The Importance of Genetic Screening on the Syndromes of Colorectal Cancer and Gastric Cancer: A 2024 Update
Abstract
1. Introduction
2. Genetic Syndromes
3. Other GI Diseases with Genetic Implications
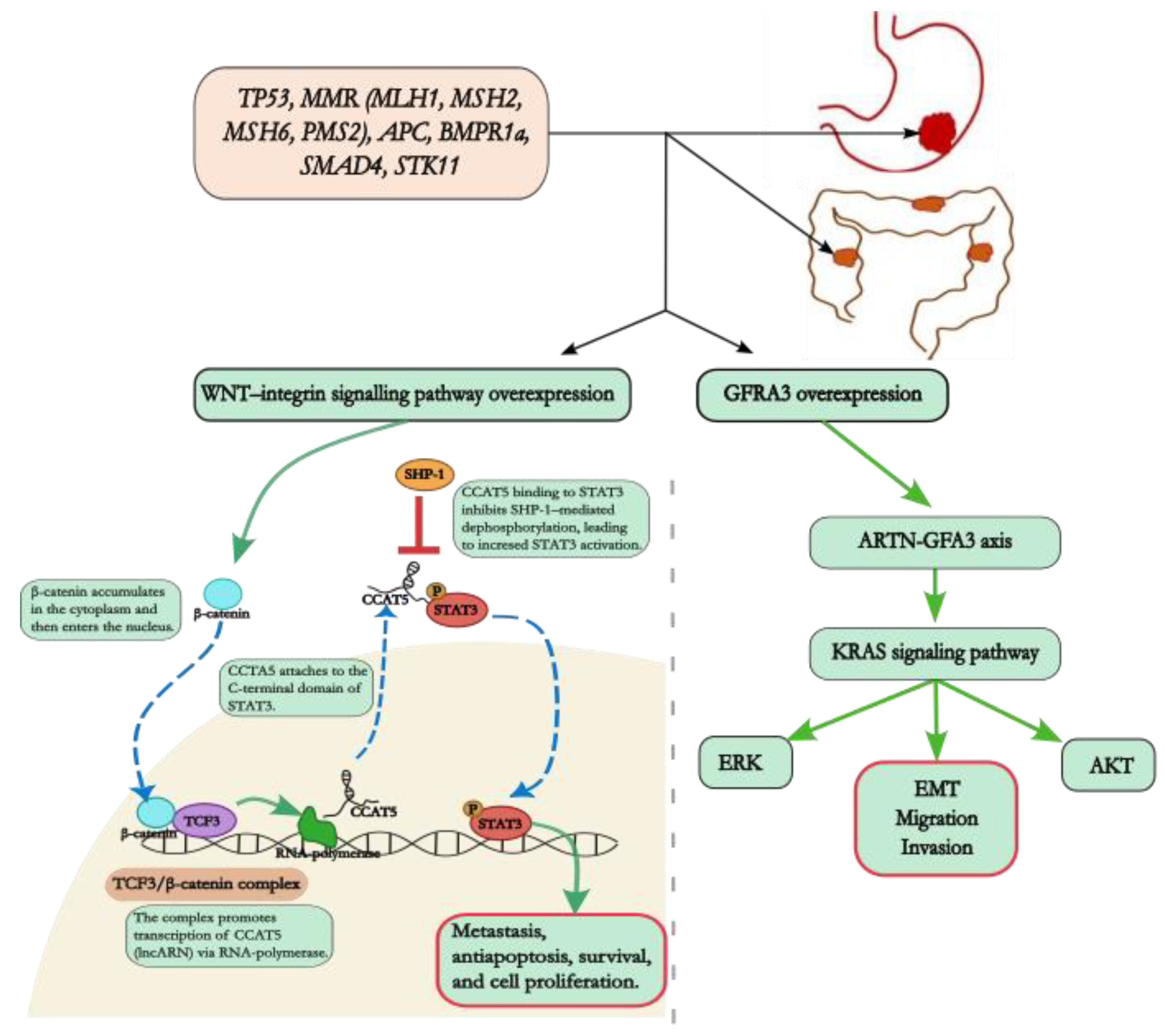
4. Genetic Mutations and Pathways in Gastric and Colorectal Adenocarcinomas: Implications for Carcinogenesis and Therapeutic Strategies
4.1. KRAS Gene
4.2. Wnt Signalling Pathway
4.3. Nucleic Acid Polymorphisms
4.4. Chemokine Ligands
4.5. Other Genes
5. Natural Products as Possible Treatment Options
6. Possible Biomarkers
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Back, T.R.; van Hooff, S.R.; Sommeijer, D.W.; Vermeulen, L. Transcriptomic subtyping of gastrointestinal malignancies. Trends Cancer 2024, 10, P842–P856. [Google Scholar] [CrossRef] [PubMed]
- Burz, C.; Pop, V.; Silaghi, C.; Lupan, I.; Samasca, G. Prognosis and Treatment of Gastric Cancer: A 2024 Update. Cancers 2024, 16, 1708. [Google Scholar] [CrossRef] [PubMed]
- Baidoun, F.; Elshiwy, K.; Elkeraie, Y.; Merjaneh, Z.; Khoudari, G.; Sarmini, M.T.; Gad, M.; Al-Husseini, M.; Saad, A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr. Drug Targets 2021, 22, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Hakami, Z.H. Biomarker discovery and validation for gastrointestinal tumors: A comprehensive review of colorectal, gastric, and liver cancers. Pathol. Res. Pract. 2024, 255, 155216. [Google Scholar] [CrossRef]
- Thrift, A.P.; Wenker, T.N.; El-Serag, H.B. Global burden of gastric cancer: Epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 2023, 20, 338–349. [Google Scholar] [CrossRef]
- Triantafillidis, J.K.; Georgiou, K.; Konstadoulakis, M.M.; Papalois, A.E. Early-onset gastrointestinal cancer: An epidemiological reality with great significance and implications. World J. Gastrointest. Oncol. 2024, 16, 583–597. [Google Scholar] [CrossRef]
- Rebuzzi, F.; Ulivi, P.; Tedaldi, G. Genetic Predisposition to Colorectal Cancer: How Many and Which Genes to Test? Int. J. Mol. Sci. 2023, 24, 2137. [Google Scholar] [CrossRef]
- Zamani, M.; Alizadeh-Tabari, S. Anxiety and depression prevalence in digestive cancers: A systematic review and meta-analysis. BMJ Support. Palliat. Care 2023, 13, e235–e243. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.; Schmid, S.; Heinimann, K.; Frick, H.; Herrmann, C.; Cerny, T.; Omlin, A. Multiple primary tumours: Challenges and approaches, a review. ESMO Open 2017, 2, e000172. [Google Scholar] [CrossRef]
- Halamkova, J.; Kazda, T.; Pehalova, L.; Gonec, R.; Kozakova, S.; Bohovicova, L.; Krakorova, D.A.; Slaby, O.; Demlova, R.; Svoboda, M.; et al. Second primary malignancies in colorectal cancer patients. Sci. Rep. 2021, 11, 2759. [Google Scholar] [CrossRef]
- Yang, X.B.; Zhang, L.H.; Xue, J.N.; Wang, Y.C.; Yang, X.; Zhang, N.; Liu, D.; Wang, Y.Y.; Xun, Z.Y.; Li, Y.R.; et al. High incidence combination of multiple primary malignant tumors of the digestive system. World J. Gastroenterol. 2022, 28, 5982–5992. [Google Scholar] [CrossRef] [PubMed]
- Marano, L. Dual primary gastric and colorectal cancer: A complex challenge in surgical oncology. World J. Gastrointest. Oncol. 2023, 15, 2049–2052. [Google Scholar] [CrossRef] [PubMed]
- Azer, S.A. Dual primary gastric and colorectal cancer: The known hereditary causes and underlying mechanisms. World J. Gastrointest. Oncol. 2024, 16, 2264–2270. [Google Scholar] [CrossRef] [PubMed]
- Fernández Aceñero, M.J.; Díaz Del Arco, C. Hereditary Gastrointestinal Tumor Syndromes: When Risk Comes with Your Genes. Curr. Issues Mol. Biol. 2024, 46, 6440–6471. [Google Scholar] [CrossRef]
- Gosangi, B.; Dixe de Oliveira Santo, I.; Keraliya, A.; Wang, Y.; Irugu, D.; Thomas, R.; Khandelwal, A.; Rubinowitz, A.N.; Bader, A.S. Li-Fraumeni Syndrome: Imaging Features and Guidelines. Radiographics 2024, 44, e230202. [Google Scholar] [CrossRef]
