Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice Samples and Tissue Preparation
2.2. TdT-Mediated dUTP-X Nicked-End Labeling (TUNEL) Assay
2.3. Quantitative Real-Time PCR
2.4. In Situ Hybridization
2.5. Western Blot Analysis
2.6. Measurement of Total Nitric Oxide (NO)/Nitrite/Nitrate
2.7. Enzyme Immunoassay (EIA)
2.8. GEO Data Analysis
2.9. Bioinformatics
2.10. Statistical Analysis
3. Results
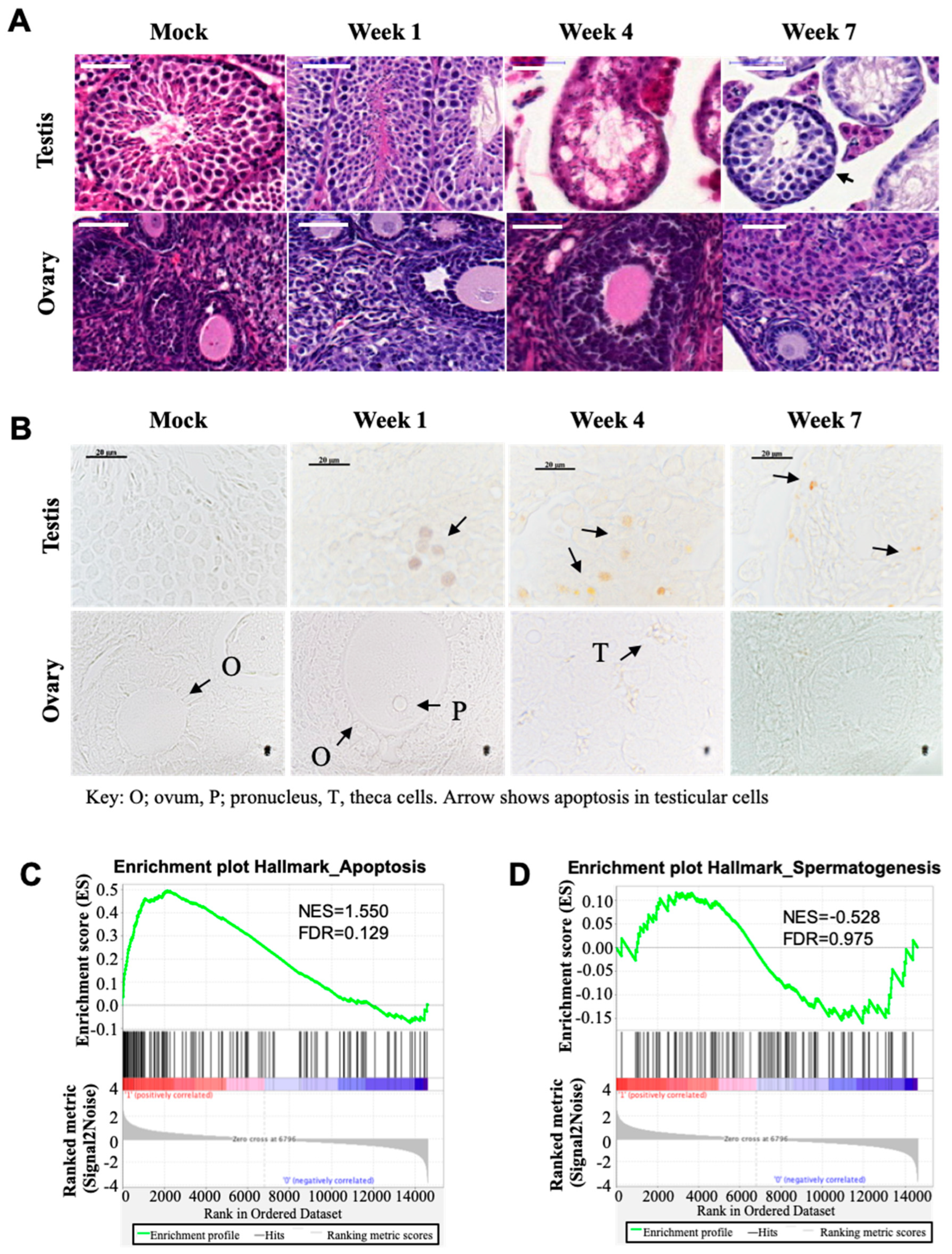
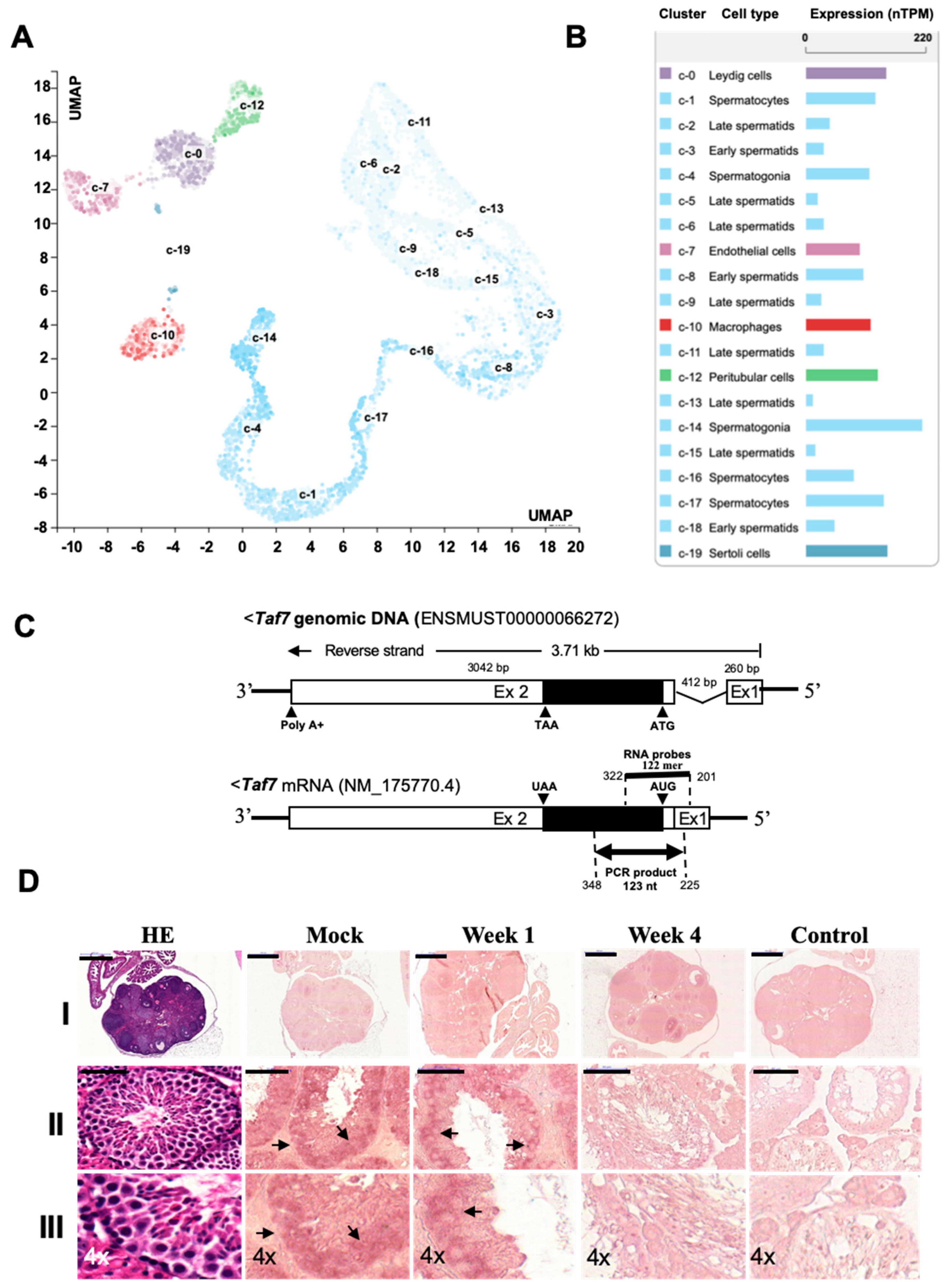
3.1. Busulfan Induces Apoptosis Signals and Reduces Spermatogenesis in Testicular Cells
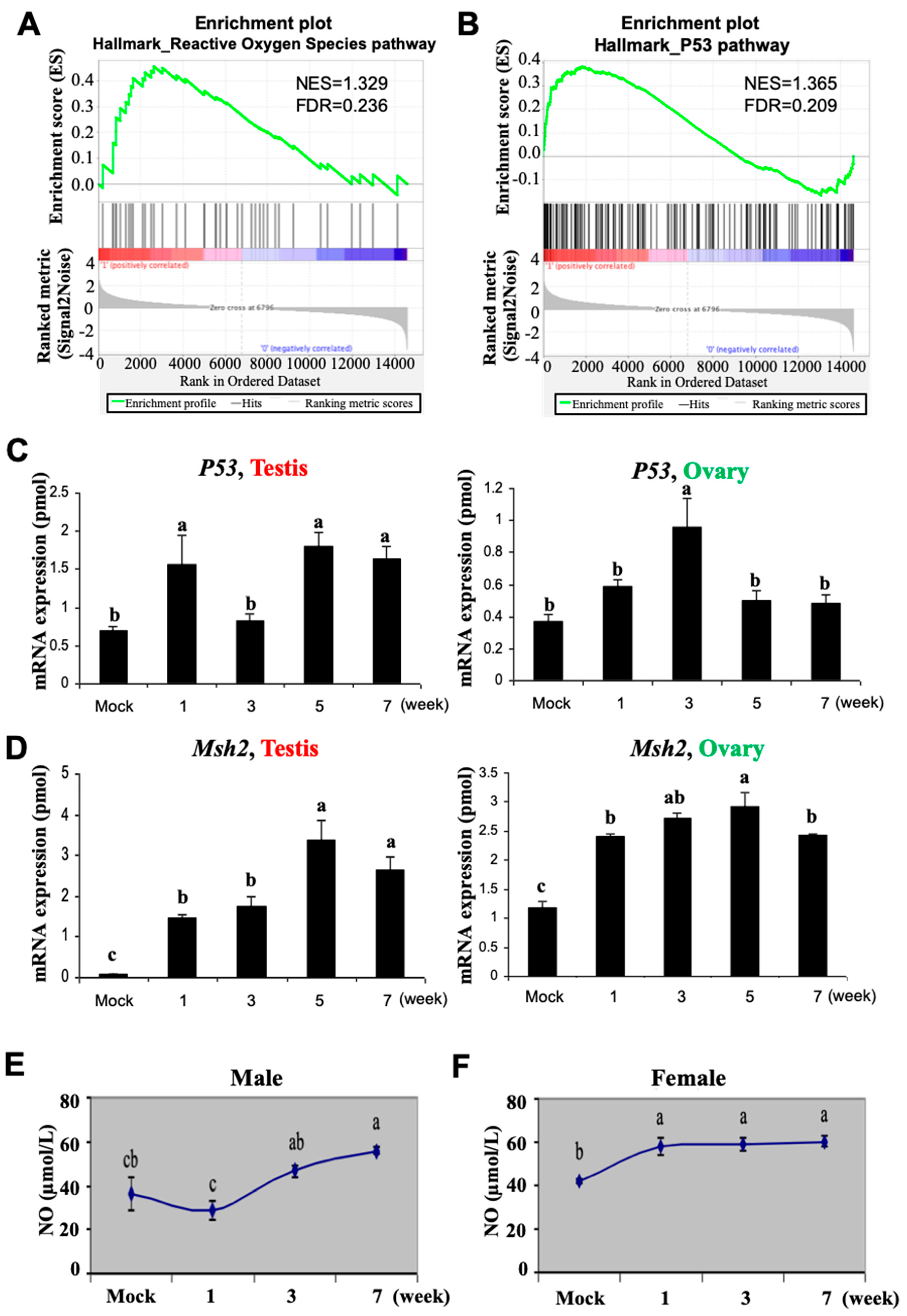
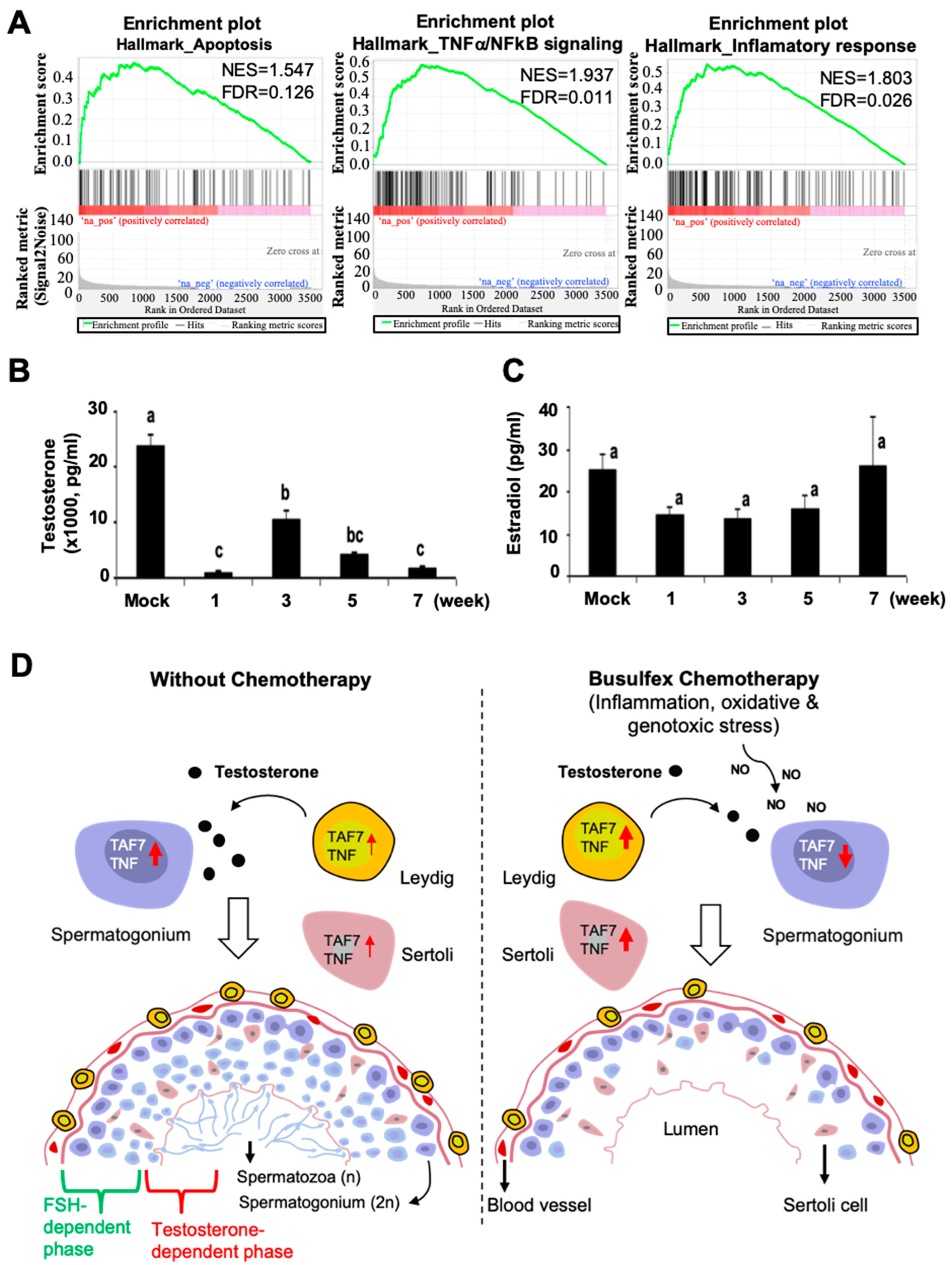
3.2. Busulfan Activates Oxidative Stress, DNA Repair, and Inflammatory Responses in Testicular Cells
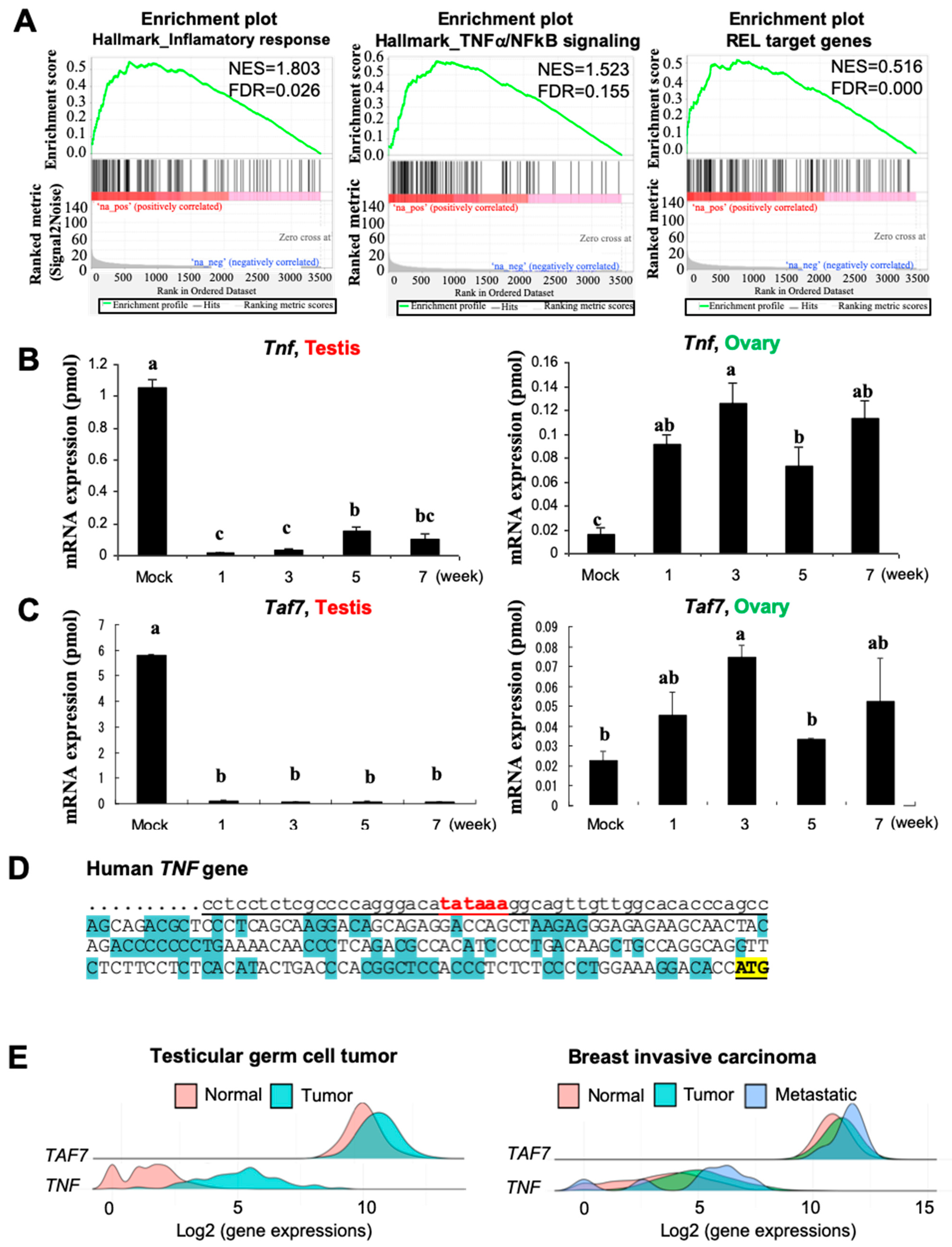
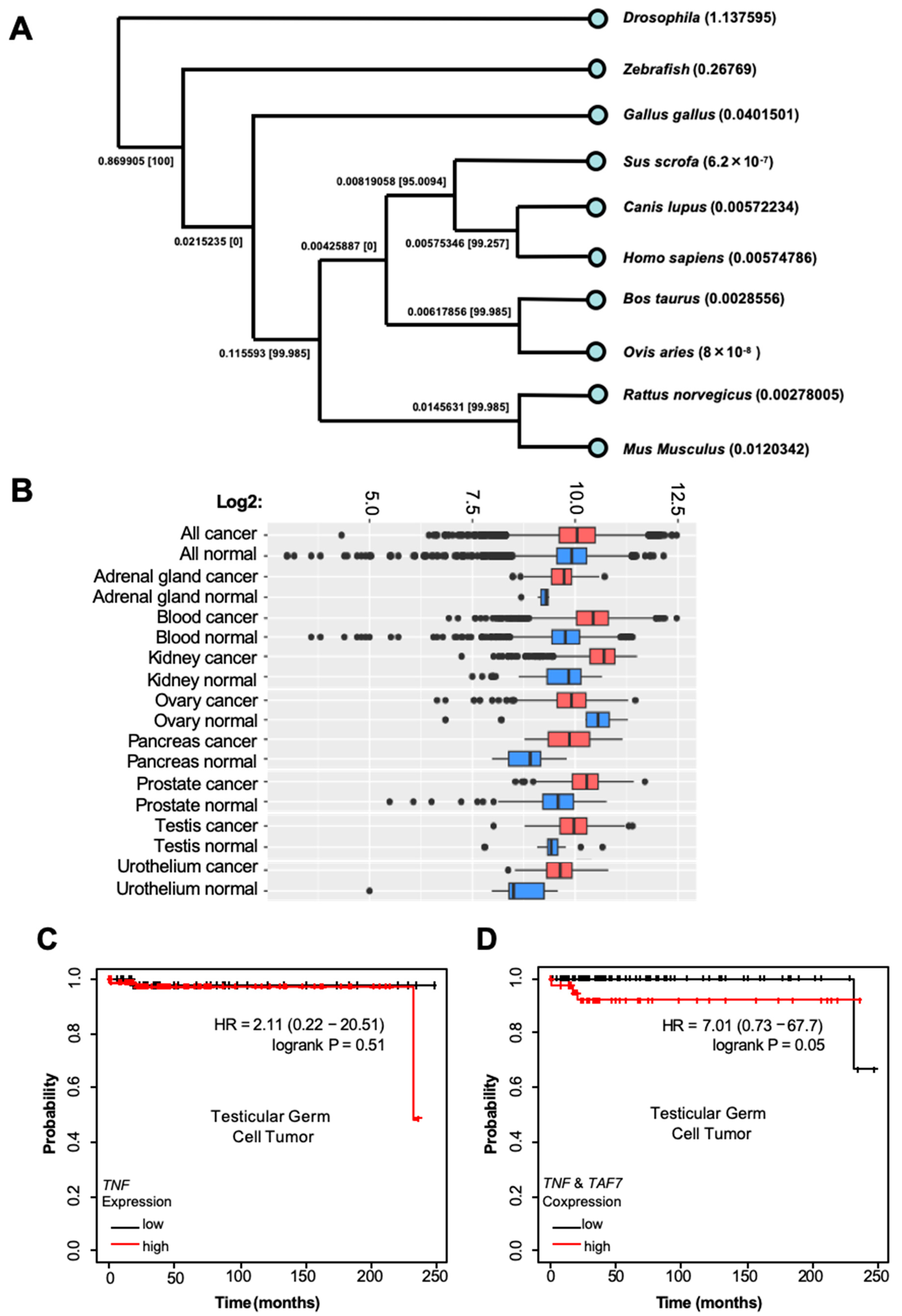
3.3. Busulfan Downregulates the Expression of TAF7 and TNF in Testicular Cells
3.4. TAF7 Is Localized in Spermatogonia Stem Cells and Expressed at Lower Levels in Female Oocytes
3.5. Busulfan Upregulates the Inflammatory Response for Downregulation of Testosterone in Sertoli Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiffman, K.S.; Bensinger, W.I.; Appelbaum, F.R.; Rowley, S.; Lilleby, K.; Clift, R.A.; Weaver, C.H.; Demirer, T.; Sanders, J.E.; Petersdorf, S.; et al. Phase II study of high-dose busulfan, melphalan and thiotepa with autologous peripheral blood stem cell support in patients with malignant disease. Bone Marrow Transplant. 1996, 17, 943–950. [Google Scholar] [PubMed]
- Santos, F.P.; Kantarjian, H.; Quintas-Cardama, A.; Cortes, J. Evolution of therapies for chronic myelogenous leukemia. Cancer J. 2011, 17, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Yuan, G.; Li, Q.; Cui, Z.; Li, W. Long-term outcomes of frontline imatinib therapy for chronic myeloid leukemia in China. Front. Oncol. 2023, 13, 1172910. [Google Scholar] [CrossRef] [PubMed]
- Lesurtel, M.; Graf, R.; Aleil, B.; Walther, D.J.; Tian, Y.; Jochum, W.; Gachet, C.; Bader, M.; Clavien, P.A. Platelet-derived serotonin mediates liver regeneration. Science 2006, 312, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Brisse, H.; Orbach, D.; Lassau, N.; Servois, V.; Doz, F.; Debray, D.; Helfre, S.; Hartmann, O.; Neuenschwander, S. Portal vein thrombosis during antineoplastic chemotherapy in children: Report of five cases and review of the literature. Eur. J. Cancer 2004, 40, 2659–2666. [Google Scholar] [CrossRef]
