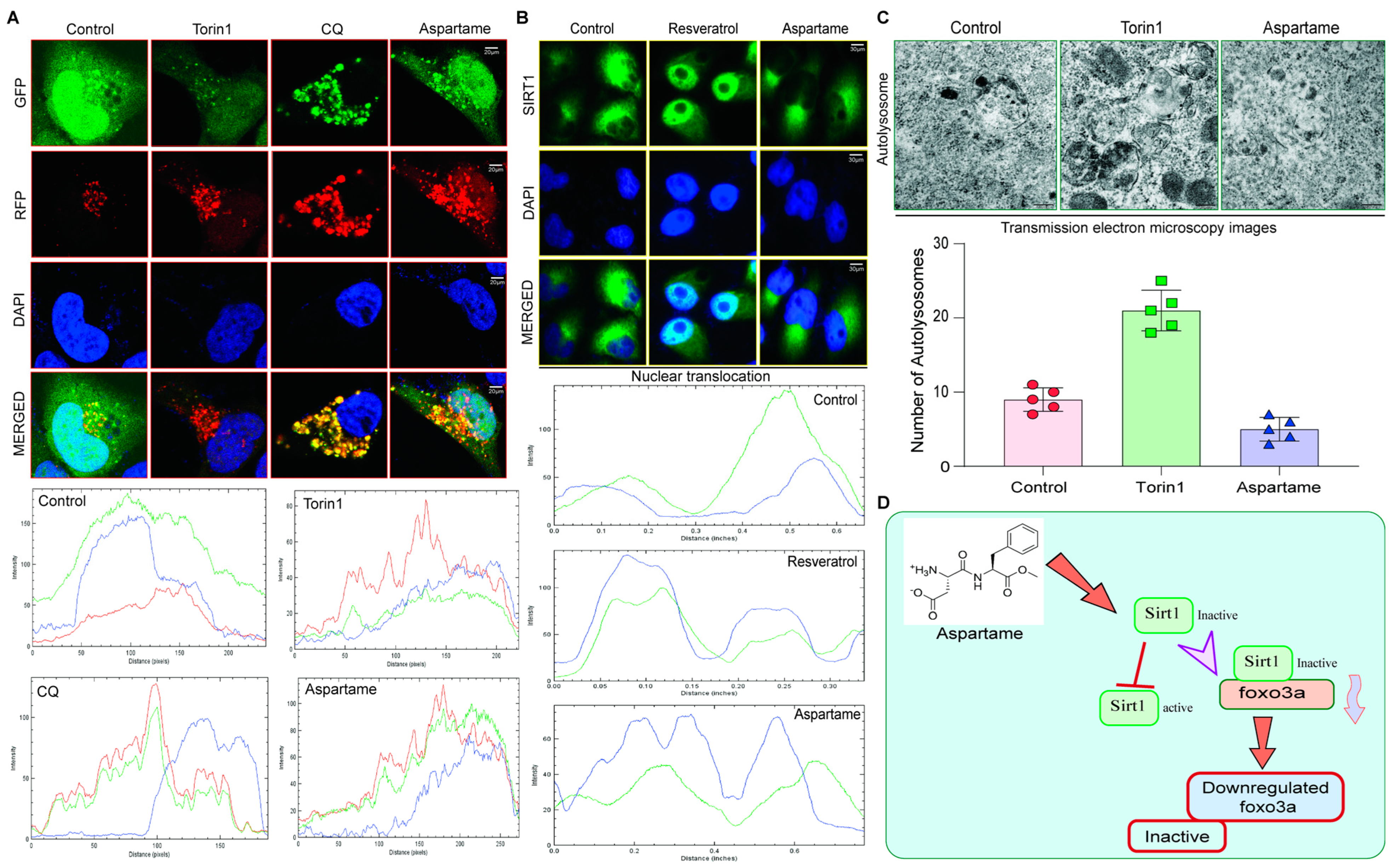
3.5. Aspartame Triggered the Autophagy Flux Reduction by Inhibiting Nuclear Translocation of SIRT1 in Neuronal Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iyaswamy, A.; Kammella, A.K.; Thavasimuthu, C.; Wankupar, W.; Dapkupar, W.; Shanmugam, S.; Rajan, R.; Rathinasamy, S. Oxidative stress evoked damages leading to attenuated memory and inhibition of NMDAR-CaMKII-ERK/CREB signalling on consumption of aspartame in rat model. J. Food Drug Anal. 2018, 26, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Ashok, I.; Sheeladevi, R.; Wankhar, D. Acute effect of aspartame-induced oxidative stress in Wistar albino rat brain. J. Biomed. Res. 2015, 29, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Ashok, I.; Sheeladevi, R. Oxidant stress evoked damage in rat hepatocyte leading to triggered nitric oxide synthase (NOS) levels on long term consumption of aspartame. J. Food Drug Anal. 2015, 23, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Ashok, I.; Sheeladevi, R. Biochemical responses and mitochondrial mediated activation of apoptosis on long-term effect of aspartame in rat brain. Redox. Biol. 2014, 2, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Iyyaswamy, A.; Rathinasamy, S. Effect of chronic exposure to aspartame on oxidative stress in the brain of albino rats. J. Biosci. 2012, 37, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Ashok, I.; Poornima, P.S.; Wankhar, D.; Ravindran, R.; Sheeladevi, R. Oxidative stress evoked damages on rat sperm and attenuated antioxidant status on consumption of aspartame. Int. J. Impot. Res. 2017, 29, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Stegink, L.D.; Filer, L.J., Jr.; Bell, E.F.; Ziegler, E.E.; Tephly, T.R. Effect of repeated ingestion of aspartame-sweetened beverage on plasma amino acid, blood methanol, and blood formate concentrations in normal adults. Metabolism 1989, 38, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Trocho, C.; Pardo, R.; Rafecas, I.; Virgili, J.; Remesar, X.; Fernandez-Lopez, J.A.; Alemany, M. Formaldehyde derived from dietary aspartame binds to tissue components in vivo. Life Sci. 1998, 63, 337–349. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- d’Amora, M.; Giordani, S. The Utility of Zebrafish as a Model for Screening Developmental Neurotoxicity. Front. Neurosci. 2018, 12, 976. [Google Scholar] [CrossRef]
- Teame, T.; Zhang, Z.; Ran, C.; Zhang, H.; Yang, Y.; Ding, Q.; Xie, M.; Gao, C.; Ye, Y.; Duan, M.; et al. The use of zebrafish (Danio rerio) as biomedical models. Anim. Front. 2019, 9, 68–77. [Google Scholar] [CrossRef] [PubMed]