- Amadou, A.; Achatz, M.I.W.; Hainaut, P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: Temporal phases of Li-Fraumeni syndrome. Curr. Opin. Oncol. 2018, 30, 23–29. [Google Scholar] [CrossRef]
- Bougeard, G.; Renaux-Petel, M.; Flaman, J.M.; Charbonnier, C.; Fermey, P.; Belotti, M.; Gauthier-Villars, M.; Stoppa-Lyonnet, D.; Consolino, E.; Brugières, L.; et al. Revisiting Li-Fraumeni Syndrome from TP53 Mutation Carriers. J. Clin. Oncol. 2015, 33, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, M.L.; Best, A.; Mai, P.L.; Khincha, P.P.; Loud, J.T.; Peters, J.A.; Achatz, M.I.; Chojniak, R.; da Costa, A.B.; Santiago, K.M.; et al. Baseline Surveillance in Li-Fraumeni Syndrome Using Whole-Body Magnetic Resonance Imaging: A Meta-analysis. JAMA Oncol. 2017, 3, 1634–1639. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Leslie, S.W.; McHugh, T.W. Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Rivero-García, P.; Chavarri-Guerra, Y.; Rodríguez Olivares, J.L.; Weitzel, J.N.; Herzog, J.; Candanedo-González, F.; Ríos-Valencia, J.; Mutchinick, O.M.; Arteaga-Vázquez, J. Lynch syndrome in Mexican-Mestizo families: Genotype, phenotypes, and challenges in cascade testing among relatives at risk. Heliyon 2024, 10, e31855. [Google Scholar] [CrossRef]
- Sfakianaki, M.; Tzardi, M.; Tsantaki, K.; Koutoulaki, C.; Messaritakis, I.; Datseri, G.; Moustou, E.; Mavroudis, D.; Souglakos, J. Evaluation of Microsatellite Instability Molecular Analysis versus Immuno-Histochemical Interpretation in Malignant Neoplasms with Different Localizations. Cancers 2023, 15, 353. [Google Scholar] [CrossRef]
- Williams, M.H.; Hadjinicolaou, A.V.; Norton, B.C.; Kader, R.; Lovat, L.B. Lynch syndrome: From detection to treatment. Front. Oncol. 2023, 13, 1166238. [Google Scholar] [CrossRef] [PubMed]
- Menon, G.; Carr, S.; Kasi, A. Familial Adenomatous Polyposis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Sasaki, K.; Kawai, K.; Nozawa, H.; Ishihara, S.; Ishida, H.; Ishibashi, K.; Mori, Y.; Shichijo, S.; Tani, Y.; Takeuchi, Y.; et al. Risk of gastric adenoma and adenocarcinoma in patients with familial adenomatous polyposis in Japan: A nationwide multicenter study. J. Gastroenterol. 2024, 59, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Lauricella, S.; Rausa, E.; Pellegrini, I.; Ricci, M.T.; Signoroni, S.; Palassini, E.; Cavalcoli, F.; Pasanisi, P.; Colombo, C.; Vitellaro, M. Current management of familial adenomatous polyposis. Expert. Rev. Anticancer Ther. 2024, 24, 363–377. [Google Scholar] [CrossRef]
- Alhassan, N.; Helmi, H.; Alzamil, A.; Alshammari, A.; Altamimi, A.; Alshammari, S.; Traiki, T.B.; Albanyan, S.; AlKhayal, K.; Zubaidi, A.; et al. Surveillance Compliance and Quality of Life Assessment Among Surgical Patients with Familial Adenomatous Polyposis Syndrome. J. Epidemiol. Glob. Health 2024, 14, 86–93. [Google Scholar] [CrossRef]
- Forte, G.; Buonadonna, A.L.; Fasano, C.; Sanese, P.; Cariola, F.; Manghisi, A.; Guglielmi, A.F.; Signorile, M.L.; De Marco, K.; Grossi, V.; et al. Clinical and Molecular Characterization of SMAD4 Splicing Variants in Patients with Juvenile Polyposis Syndrome. Int. J. Mol. Sci. 2024, 25, 7939. [Google Scholar] [CrossRef]
- Papadopulos, M.E.; Plazzer, J.P.; Macrae, F.A. Genotype-phenotype correlation of BMPR1a disease causing variants in juvenile polyposis syndrome. Hered. Cancer Clin. Pract. 2023, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Umeno, J.; Jimbo, K.; Arai, M.; Iwama, I.; Kashida, H.; Kudo, T.; Koizumi, K.; Sato, Y.; Sekine, S.; et al. Clinical Guidelines for Diagnosis and Management of Juvenile Polyposis Syndrome in Children and Adults-Secondary Publication. J. Anus Rectum Colon. 2023, 7, 115–125. [Google Scholar] [CrossRef]
- Aslan, P.G.; Çağlayan, A.O.; Bora, E.; Koç, A.; Yücel, H.; Ülgenalp, A.; Öztürk, Y.; Şeker, G.; Akarsu, M. Clinical and Molecular Analysis in Patients with Peutz–Jeghers Syndrome. Turk. J. Gastroenterol. 2024, 35, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Zhang, J.; Liu, C.; Deng, A.; Li, J. Genetic variation at a splicing branch point in intron 7 of STK11: A rare variant decreasing its expression in a Chinese family with Peutz–Jeghers syndrome. World J. Surg. Oncol. 2024, 22, 202. [Google Scholar] [CrossRef] [PubMed]