- Grigg, A.; Gibson, R.; Bardy, P.; Szer, J. Acute portal vein thrombosis after autologous stem cell transplantation. Bone Marrow Transplant. 1996, 18, 949–953. [Google Scholar]
- Jain, R.; Gupta, K.; Bhatia, A.; Bansal, A.; Bansal, D. Hepatic Sinusoidal-obstruction Syndrome and Busulfan-induced Lung Injury in a Post-autologous Stem Cell Transplant Recipient. Indian. Pediatr. 2017, 54, 765–770. [Google Scholar] [CrossRef]
- Poorvu, P.D.; Frazier, A.L.; Feraco, A.M.; Manley, P.E.; Ginsburg, E.S.; Laufer, M.R.; LaCasce, A.S.; Diller, L.R.; Partridge, A.H. Cancer Treatment-Related Infertility: A Critical Review of the Evidence. JNCI Cancer Spectr. 2019, 3, pkz008. [Google Scholar] [CrossRef]
- Gosden, R.; Spears, N. Programmed cell death in the reproductive system. Br. Med. Bull. 1997, 53, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Wambi, J.S.; Jordan, V.C. Estrogen regulation of apoptosis: How can one hormone stimulate and inhibit? Breast Cancer Res. 2009, 11, 206. [Google Scholar] [CrossRef]
- Li, D.; Chen, J.; Ai, Y.; Gu, X.; Li, L.; Che, D.; Jiang, Z.; Li, L.; Chen, S.; Huang, H.; et al. Estrogen-Related Hormones Induce Apoptosis by Stabilizing Schlafen-12 Protein Turnover. Mol. Cell 2019, 75, 1103–1116.e9. [Google Scholar] [CrossRef]
- Zohni, K.; Zhang, X.; Tan, S.L.; Chan, P.; Nagano, M.C. The efficiency of male fertility restoration is dependent on the recovery kinetics of spermatogonial stem cells after cytotoxic treatment with busulfan in mice. Hum. Reprod. 2012, 27, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Kerbauy, M.N.; Mariano, L.; Seber, A.; Rocha, V. The impact of low dose busulfan on gonodal function after allogeneic hematopoietic stem cell transplantation for aplastic anemia. Bone Marrow Transplant. 2020, 55, 1169–1171. [Google Scholar] [CrossRef] [PubMed]