- McCollum, C.W.; Ducharme, N.A.; Bondesson, M.; Gustafsson, J.A. Developmental toxicity screening in zebrafish. Birth Defects Res. C Embryo Today 2011, 93, 67–114. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Murakami, S.; Ashikawa, Y.; Sasagawa, S.; Umemoto, N.; Shimada, Y.; Tanaka, T. Zebrafish as a systems toxicology model for developmental neurotoxicity testing. Congenit. Anom. 2015, 55, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dong, G.; Han, G.; Du, L.; Li, M. Zebrafish Behavioral Phenomics Links Artificial Sweetener Aspartame to Behavioral Toxicity and Neurotransmitter Homeostasis. J. Agric. Food Chem. 2021, 69, 15393–15402. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, K.H.; Kim, J.; Choi, I.; Cho, K.H. Modified high-density lipoproteins by artificial sweetener, aspartame, and saccharin, showed loss of anti-atherosclerotic activity and toxicity in zebrafish. Cardiovasc. Toxicol. 2015, 15, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Maiese, K. Targeting the core of neurodegeneration: FoxO, mTOR, and SIRT1. Neural Regen. Res. 2021, 16, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Ubaid, S. Role of Silent Information Regulator 1 (SIRT1) in Regulating Oxidative Stress and Inflammation. Inflammation 2020, 43, 1589–1598. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Kareem, O.; Goyal, R.K.; Mumtaz, S.M.; Tonk, R.K.; Gupta, R.; Pottoo, F.H. Role of Forkhead Transcription Factors of the O Class (FoxO) in Development and Progression of Alzheimer’s Disease. CNS Neurol. Disord. Drug Targets 2020, 19, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Damas-Souza, D.M.; Nunes, R.; Carvalho, H.F. An improved acridine orange staining of DNA/RNA. Acta Histochem. 2019, 121, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Amadei, G.; Handford, C.E.; Qiu, C.; De Jonghe, J.; Greenfeld, H.; Tran, M.; Martin, B.K.; Chen, D.Y.; Aguilera-Castrejon, A.; Hanna, J.H.; et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 2022, 610, 143–153. [Google Scholar] [CrossRef]
- Ling, D.; Chen, H.; Chan, G.; Lee, S.M. Quantitative measurements of zebrafish heartrate and heart rate variability: A survey between 1990–2020. Comput. Biol. Med. 2022, 142, 105045. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, C.Y.; Oh, H.; Ryu, B.; Kim, U.; Lee, J.M.; Jung, C.R.; Park, J.H. Trimethyltin chloride induces reactive oxygen species-mediated apoptosis in retinal cells during zebrafish eye development. Sci. Total Environ. 2019, 653, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, H.; Wang, R.; Li, T.; Gu, L.; Sun, L. Accumulation and Distribution of Fluorescent Microplastics in the Early Life Stages of Zebrafish. J. Vis. Exp. 2021, 173, e62117. [Google Scholar] [CrossRef]
- Li, D.; Sun, W.; Chen, H.; Lei, H.; Li, X.; Liu, H.; Huang, G.Y.; Shi, W.J.; Ying, G.G.; Luo, Y.; et al. Cyclophosphamide affects eye development and locomotion in zebrafish (Danio rerio). Sci. Total Environ. 2022, 805, 150460. [Google Scholar] [CrossRef]