- Savelyeva, T.A.; Ponomarenko, A.A.; Shelygin, Y.A.; Kuzminov, A.M.; Vyshegorodtsev, D.V.; Loginova, A.N.; Pikunov, D.Y.; Goncharova, E.P.; Likutov, A.A.; Mainovskaya, O.A.; et al. The course and clinical manifestations of Peutz–Jeghers syndrome in the Russian population. Ter. Arkh. 2023, 95, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Kanneganti, P.; Kumar, B.; Upadhyaya, V.D.; Mandelia, A.; Naik, P.B.; Kumar, T.; Agarwal, N. Peutz–Jeghers syndrome: Management for recurrent intussusceptions. Pediatr. Surg. Int. 2024, 40, 148. [Google Scholar] [CrossRef] [PubMed]
- Seppälä, T.T.; Burkhart, R.A.; Katona, B.W. Hereditary colorectal, gastric, and pancreatic cancer: Comprehensive review. BJS Open. 2023, 7, zrad023. [Google Scholar] [CrossRef] [PubMed]
- Gené, M.; Cuatrecasas, M.; Amat, I.; Veiga, J.A.; Aceñero, M.J.F.; Chimisana, V.F.; Tarragona, J.; Jurado, I.; Fernández-Victoria, R.; Ciarpaglini, C.M.; et al. Alterations in p53, Microsatellite Stability and Lack of MUC5AC Expression as Molecular Features of Colorectal Carcinoma Associated with Inflammatory Bowel Disease. Int. J. Mol. Sci. 2023, 24, 8655. [Google Scholar] [CrossRef] [PubMed]
- Haumaier, F.; Dregelies, T.; Sterlacci, W.; Atreya, R.; Vieth, M. Methylation Analysis of Colitis-Associated Colorectal Carcinomas. Discov. Med. 2024, 36, 1363–1369. [Google Scholar] [CrossRef]
- Kabir, M.; Thomas-Gibson, S.; Ahmad, A.; Kader, R.; Al-Hillawi, L.; Mcguire, J.; David, L.; Shah, K.; Rao, R.; Vega, R.; et al. Cancer Biology or Ineffective Surveillance? A Multicentre Retrospective Analysis of Colitis-Associated Post-Colonoscopy Colorectal Cancers. J. Crohns Colitis 2024, 18, 686–694. [Google Scholar] [CrossRef]
- Cai, Y.; Li, S.; Yang, Y.; Duan, S.; Fan, G.; Bai, J.; Zheng, Q.; Gu, Y.; Li, X.; Liu, R. Intestinal epithelial damage-derived mtDNA activates STING-IL12 axis in dendritic cells to promote colitis. Theranostics 2024, 14, 4393–4410. [Google Scholar] [CrossRef]
- Munteanu, A.; Patrascu, S.; Bordu, S.; Laskou, S.; Surlin, V.; Radu, P. Clinical and Morphological Characteristics of Gastrointestinal Stromal Tumor. Chirurgia 2023, 118, 618–623. [Google Scholar] [CrossRef]
- Olivera-Salazar, R.; Salcedo Cabañas, G.; Vega-Clemente, L.; Alonso-Martín, D.; Castellano Megías, V.M.; Volward, P.; García-Olmo, D.; García-Arranz, M. Pilot. Study by Liquid Biopsy in Gastrointestinal Stromal Tumors: Analysis of PDGFRA D842V Mutation and Hypermethylation of SEPT9 Presence by Digital Droplet PCR. Int. J. Mol. Sci. 2024, 25, 6783. [Google Scholar] [CrossRef]
- Wu, C.; Wen, F.; Lin, F.; Zeng, Y.; Lin, X.; Hu, X.; Zhang, X.; Zhang, X.; Wang, X. Predictive performance of [(18)F]F-fibroblast activation protein inhibitor (FAPI)-42 positron emission tomography/computed tomography (PET/CT) in evaluating response of recurrent or metastatic gastrointestinal stromal tumors: Complementary or alternative to [(18)F]fluorodeoxyglucose (FDG) PET/CT? Quant. Imaging Med. Surg. 2024, 14, 5333–5345. [Google Scholar] [CrossRef]
- Yermekova, S.; Orazgaliyeva, M.; Goncharova, T.; Rakhimbekova, F.; Kaidarova, D.; Shatkovskaya, O. Characteristic Mutational Damages in Gastric and Colorectal Adenocarcinomas. Asian Pac. J. Cancer Prev. 2023, 24, 3939–3947. [Google Scholar] [CrossRef]
- Whitley, M.J.; Tran, T.H.; Rigby, M.; Yi, M.; Dharmaiah, S.; Waybright, T.J.; Ramakrishnan, N.; Perkins, S.; Taylor, T.; Messing, S.; et al. Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties. Sci. Adv. 2024, 10, eadj4137. [Google Scholar] [CrossRef] [PubMed]