- Mobarak, H.; Rahbarghazi, R.; Nouri, M.; Heidarpour, M.; Mahdipour, M. Intratesticular versus intraperitoneal injection of Busulfan for the induction of azoospermia in a rat model. BMC Pharmacol. Toxicol. 2022, 23, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol. Sin. 2008, 29, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Sethi, J.K.; Hotamisligil, G.S. Metabolic Messengers: Tumour necrosis factor. Nat. Metab. 2021, 3, 1302–1312. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Capece, D.; Verzella, D.; Flati, I.; Arboretto, P.; Cornice, J.; Franzoso, G. NF-kappaB: Blending metabolism, immunity, and inflammation. Trends Immunol. 2022, 43, 757–775. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-kappaB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Parameswaran, N.; Patial, S. Tumor necrosis factor-alpha signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87–103. [Google Scholar] [CrossRef]
- Siegmund, D.; Wajant, H. TNF and TNF receptors as therapeutic targets for rheumatic diseases and beyond. Nat. Rev. Rheumatol. 2023, 19, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Chun, N.; Ang, R.L.; Chan, M.; Fairchild, R.L.; Baldwin, W.M., 3rd; Horwitz, J.K.; Gelles, J.D.; Chipuk, J.E.; Kelliher, M.A.; Pavlov, V.I.; et al. T cell-derived tumor necrosis factor induces cytotoxicity by activating RIPK1-dependent target cell death. JCI Insight 2021, 6, e148643. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xie, C.; Chen, Y.; Wang, J.; Chen, X.; Lu, Z.; June, R.R.; Zheng, S.G. Differential roles of TNFalpha-TNFR1 and TNFalpha-TNFR2 in the differentiation and function of CD4(+)Foxp3(+) induced Treg cells in vitro and in vivo periphery in autoimmune diseases. Cell Death Dis. 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- van Loo, G.; Bertrand, M.J.M. Death by TNF: A road to inflammation. Nat. Rev. Immunol. 2023, 23, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; McKay, B.S.; Allen, J.B.; Jaffe, G.J. Effect of NF-kappa B inhibition on TNF-alpha-induced apoptosis in human RPE cells. Invest. Ophthalmol. Vis. Sci. 2004, 45, 2438–2446. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, S.; Mandrekar, P. Cellular stress response and innate immune signaling: Integrating pathways in host defense and inflammation. J. Leukoc. Biol. 2013, 94, 1167–1184. [Google Scholar] [CrossRef]