- Xi, Y.; Chen, D.; Sun, L.; Li, Y.; Li, L. Characterization of zebrafish mutants with defects in bone calcification during development. Biochem. Biophys. Res. Commun. 2013, 440, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Iyaswamy, A.; Thakur, A.; Guan, X.J.; Krishnamoorthi, S.; Fung, T.Y.; Lu, K.; Gaurav, I.; Yang, Z.; Su, C.F.; Lau, K.F.; et al. Fe65-engineered neuronal exosomes encapsulating corynoxine-B ameliorate cognition and pathology of Alzheimer’s disease. Signal. Transduct. Target Ther. 2023, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthi, S.; Iyaswamy, A.; Sreenivasmurthy, S.G.; Thakur, A.; Vasudevan, K.; Kumar, G.; Guan, X.J.; Lu, K.; Gaurav, I.; Su, C.F.; et al. PPARa Ligand Caudatin Improves Cognitive Functions and Mitigates Alzheimer’s Disease Defects By Inducing Autophagy in Mice Models. J. Neuroimmune Pharmacol. 2023, 18, 509–528. [Google Scholar] [CrossRef] [PubMed]
- Iyaswamy, A.; Vasudevan, K.; Jayaraman, S.; Jaganathan, R.; Thakur, A.; Chang, R.C.; Yang, C. Editorial: Advances in Alzheimer’s disease diagnostics, brain delivery systems, and therapeutics. Front. Mol. Biosci. 2023, 10, 1162879. [Google Scholar] [CrossRef]
- Tong, B.C.; Huang, A.S.; Wu, A.J.; Iyaswamy, A.; Ho, O.K.; Kong, A.H.; Sreenivasmurthy, S.G.; Zhu, Z.; Su, C.; Liu, J.; et al. Tetrandrine ameliorates cognitive deficits and mitigates tau aggregation in cell and animal models of tauopathies. J. Biomed. Sci. 2022, 29, 85. [Google Scholar] [CrossRef]
- Gaurav, I.; Thakur, A.; Kumar, G.; Long, Q.; Zhang, K.; Sidu, R.K.; Thakur, S.; Sarkar, R.K.; Kumar, A.; Iyaswamy, A.; et al. Delivery of Apoplastic Extracellular Vesicles Encapsulating Green-Synthesized Silver Nanoparticles to Treat Citrus Canker. Nanomaterials 2023, 13, 1306. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.S.; de la Calle-Mustienes, E.; Gomez-Skarmeta, J.L.; Lister, R.; Bogdanovic, O. Depletion of Foxk transcription factors causes genome-wide transcriptional misregulation and developmental arrest in zebrafish embryos. MicroPubl. Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, A.K.; Davuluri, S.; Tiwary, K.; Narayanan, S.; Oguru, S.; Basavaraju, K.; Dayalan, D.; Thirumurugan, K.; Acharya, K.K. Systematic comparison of the protein-protein interaction databases from a user’s perspective. J. Biomed. Inform. 2020, 103, 103380. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Galon, J.; Bindea, G. Automated exploration of gene ontology term and pathway networks with ClueGO-REST. Bioinformatics 2019, 35, 3864–3866. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Tambalo, M.; Mitter, R.; Wilkinson, D.G. A single cell transcriptome atlas of the developing zebrafish hindbrain. Development 2020, 147, dev184143. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Liang, L.; Banerjee, S.; Zhang, K. Single-Cell Transcriptomics Reveals Evidence of Endothelial Dysfunction in the Brains of COVID-19 Patients with Implications for Glioblastoma Progression. Brain Sci. 2023, 13, 762. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Christianson, D.W. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat. Chem. Biol. 2016, 12, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, K.; Pilarz, A.; Rogut, A.; Maj, P.; Szymanska, J.; Olejnik, L.; Szymanski, P. Aspartame-True or False? Narrative Review of Safety Analysis of General Use in Products. Nutrients 2021, 13, 1957. [Google Scholar] [CrossRef] [PubMed]