- Venegas-Rodríguez, J.L.; Hernández-Sandoval, J.A.; Gutiérrez-Angulo, M.; Moreno-Ortiz, J.M.; González-Mercado, A.; Peregrina-Sandoval, J.; Ramírez-Plascencia, H.H.F.; Flores-López, B.A.; Alvizo-Rodríguez, C.R.; Valenzuela-Pérez, J.A.; et al. KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations. Cancers 2024, 16, 2323. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Jin, G.X.; Dong, X.Q. ARTN-GFRA3 axis induces epithelial–mesenchymal transition phenotypes, migration, and invasion of gastric cancer cells via KRAS signalling. Neoplasma 2024, 71, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Mohan, C.D.; Rangappa, S.; Zarrabi, A.; Hushmandi, K.; Kumar, A.P.; Sethi, G.; Rangappa, K.S. Noncoding RNAs as regulators of STAT3 pathway in gastrointestinal cancers: Roles in cancer progression and therapeutic response. Med. Res. Rev. 2023, 43, 1263–1321. [Google Scholar] [CrossRef]
- Raso, M.G.; Toro, E.B.; Evans, K.; Rizvi, Y.; Lazcano, R.; Akcakanat, A.; Sini, P.; Trapani, F.; Madlener, E.J.; Waldmeier, L.; et al. Heterogeneous Profile of ROR1 Protein Expression across Tumor Types. Cancers 2024, 16, 1874. [Google Scholar] [CrossRef]
- Liu, C.; Shen, A.; Song, J.; Cheng, L.; Zhang, M.; Wang, Y.; Liu, X. LncRNA-CCAT5-mediated crosstalk between Wnt/beta-Catenin and STAT3 signalling suggests novel therapeutic approaches for metastatic gastric cancer with high Wnt activity. Cancer Commun. 2024, 44, 76–100. [Google Scholar] [CrossRef]
- Deng, H.; Gao, J.; Cao, B.; Qiu, Z.; Li, T.; Zhao, R.; Li, H.; Wei, B. LncRNA CCAT2 promotes malignant progression of metastatic gastric cancer through regulating CD44 alternative splicing. Cell. Oncol. 2023, 46, 1675–1690. [Google Scholar] [CrossRef]
- Young, I.C.; Brabletz, T.; Lindley, L.E.; Abreu, M.; Nagathihalli, N.; Zaika, A.; Briegel, K.J. Multicancer analysis reveals universal association of oncogenic LBH expression with DNA hypomethylation and WNT-Integrin signalling pathways. Cancer Gene Ther. 2023, 30, 1234–1248. [Google Scholar] [CrossRef]
- Beihaghi, M.; Sahebi, R.; Beihaghi, M.R.; Nessiani, R.K.; Yarasmi, M.R.; Gholamalizadeh, S.; Shahabnavaie, F.; Shojaei, M. Evaluation of rs10811661 polymorphism in CDKN2A/B in colon and gastric cancer. BMC Cancer 2023, 23, 985. [Google Scholar] [CrossRef]
- Zhang, C.; Cui, Y. N6-methyladenosine-induced METTL1 promotes tumor proliferation via CDK4. Biol. Chem. 2023, 405, 217–228. [Google Scholar] [CrossRef]
- Oh, J.; Oh, J.M.; Cho, S.Y. METTL3-mediated downregulation of splicing factor SRSF11 is associated with carcinogenesis and poor survival of cancer patients. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 2561–2570. [Google Scholar] [CrossRef] [PubMed]
- Chivukula, S.; Malkhed, V. The Role of CDK20 Protein in Carcinogenesis. Curr. Drug Targets 2023, 24, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qu, W.; Tu, J.; Qi, H. Implications of Advances in Studies of O6-Methylguanine-DNA- Methyltransferase for Tumor Prognosis and Treatment. Front. Biosci. 2023, 28, 197. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Bosiacki, M.; Barczak, K.; Łagocka, R.; Chlubek, D.; Baranowska-Bosiacka, I. The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers. Cells 2023, 12, 1406. [Google Scholar] [CrossRef]
- Korbecki, J.; Bosiacki, M.; Chlubek, D.; Baranowska-Bosiacka, I. Bioinformatic Analysis of the CXCR2 Ligands in Cancer Processes. Int. J. Mol. Sci. 2023, 24, 13287. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Li, Q.; Wang, C.; Bao, X.; Sun, F.; Qian, X.; Sun, W. Constructing a prognostic model for colon cancer: Insights from immunity-related genes. BMC Cancer 2024, 24, 758. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, Y.; Tao, Q.; Chen, Y.; Zeng, C. Causal effects of circulating glutamine on colitis, IBD, and digestive system cancers: A Mendelian randomization study. J. Cancer 2024, 15, 3738–3749. [Google Scholar] [CrossRef]
- de Oliveira Santos, R.; da Silva Cardoso, G.; da Costa Lima, L.; de Sousa Cavalcante, M.L.; Silva, M.S.; Cavalcante, A.K.M.; Severo, J.S.; de Melo Sousa, F.B.; Pacheco, G.; Alves, E.H.P.; et al. L-Glutamine and Physical Exercise Prevent Intestinal Inflammation and Oxidative Stress Without Improving Gastric Dysmotility in Rats with Ulcerative Colitis. Inflammation 2021, 44, 617–632. [Google Scholar] [CrossRef]