- Jeong, Y.; Lim, J.W.; Kim, H. Lycopene Inhibits Reactive Oxygen Species-Mediated NF-kappaB Signaling and Induces Apoptosis in Pancreatic Cancer Cells. Nutrients 2019, 11, 762. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, P.; Guo, S.; Schrodi, S.J.; He, D. Apoptosis, Autophagy, NETosis, Necroptosis, and Pyroptosis Mediated Programmed Cell Death as Targets for Innovative Therapy in Rheumatoid Arthritis. Front. Immunol. 2021, 12, 809806. [Google Scholar] [CrossRef]
- Masli, S.; Turpie, B. Anti-inflammatory effects of tumour necrosis factor (TNF)-alpha are mediated via TNF-R2 (p75) in tolerogenic transforming growth factor-beta-treated antigen-presenting cells. Immunology 2009, 127, 62–72. [Google Scholar] [CrossRef]
- Spender, L.C.; O’Brien, D.I.; Simpson, D.; Dutt, D.; Gregory, C.D.; Allday, M.J.; Clark, L.J.; Inman, G.J. TGF-beta induces apoptosis in human B cells by transcriptional regulation of BIK and BCL-XL. Cell Death Differ. 2009, 16, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, K.C.; Ehata, S.; Komuro, A.; Takeuchi, K.; Miyazono, K. TGF-beta-induced apoptosis of B-cell lymphoma Ramos cells through reduction of MS4A1/CD20. Oncogene 2013, 32, 2096–2106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Zhang, Y.M.; Zhang, L.Y.; Zhou, T.; Li, Y.Y.; Zhou, G.C.; Miao, Z.M.; Shang, M.; He, J.P.; Ding, N.; et al. Duality of Interactions Between TGF-beta and TNF-alpha During Tumor Formation. Front. Immunol. 2021, 12, 810286. [Google Scholar] [CrossRef]
- Chiang, C.M.; Roeder, R.G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 1995, 267, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qi, Y.; Wu, Z.; Wang, X.; Li, J.; Zhao, D.; Hou, H.; Li, Y.; Yu, Z.; Liu, W.; et al. Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372, eaba8490. [Google Scholar] [CrossRef]
- Munz, C.; Psichari, E.; Mandilis, D.; Lavigne, A.C.; Spiliotaki, M.; Oehler, T.; Davidson, I.; Tora, L.; Angel, P.; Pintzas, A. TAF7 (TAFII55) plays a role in the transcription activation by c-Jun. J. Biol. Chem. 2003, 278, 21510–21516. [Google Scholar] [CrossRef]
