Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types: A Pan-Cancer Analysis
Abstract
1. Introduction
2. Materials and Methods
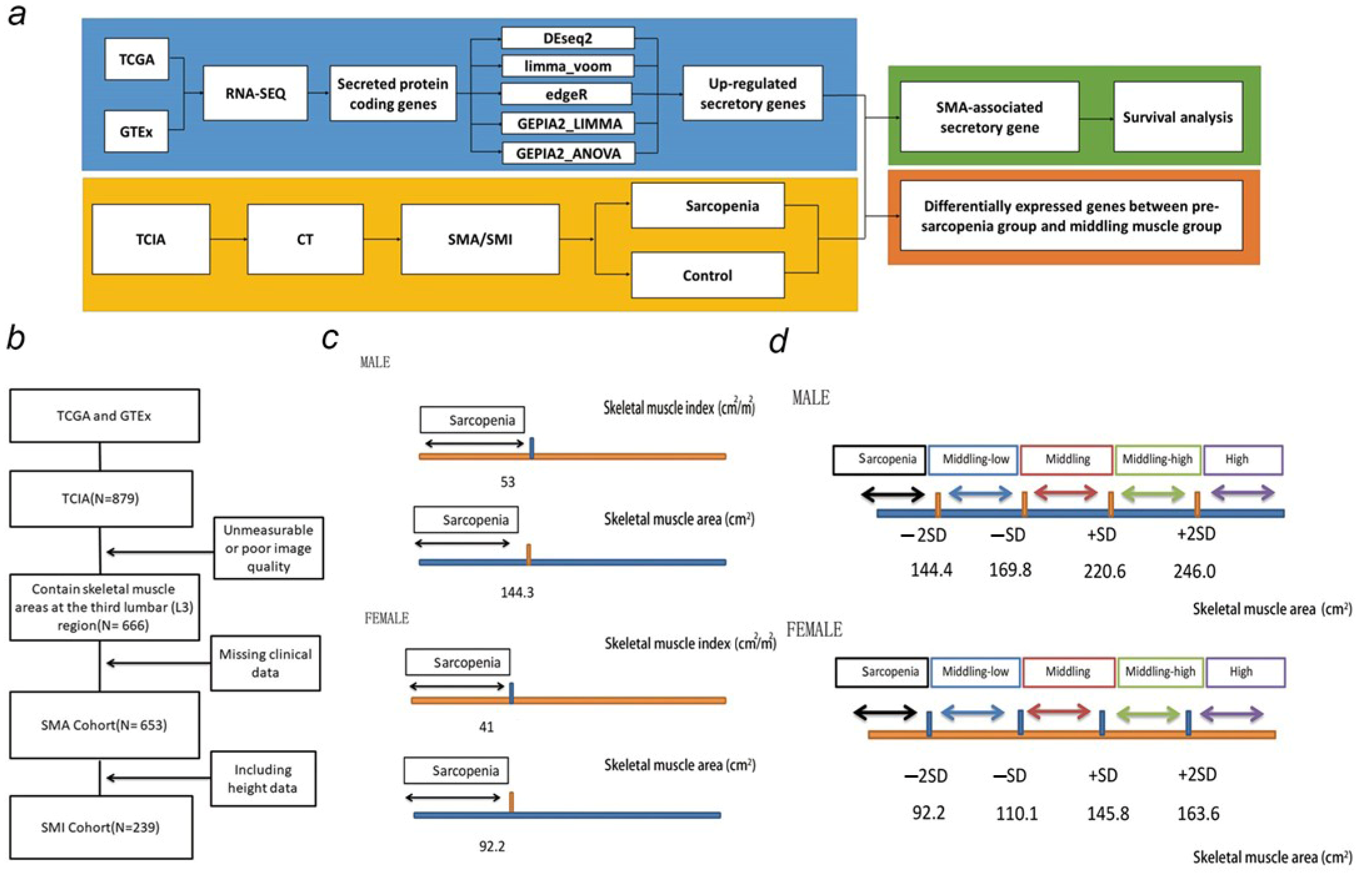