- Stary, D.; Bajda, M. Taurine and Creatine Transporters as Potential Drug Targets in Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 3788. [Google Scholar] [CrossRef]
- Stary, D.; Bajda, M. Structural Studies of the Taurine Transporter: A Potential Biological Target from the GABA Transporter Subfamily in Cancer Therapy. Int. J. Mol. Sci. 2024, 25, 7339. [Google Scholar] [CrossRef]
- Moreno-Sanchez, P.M.; Scafidi, A.; Salvato, I. SUSD2-IL-2 receptor interaction hinders antitumoral CD8(+) T-cell activity: Implications for cancer immunotherapy. Allergy 2023, 78, 3035–3037. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Xian, N.; Zhao, F.; Zhou, Y.; Qin, S. The dual role of SUSD2 in cancer development. Eur. J. Pharmacol. 2024, 977, 176754. [Google Scholar] [CrossRef] [PubMed]
- Capuano, A.; Vescovo, M.; Canesi, S.; Pivetta, E.; Doliana, R.; Nadin, M.G.; Yamamoto, M.; Tsukamoto, T.; Nomura, S.; Pilozzi, E.; et al. The extracellular matrix protein EMILIN-1 impacts on the microenvironment by hampering gastric cancer development and progression. Gastric Cancer 2024, 27, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Liao, M.; Zhang, H.; Li, Y.; Bai, J.; Zhang, J.; Li, L.; Zhang, L. DARS expression in BCR/ABL1-negative myeloproliferative neoplasms and its association with the immune microenvironment. Sci. Rep. 2024, 14, 16711. [Google Scholar] [CrossRef]
- Lim, L.M.; Lee, Y.C.; Lin, T.W.; Hong, Z.X.; Hsu, W.C.; Ke, H.L.; Hwang, D.Y.; Chung, W.Y.; Li, W.M.; Lin, H.H.; et al. NTRK3 exhibits a pro-oncogenic function in upper tract urothelial carcinomas. Kaohsiung J. Med. Sci. 2024, 40, 445–455. [Google Scholar] [CrossRef]
- Hu, R.; Cao, Y.; Wang, Y.; Zhao, T.; Yang, K.; Fan, M.; Guan, M.; Hou, Y.; Ying, J.; Ma, X.; et al. TMEM120B strengthens breast cancer cell stemness and accelerates chemotherapy resistance via beta1-integrin/FAK-TAZ-mTOR signalling axis by binding to MYH9. Breast Cancer Res. 2024, 26, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ou, S.; Liang, T.; Li, M.; Xiao, P.; Cheng, J.; Zhou, J.; Yuan, L. MAEL facilitates metabolic reprogramming and breast cancer progression by promoting the degradation of citrate synthase and fumarate hydratase via chaperone-mediated autophagy. FEBS J. 2023, 290, 3614–3628. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, J.; Niu, N.; Li, X.; Liu, Y.; Huo, L.; Euscher, E.D.; Wang, H.; Bell, D.; Sood, A.K.; et al. SOX17: A Highly Sensitive and Specific Immunomarker for Ovarian and Endometrial Carcinomas. Mod. Pathol. 2023, 36, 100001. [Google Scholar] [CrossRef]
- Alshehri, S.A.; Almarwani, W.A.; Albalawi, A.Z.; Al-Atwi, S.M.; Aljohani, K.K.; Alanazi, A.A.; Ebrahim, M.A.; Hassan, H.M.; Al-Gayyar, M.M. Role of Arctiin in Fibrosis and Apoptosis in Experimentally Induced Hepatocellular Carcinoma in Rats. Cureus 2024, 16, e51997. [Google Scholar] [CrossRef]
- Chowdhury, R.; Bhuia, M.S.; Wilairatana, P.; Afroz, M.; Hasan, R.; Ferdous, J.; Rakib, A.I.; Sheikh, S.; Mubarak, M.S.; Islam, M.T. An insight into the anticancer potentials of lignan arctiin: A comprehensive review of molecular mechanisms. Heliyon 2024, 10, e32899. [Google Scholar] [CrossRef] [PubMed]
- Davarzani, Z.; Salehi, P.; Asghari, S.M.; Bararjanian, M.; Mohsen, A.H.; Harati, H.D. Design, Synthesis, and Functional Studies on Noscapine and Cotarnine Amino Acid Derivatives as Antitumour Agents. ACS Omega 2023, 8, 45502–45509. [Google Scholar] [CrossRef] [PubMed]
- Calaf, G.M.; Crispin, L.A.; Quisbert-Valenzuela, E.O. Noscapine and Apoptosis in Breast and Other Cancers. Int. J. Mol. Sci. 2024, 25, 3536. [Google Scholar] [CrossRef]
- Ijaz, S.; Iqbal, J.; Abbasi, B.A.; Tufail, A.; Yaseen, T.; Uddin, S.; Usman, K.; Ullah, R.; Bibi, H.; Inam, P.; et al. Current stage of preclinical and clinical development of guggulsterone in cancers: Challenges and promises. Cell Biol. Int. 2024, 48, 128–142. [Google Scholar] [CrossRef]