- Pal, S.; Paladhi, P.; Dutta, S.; Bose, G.; Ghosh, P.; Chattopadhyay, R.; Chakravarty, B.; Saha, I.; Ghosh, S. Novel variations in spermatogenic transcription regulators RFX2 and TAF7 increase risk of azoospermia. J. Assist. Reprod. Genet. 2021, 38, 3195–3212. [Google Scholar] [CrossRef]
- Nakagawa, T.; Hosogane, M.; Nakagawa, M.; Morohoshi, A.; Funayama, R.; Nakayama, K. Transforming Growth Factor beta-Induced Proliferative Arrest Mediated by TRIM26-Dependent TAF7 Degradation and Its Antagonism by MYC. Mol. Cell Biol. 2018, 38, e00449-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Duan, B.; Li, F.; Jing, X.; Li, R.; Cai, S.; Cao, L.; Jiang, Q.; Zhou, J.; Zhou, J.; et al. SETD7 Promotes Cell Proliferation and Migration via Methylation-mediated TAF7 in Clear Cell Renal Cell Carcinoma. Int. J. Biol. Sci. 2024, 20, 3008–3027. [Google Scholar] [CrossRef] [PubMed]
- Rezaeian, A.H.; Isokane, T.; Nishibori, M.; Chiba, M.; Hiraiwa, N.; Yoshizawa, M.; Yasue, H. alphaCGRP and betaCGRP transcript amount in mouse tissues of various developmental stages and their tissue expression sites. Brain Dev. 2009, 31, 682–693. [Google Scholar] [CrossRef]
- Rezaeian, A.H.; Katafuchi, T.; Yoshizawa, M.; Hiraiwa, N.; Saito, T.; Nishibori, M.; Hamano, K.; Minamino, N.; Yasue, H. Genomic organization, expression and evolution of porcine CRSP1, 2, and 3. Cytogenet. Genome Res. 2008, 121, 41–49. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Shen, Y.; White, E. p53-dependent apoptosis pathways. Adv. Cancer Res. 2001, 82, 55–84. [Google Scholar] [CrossRef]
- Simon, H.U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef]
- Liu, Y.; Su, Z.; Tavana, O.; Gu, W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell 2024, 42, 946–967. [Google Scholar] [CrossRef]
- Sies, H.; Belousov, V.V.; Chandel, N.S.; Davies, M.J.; Jones, D.P.; Mann, G.E.; Murphy, M.P.; Yamamoto, M.; Winterbourn, C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022, 23, 499–515. [Google Scholar] [CrossRef]
- Rodriguez-Pastrana, I.; Birli, E.; Coutts, A.S. p53-dependent DNA repair during the DNA damage response requires actin nucleation by JMY. Cell Death Differ. 2023, 30, 1636–1647. [Google Scholar] [CrossRef] [PubMed]
- Barnabei, L.; Laplantine, E.; Mbongo, W.; Rieux-Laucat, F.; Weil, R. NF-kappaB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021, 12, 716469. [Google Scholar] [CrossRef]