- Baronio, D.; Chen, Y.C.; Decker, A.R.; Enckell, L.; Fernandez-Lopez, B.; Semenova, S.; Puttonen, H.A.J.; Cornell, R.A.; Panula, P. Vesicular monoamine transporter 2 (SLC18A2) regulates monoamine turnover and brain development in zebrafish. Acta Physiol. 2022, 234, e13725. [Google Scholar] [CrossRef] [PubMed]
- Rollwitz, E.; Jastroch, M. Plate-Based Respirometry to Assess Thermal Sensitivity of Zebrafish Embryo Bioenergetics in situ. Front. Physiol. 2021, 12, 746367. [Google Scholar] [CrossRef]
- Carneiro, M.; Gutierrez-Praena, D.; Osorio, H.; Vasconcelos, V.; Carvalho, A.P.; Campos, A. Proteomic analysis of anatoxin-a acute toxicity in zebrafish reveals gender specific responses and additional mechanisms of cell stress. Ecotoxicol. Environ. Saf. 2015, 120, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gao, F.; Wang, K.; Liu, X.; Zhang, Z. MiR-34a inhibitor protects mesenchymal stem cells from hyperglycaemic injury through the activation of the SIRT1/FoxO3a autophagy pathway. Stem Cell Res. Ther. 2021, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Santo, E.E.; Paik, J. FOXO in Neural Cells and Diseases of the Nervous System. Curr. Top Dev. Biol. 2018, 127, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Daitoku, H.; Hatta, M.; Matsuzaki, H.; Aratani, S.; Ohshima, T.; Miyagishi, M.; Nakajima, T.; Fukamizu, A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10042–10047. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.; Chen, Y.R.; Kojima, N.; Ogata, K.; Fukamizu, A.; Miyajima, A. Foxo1 links insulin signaling to C/EBPalpha and regulates gluconeogenesis during liver development. EMBO J. 2007, 26, 3607–3615. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef]
- Parker, J.A.; Vazquez-Manrique, R.P.; Tourette, C.; Farina, F.; Offner, N.; Mukhopadhyay, A.; Orfila, A.M.; Darbois, A.; Menet, S.; Tissenbaum, H.A.; et al. Integration of beta-catenin, sirtuin, and FOXO signaling protects from mutant huntingtin toxicity. J. Neurosci. 2012, 32, 12630–12640. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Zheng, H. Role of FoxO transcription factors in aging and age-related metabolic and neurodegenerative diseases. Cell Biosci. 2021, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Herskovits, A.Z.; Guarente, L. SIRT1 in neurodevelopment and brain senescence. Neuron 2014, 81, 471–483. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, H.; Wang, Q.; Chen, S.; Wang, R.; Wang, Z.; Yang, C.; Chen, A.; Zhao, J.; Zhou, Z.; et al. Sirt1 overexpression improves senescence-associated pulmonary fibrosis induced by vitamin D deficiency through downregulating IL-11 transcription. Aging Cell 2022, 21, e13680. [Google Scholar] [CrossRef] [PubMed]
- Langley, B.; Sauve, A. Sirtuin deacetylases as therapeutic targets in the nervous system. Neurotherapeutics 2013, 10, 605–620. [Google Scholar] [CrossRef]