- Gupta, M.; Singh, D.; Rastogi, S.; Siddique, H.R.; Al-Dayan, N.; Ahmad, A.; Sikander, M.; Sarwat, M. Anticancer activity of guggulsterone by modulating apoptotic markers: A systematic review and meta-analysis. Front. Pharmacol. 2023, 14, 1155163. [Google Scholar] [CrossRef] [PubMed]
- Koç, T.Y.; Doğan, S.; Karadayı, M. Potential Using of Resveratrol and Its Derivatives in Medicine. Eurasian J. Med. 2024, 56, 136–141. [Google Scholar] [CrossRef]
- Asemi, R.; Rajabpoor Nikoo, N.; Asemi, Z.; Shafabakhsh, R.; Hajijafari, M.; Sharifi, M.; Homayoonfal, M.; Davoodvandi, A.; Hakamifard, A. Modulation of long noncoding RNAs by resveratrol as a potential therapeutic approach in cancer: A comprehensive review. Pathol. Res. Pract. 2023, 246, 154507. [Google Scholar] [CrossRef]
- Thoyajakshi, R.S.; Megha, G.T.; Ravi Kumar, H.; Mathad, S.N.; Khan, A.; Nagaraju, S.; Mahmoud, M.H.; Ansari, A. Garcinol: A novel and potent inhibitor of hyaluronidase enzyme. Int. J. Biol. Macromol. 2024, 266, 131145. [Google Scholar] [CrossRef] [PubMed]
- Patwa, N.; Chauhan, R.; Chauhan, A.; Kumar, M.; Ramniwas, S.; Mathkor, D.M.; Saini, A.K.; Tuli, H.S.; Haque, S.; Slama, P. Garcinol in gastrointestinal cancer prevention: Recent advances and future prospects. J. Cancer Res. Clin. Oncol. 2024, 150, 370. [Google Scholar] [CrossRef]
- Saeed, R.F.; Awan, U.A.; Aslam, S.; Qazi, A.S.; Bhatti, M.Z.; Akhtar, N. Micronutrients Importance in Cancer Prevention-Minerals. Cancer Treat. Res. 2024, 191, 145–161. [Google Scholar] [CrossRef]
- Saeed, R.F.; Naz, S.; Awan, U.A.; Gul, S.; Subhan, F.; Saeed, S. Micronutrients Importance in Cancer Prevention-Vitamins. Cancer Treat. Res. 2024, 191, 119–144. [Google Scholar] [CrossRef]
- Xiong, D.; Han, T.; Li, Y.; Hong, Y.; Li, S.; Li, X.; Tao, W.; Huang, Y.S.; Chen, W.; Li, C. TOTEM: A multicancer detection and localization approach using circulating tumor DNA methylation markers. BMC Cancer 2024, 24, 840. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yu, J.; Xu, Q.; Peng, Y.; Li, H.; Lu, Y.; Ouyang, M. Disulfidptosis-related long noncoding RNA signature predicts the prognosis, tumor microenvironment, immunotherapy, and antitumour drug options in colon adenocarcinoma. Apoptosis 2024, 29, 2074–2090. [Google Scholar] [CrossRef] [PubMed]
- Beton-Mysur, K.; Brozek-Pluska, B. Raman Spectroscopy and Imaging Studies of Human Digestive Tract Cells and Tissues-Impact of Vitamin C and E Supplementation. Molecules 2022, 28, 137. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Seeing data as t-SNE and UMAP do. Nat. Methods 2024, 21, 930–933. [Google Scholar] [CrossRef]
- Sugai, T.; Uesugi, N.; Osakabe, M.; Yamamoto, R.; Hamada, K.; Honda, M.; Yanagawa, N.; Suzuki, H. The molecular profile of gastric intraepithelial foveolar type neoplasia based on somatic copy number alterations and multiple mutation analysis. Gastric Cancer 2024, 27, 1220–1228. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Wang, J. Functional Proteomic Profiling Analysis in Four Major Types of Gastrointestinal Cancers. Biomolecules 2023, 13, 701. [Google Scholar] [CrossRef]
- Pang, N.; Yang, W.; Yang, G.; Yang, C.; Tong, K.; Yu, R.; Jiang, F. The utilization of blood serum ATR-FTIR spectroscopy for the identification of gastric cancer. Discov. Oncol. 2024, 15, 350. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Larsen, V.; Morton, V.; Norat, T.; Moreira, A.; Potts, J.F.; Reeves, T.; Bakolis, I. Dietary patterns derived from principal component analysis (PCA) and risk of colorectal cancer: A systematic review and meta-analysis. Eur. J. Clin. Nutr. 2019, 73, 366–386. [Google Scholar] [CrossRef]
- Roshani, M.; Molavizadeh, D.; Sadeghi, S.; Jafari, A.; Dashti, F.; Mirazimi, S.M.A.; Asouri, S.A.; Rajabi, A.; Hamblin, M.R.; Anoushirvani, A.A.; et al. Emerging roles of miR-145 in gastrointestinal cancers: A new paradigm. Biomed. Pharmacother. 2023, 166, 115264. [Google Scholar] [CrossRef]