- Yang, D.; Dai, F.; Yuan, M.; Zheng, Y.; Liu, S.; Deng, Z.; Tan, W.; Chen, L.; Zhang, Q.; Zhao, X.; et al. Role of Transforming Growth Factor-beta1 in Regulating Fetal-Maternal Immune Tolerance in Normal and Pathological Pregnancy. Front. Immunol. 2021, 12, 689181. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.Z.; Loizidou, M.; Ahmed, M.; Charles, I.G. The role of nitric oxide in cancer. Cell Res. 2002, 12, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.O.; Weitzberg, E. Nitric oxide signaling in health and disease. Cell 2022, 185, 2853–2878. [Google Scholar] [CrossRef] [PubMed]
- Pointud, J.C.; Mengus, G.; Brancorsini, S.; Monaco, L.; Parvinen, M.; Sassone-Corsi, P.; Davidson, I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 2003, 116, 1847–1858. [Google Scholar] [CrossRef]
- Digre, A.; Lindskog, C. The human protein atlas-Integrated omics for single cell mapping of the human proteome. Protein Sci. 2023, 32, e4562. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.; Smith, L.B.; Rebourcet, D. Sertoli cells as key drivers of testis function. Semin. Cell Dev. Biol. 2022, 121, 2–9. [Google Scholar] [CrossRef]
- Choi, Y.J.; Ok, D.W.; Kwon, D.N.; Chung, J.I.; Kim, H.C.; Yeo, S.M.; Kim, T.; Seo, H.G.; Kim, J.H. Murine male germ cell apoptosis induced by busulfan treatment correlates with loss of c-kit-expression in a Fas/FasL- and p53-independent manner. FEBS Lett. 2004, 575, 41–51. [Google Scholar] [CrossRef]
- Anand, S.; Bhartiya, D.; Sriraman, K.; Mallick, A. Underlying Mechanisms that Restore Spermatogenesis on Transplanting Healthy Niche Cells in Busulphan Treated Mouse Testis. Stem Cell Rev. Rep. 2016, 12, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Toyokuni, S.; Morimoto, T.; Matsui, S.; Honjo, T.; Shinohara, T. Functional assessment of self-renewal activity of male germline stem cells following cytotoxic damage and serial transplantation. Biol. Reprod. 2003, 68, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- La, H.M.; Liao, J.; Legrand, J.M.D.; Rossello, F.J.; Chan, A.L.; Vaghjiani, V.; Cain, J.E.; Papa, A.; Lee, T.L.; Hobbs, R.M. Distinctive molecular features of regenerative stem cells in the damaged male germline. Nat. Commun. 