- Jiao, F.; Gong, Z. The Beneficial Roles of SIRT1 in Neuroinflammation-Related Diseases. Oxid. Med. Cell Longev. 2020, 2020, 6782872. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, I.; Minakaki, G.; Khan, M.A.; Balta, E.A.; Schlotzer-Schrehardt, U.; Schwarz, T.J.; Beckervordersandforth, R.; Winner, B.; Webb, A.E.; DePinho, R.A.; et al. FoxO Function Is Essential for Maintenance of Autophagic Flux and Neuronal Morphogenesis in Adult Neurogenesis. Neuron 2018, 99, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.J.; Deng, Z.Q.; Liu, J.; Su, C.F.; Tong, B.C.; Zhu, Z.; Sreenivasmurthy, S.G.; Kan, Y.X.; Lu, K.J.; Chu, C.P.; et al. Corynoxine promotes TFEB/TFE3-mediated autophagy and alleviates Abeta pathology in Alzheimer’s disease models. Acta Pharmacol. Sin. 2024. [Google Scholar] [CrossRef] [PubMed]
- Saputra, F.; Lai, Y.H.; Fernandez, R.A.T.; Macabeo, A.P.G.; Lai, H.T.; Huang, J.C.; Hsiao, C.D. Acute and Sub-Chronic Exposure to Artificial Sweeteners at the Highest Environmentally Relevant Concentration Induce Less Cardiovascular Physiology Alterations in Zebrafish Larvae. Biology 2021, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4, 1070. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasmurthy, S.G.; Iyaswamy, A.; Krishnamoorthi, S.; Reddi, R.N.; Kammala, A.K.; Vasudevan, K.; Senapati, S.; Zhu, Z.; Su, C.F.; Liu, J.; et al. Bromo-protopine, a novel protopine derivative, alleviates tau pathology by activating chaperone-mediated autophagy for Alzheimer’s disease therapy. Front. Mol. Biosci. 2022, 9, 1030534. [Google Scholar] [CrossRef] [PubMed]



| Ligand | Binding Energy (Kcal/mol) | Binding Residues |
|---|---|---|
| SIRT1 vs. Aspartame | −7.4 | ARG-274, GLN-345, ASN-346, ILE-347, SER-442, LEU-443 |
| SIRT1 vs. Selisistat | −7.0 | GLU-214, ASP-298 |
| FOXO3a vs. Aspartame | −5.4 | SER-200, ASN-201 |
| FOXO3a vs. Carbenoxolone | −7.2 | ARG-222, LYS-230 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandaram, A.; Paul, J.; Wankhar, W.; Thakur, A.; Verma, S.; Vasudevan, K.; Wankhar, D.; Kammala, A.K.; Sharma, P.; Jaganathan, R.; et al. Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells. Biomedicines 2024, 12, 855. https://doi.org/10.3390/biomedicines12040855
Pandaram A, Paul J, Wankhar W, Thakur A, Verma S, Vasudevan K, Wankhar D, Kammala AK, Sharma P, Jaganathan R, et al. Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells. Biomedicines. 2024; 12(4):855. https://doi.org/10.3390/biomedicines12040855
Chicago/Turabian StylePandaram, Athiram, Jeyakumari Paul, Wankupar Wankhar, Abhimanyu Thakur, Sakshi Verma, Karthick Vasudevan, Dapkupar Wankhar, Ananth Kumar Kammala, Priyanshu Sharma, Ravindran Jaganathan, and et al. 2024. "Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells" Biomedicines 12, no. 4: 855. https://doi.org/10.3390/biomedicines12040855
APA StylePandaram, A., Paul, J., Wankhar, W., Thakur, A., Verma, S., Vasudevan, K., Wankhar, D., Kammala, A. K., Sharma, P., Jaganathan, R., Iyaswamy, A., & Rajan, R. (2024). Aspartame Causes Developmental Defects and Teratogenicity in Zebra Fish Embryo: Role of Impaired SIRT1/FOXO3a Axis in Neuron Cells. Biomedicines, 12(4), 855. https://doi.org/10.3390/biomedicines12040855