- Gu, C.; Li, X. Prediction of disease-related miRNAs by voting with multiple classifiers. BMC Bioinform. 2023, 24, 177. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Ying, J.; Chen, H.; Wu, Z.; Huang, C.; Zhang, C.; Chen, Z.; Chen, H. PPIH acts as a potential predictive biomarker for patients with common solid tumors. BMC Cancer 2024, 24, 681. [Google Scholar] [CrossRef]
- Sanderson, E.; Glymour, M.M.; Holmes, M.V.; Kang, H.; Morrison, J.; Munafò, M.R.; Palmer, T.; Schooling, C.M.; Wallace, C.; Zhao, Q.; et al. Mendelian randomization. Nat. Rev. Methods Primers 2022, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yu, W.; Gu, Y.; Xia, J.; Sun, G. Height and cancer risk in East Asians: Evidence from a prospective cohort study and Mendelian randomization analyses. Cancer Epidemiol. 2024, 92, 102647. [Google Scholar] [CrossRef]
- Fu, Q.; Li, L.; Zhuoma, N.; Ma, R.; Zhao, Z.; Quzuo, Z.; Wang, Z.; Yangzong, D.; Di, J. Causality between six psychiatric disorders and digestive tract cancers risk: A two-sample Mendelian randomization study. Sci. Rep. 2024, 14, 16689. [Google Scholar] [CrossRef]
- Yan, W.; Zhou, J.; Jiang, M.; Kong, Y.; Qin, H.; Qi, Y.; Wang, S.; Tai, J. Obstructive sleep apnea and 19 gastrointestinal diseases: A Mendelian randomization study. Front. Psychiatry 2024, 15, 1256116. [Google Scholar] [CrossRef] [PubMed]
- Hiwasa, T.; Yoshida, Y.; Kubota, M.; Li, S.Y.; Zhang, B.S.; Matsutani, T.; Mine, S.; Machida, T.; Ito, M.; Yajima, S.; et al. Serum anti-KIAA0513 antibody as a common biomarker for mortal atherosclerotic and cancerous diseases. Med. Int. 2024, 4, 45. [Google Scholar] [CrossRef]
- Zhao, D.; Cai, F.; Liu, X.; Li, T.; Zhao, E.; Wang, X.; Zheng, Z. CEACAM6 expression and function in tumor biology: A comprehensive review. Discov. Oncol. 2024, 15, 186. [Google Scholar] [CrossRef]
- Wu, G.; Wang, D.; Xiong, F.; Wang, Q.; Liu, W.; Chen, J.; Chen, Y. The emerging roles of CEACAM6 in human cancer (Review). Int. J. Oncol. 2024, 64, 27. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ni, B.; Shen, L.; Li, Z.; Zhou, L.; Wu, H.; Zhang, Y.; Liu, L.; Liu, J.; Tian, L.; et al. Systematic pancancer analysis insights into ICAM1 as an immunological and prognostic biomarker. FASEB J. 2024, 38, e23802. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhou, C.P.; Wang, D.T.; Ma, J.; Sun, X.H.; Wang, Y.; Zhang, Y.M. Unravelling the causal role of immune cells in gastrointestinal tract cancers: Insights from a Mendelian randomization study. Front. Immunol. 2024, 15, 1343512. [Google Scholar] [CrossRef]
- Märkl, B.; Reitsam, N.G.; Grochowski, P.; Waidhauser, J.; Grosser, B. The SARIFA biomarker in the context of basic research of lipid-driven cancers. NPJ Precis. Oncol. 2024, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Reitsam, N.G.; Grozdanov, V.; Löffler, C.M.L.; Muti, H.S.; Grosser, B.; Kather, J.N.; Märkl, B. Novel biomarker SARIFA in colorectal cancer: Highly prognostic, not genetically driven and histologic indicator of a distinct tumor biology. Cancer Gene Ther. 2024, 31, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Kulecka, M.; Czarnowski, P.; Bałabas, A.; Turkot, M.; Kruczkowska-Tarantowicz, K.; Żeber-Lubecka, N.; Dąbrowska, M.; Paszkiewicz-Kozik, E.; Walewski, J.; Ługowska, I.; et al. Microbial and Metabolic Gut Profiling across Seven Malignancies Identifies Fecal Faecalibacillus intestinalis and Formic Acid as Commonly Altered in Cancer Patients. Int. J. Mol. Sci. 2024, 25, 8026. [Google Scholar] [CrossRef]
| The Frequency of Their Occurrence | Related Genes | Cancer Risk Estimates | Surveillance | Management Guidelines | References | |
|---|---|---|---|---|---|---|
| LFS | 80% carriers of a germline TP53 mutation at age 70 | TP53 encodes p53 | Males > 70%, females > 90% | Earlier discovery of TP53 mutant carriers results in reduced morbidity and improved survival | Clinical therapy varies and should be divided into classes according to the kind of mutation | [15,16,17,18] |