2022, 13, 2500. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.R. Apoptosis in the ovary: Molecular mechanisms. Hum. Reprod. Update 2005, 11, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Prasad, S.; Tripathi, A.; Pandey, A.N.; Ali, I.; Singh, A.K.; Shrivastav, T.G.; Chaube, S.K. Apoptosis in mammalian oocytes: A review. Apoptosis 2015, 20, 1019–1025. [Google Scholar] [CrossRef]
- Iwamoto, T.; Hiraku, Y.; Oikawa, S.; Mizutani, H.; Kojima, M.; Kawanishi, S. DNA intrastrand cross-link at the 5′-GA-3′ sequence formed by busulfan and its role in the cytotoxic effect. Cancer Sci. 2004, 95, 454–458. [Google Scholar] [CrossRef]
- Guichard, N.; Bonnabry, P.; Rudaz, S.; Fleury-Souverain, S. Stability of busulfan solutions in polypropylene syringes and infusion bags as determined with an original assay. Am. J. Health Syst. Pharm. 2017, 74, 1887–1894. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L. DNA crosslinking damage and cancer—A tale of friend and foe. Transl. Cancer Res. 2013, 2, 144–154. [Google Scholar] [CrossRef]
- Amunugama, R.; Walter, J.C. A new varietal of DNA interstrand crosslink repair. Cell Res. 2020, 30, 459–460. [Google Scholar] [CrossRef]
- Hanawalt, P.C. Subpathways of nucleotide excision repair and their regulation. Oncogene 2002, 21, 8949–8956. [Google Scholar] [CrossRef]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Speer, R.M.; Ulibarri, J.; Liu, K.J.; Mao, P. Transcription-coupled nucleotide excision repair: New insights revealed by genomic approaches. DNA Repair 2021, 103, 103126. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Li, Q.; Yu, J.J.; Mu, C.; Yunmbam, M.K.; Slavsky, D.; Cross, C.L.; Bostick-Bruton, F.; Reed, E. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer. Res. 2000, 20, 645–652. [Google Scholar]
- Gegonne, A.; Weissman, J.D.; Zhou, M.; Brady, J.N.; Singer, D.S. TAF7: A possible transcription initiation check-point regulator. Proc. Natl. Acad. Sci. USA 2006, 103, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Regulation of NF-kappaB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, J.; Imrich, A.; Cushion, M.; Kinane, T.B.; Koziel, H. Pneumocystis activates human alveolar macrophage NF-kappaB signaling through mannose receptors. Infect. Immun. 2004, 72, 3147–3160. [Google Scholar] [CrossRef]