| LS | Between 5 and 10% of CRC cases are hereditary. The majority are associated with LS | MMR | Risk of CRC is 80%, risk of endometrial cancer is 60% | The most common malignancies are:
| Chemotherapy, immunotherapy, and vaccines | [19,20,21,22] |
| FAP | A prevalence of between 1:20,000 and 1:10,000 in Western countries and 1:17,400 in Japan | APC | Incidence rates in 50-year-old individuals with FAP were 22.8% and 7.6%, respectively | Poor compliance | Colonectomy and multidisciplinary and individualized approaches | [23,24,25,26] |
| JPS | An estimated incidence varying from 1:16,000 to 1:100,000 | SMAD4 or BMPR1A | 75% of cases have an autosomal-dominant genetic disease | A low risk of carcinogenesis | No curative treatment | [27,28,29] |
| PJS | PJS is a rare hereditary syndrome
| STK11 | Patients without STK11 mutations may have had a decreased risk of developing cancer and later start of symptoms | Due of the problems of polyps, children with PJS are at a significant risk of requiring multiple laparotomies | Given the diffuse involvement of the gut, substantial intestinal resection and early surgical decision-making are not recommended | [30,31,32,33] |
| CAC | CAC is the leading cause of death among long-standing IBD patients | p53 mutations | The first step in the development of serrated CRC is known as GM | The considerable proportion that appears to originate from non-adenomatous-looking mucosa that displays rapid cancer progression but fails to reveal neoplasia on biopsy | STING | [34,35,36,37,38] |
| GISTs | An incidence of 7–15 cases per million | PDGFRA D842V mutation | The stomach (n = 7, 35%), colon (n = 3, 15%), and peritoneum (n = 1, n = 5%) were the most commonly affected sites (n = 9, 45%) | The long-term prognosis is strongly correlated with the size of the tumor and the number of mitoses | Targeted therapy type, [18F] FDG PET/CT characteristics, recurring metastatic GISTs, and the nomogram could produce more accurate responses | [39,40,41] |
| Natural Products | Biological Activities | References |
|---|---|---|
| Arctiin |
| [71] |
| [72] | |
| Noscapine |
| [73] |
| [74] | |
| Guggulsterone |
| [75] |
| [76] | |
| Resveratrol |
| [77] |
| [78] | |
| Garcinol |
| [79] |
| [80] | |
| Minerals(trace elements) |
| [81] |
| Vitamins |
| [82] |
| Biomarkers | Utilizations | References |
|---|---|---|
| TOTEM |
| [83] |
| PCA |
| [84] |
| PLS-DA |
| [85] |
| t-SNE |
| [86] |
| Hierarchical clustering analysis |
| [87] |
| miRNAs |
| [91,92,93] |
| MR |
| [94,95,96,97] |
| AlphaLISA |
| [98] |
| CEACAM6 |
| [99,100] |
| ICAM1 |
| [101,102] |
| SARIFA |
| [103,104] |
| microbiome |
| [105] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lupan, I.; Silaghi, C.; Stroe, C.; Muntean, A.; Deleanu, D.; Bintintan, V.; Samasca, G. The Importance of Genetic Screening on the Syndromes of Colorectal Cancer and Gastric Cancer: A 2024 Update. Biomedicines 2024, 12, 2655. https://doi.org/10.3390/biomedicines12122655
Lupan I, Silaghi C, Stroe C, Muntean A, Deleanu D, Bintintan V, Samasca G. The Importance of Genetic Screening on the Syndromes of Colorectal Cancer and Gastric Cancer: A 2024 Update. Biomedicines. 2024; 12(12):2655. https://doi.org/10.3390/biomedicines12122655
Chicago/Turabian StyleLupan, Iulia, Ciprian Silaghi, Claudia Stroe, Adriana Muntean, Diana Deleanu, Vasile Bintintan, and Gabriel Samasca. 2024. "The Importance of Genetic Screening on the Syndromes of Colorectal Cancer and Gastric Cancer: A 2024 Update" Biomedicines 12, no. 12: 2655. https://doi.org/10.3390/biomedicines12122655
APA StyleLupan, I., Silaghi, C., Stroe, C., Muntean, A., Deleanu, D., Bintintan, V., & Samasca, G. (2024). The Importance of Genetic Screening on the Syndromes of Colorectal Cancer and Gastric Cancer: A 2024 Update. Biomedicines, 12(12), 2655. https://doi.org/10.3390/biomedicines12122655