- Beattie, E.C.; Stellwagen, D.; Morishita, W.; Bresnahan, J.C.; Ha, B.K.; Von Zastrow, M.; Beattie, M.S.; Malenka, R.C. Control of synaptic strength by glial TNFalpha. Science 2002, 295, 2282–2285. [Google Scholar] [CrossRef]
- Mauduit, C.; Siah, A.; Foch, M.; Chapet, O.; Clippe, S.; Gerard, J.P.; Benahmed, M. Differential expression of growth factors in irradiated mouse testes. Int. J. Radiat. Oncol. Biol. Phys. 2001, 50, 203–212. [Google Scholar] [CrossRef]
- Hong, C.Y.; Park, J.H.; Ahn, R.S.; Im, S.Y.; Choi, H.S.; Soh, J.; Mellon, S.H.; Lee, K. Molecular mechanism of suppression of testicular steroidogenesis by proinflammatory cytokine tumor necrosis factor alpha. Mol. Cell Biol. 2004, 24, 2593–2604. [Google Scholar] [CrossRef]
- Jin, H.; Qiu, W.B.; Mei, Y.F.; Wang, D.M.; Li, Y.G.; Tan, X.R. Testosterone alleviates tumor necrosis factor-alpha-mediated tissue factor pathway inhibitor downregulation via suppression of nuclear factor-kappa B in endothelial cells. Asian J. Androl. 2009, 11, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Mancuso, F.; Frosini, R.; Arato, I.; Calvitti, M.; Calafiore, R.; Talesa, V.N.; Luca, G. Testosterone and Follicle Stimulating Hormone-Dependent Glyoxalase 1 Up-Regulation Sustains the Viability of Porcine Sertoli Cells through the Control of Hydroimidazolone- and Argpyrimidine-Mediated NF-kappaB Pathway. Am. J. Pathol. 2018, 188, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, D.; Tu, Z.; Karnoub, A.E.; Wei, W.; Rezaeian, A.-H. Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction. Biomedicines 2024, 12, 2220. https://doi.org/10.3390/biomedicines12102220
Huang D, Tu Z, Karnoub AE, Wei W, Rezaeian A-H. Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction. Biomedicines. 2024; 12(10):2220. https://doi.org/10.3390/biomedicines12102220
Chicago/Turabian StyleHuang, Daoyuan, Zhenbo Tu, Antoine E. Karnoub, Wenyi Wei, and Abdol-Hossein Rezaeian. 2024. "Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction" Biomedicines 12, no. 10: 2220. https://doi.org/10.3390/biomedicines12102220