2.1. Data Sources and Study Population
2.2. Assessment of Muscle Cross-Sectional Area from CT
2.3. Transcriptome-Based Secretome Analysis
2.4. Statistical Analysis
3. Results
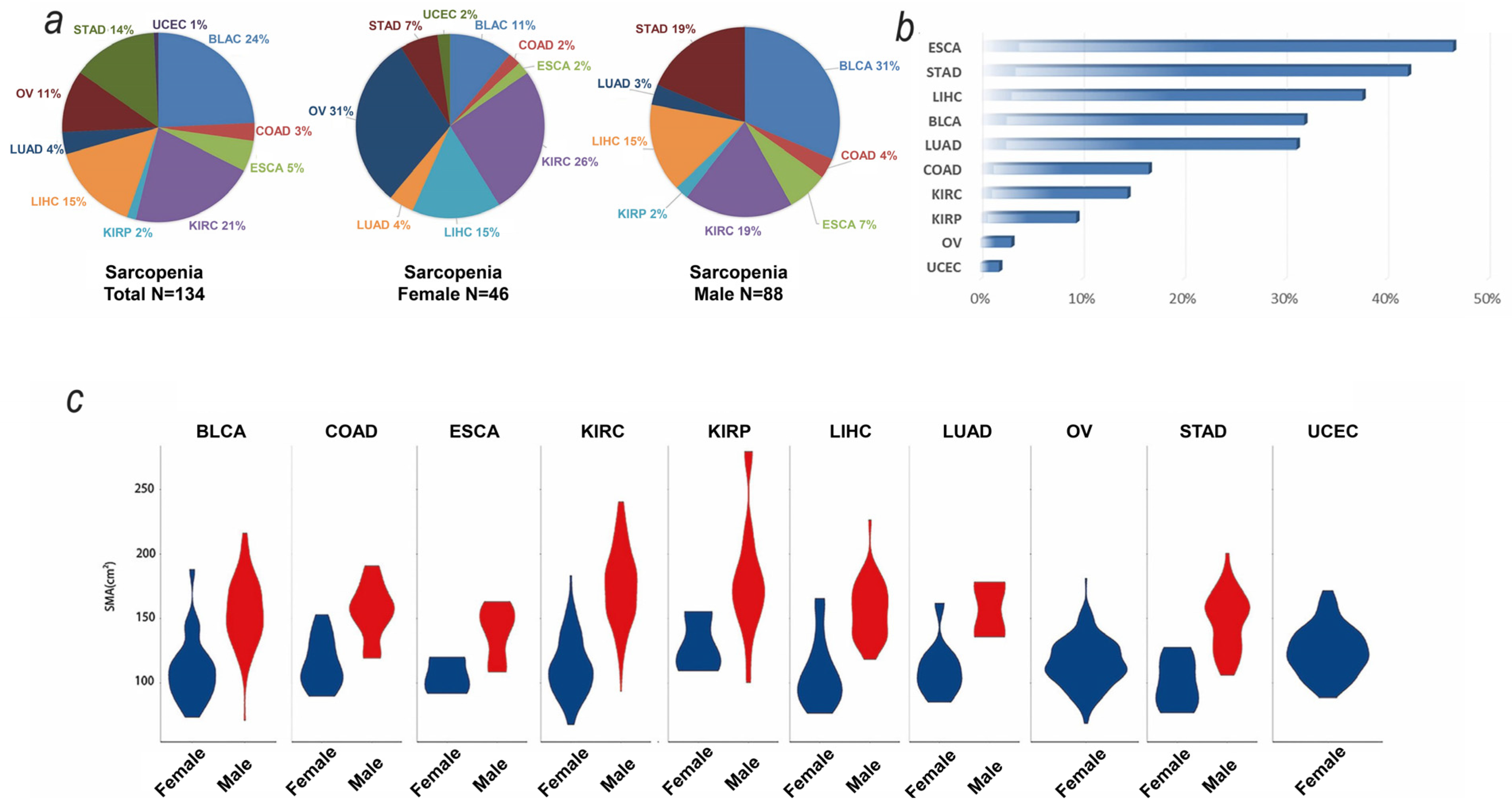
3.1. Baseline and SMA Characteristics of TCGA Samples
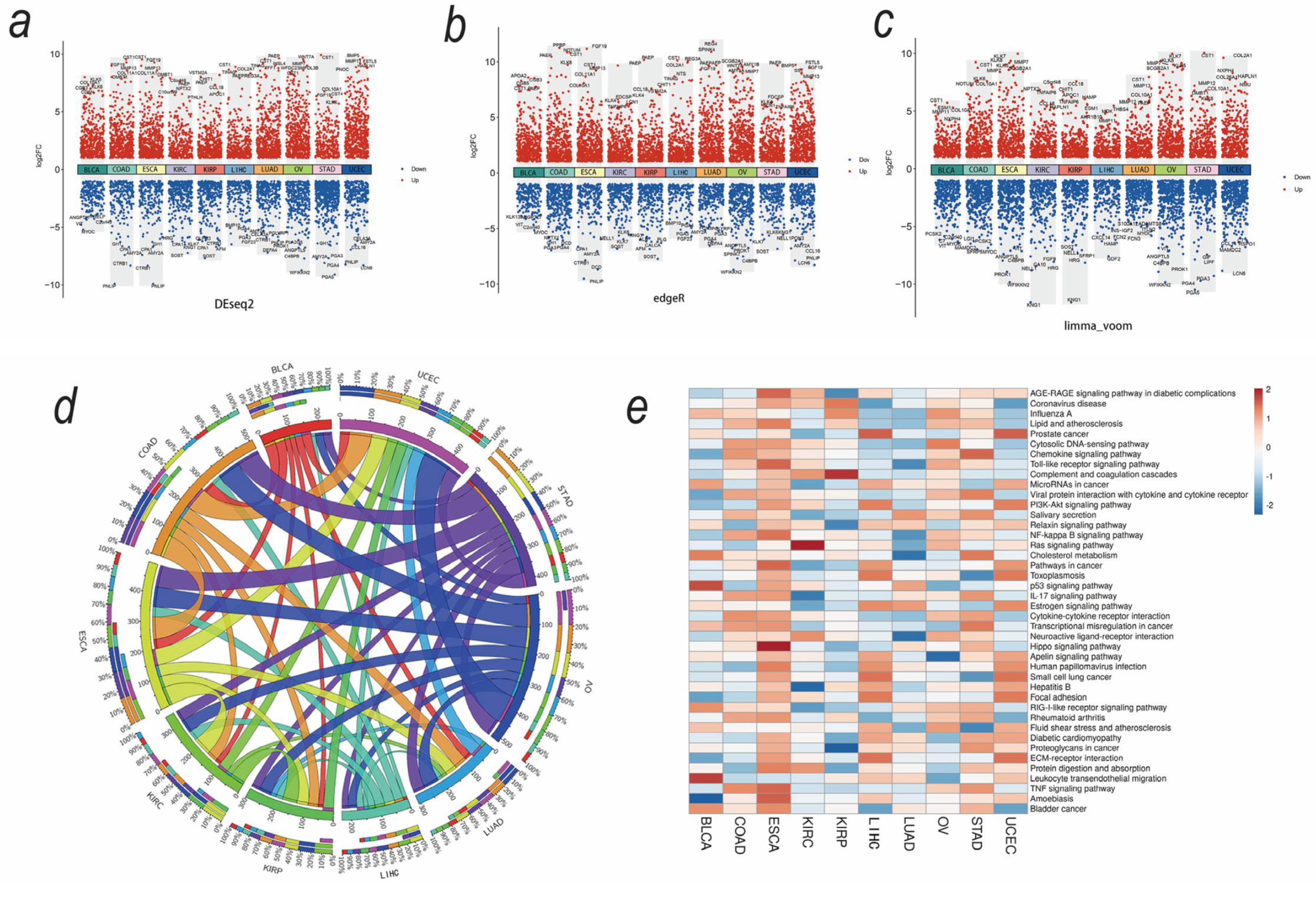
3.2. Upregulated Secreted Protein-Coding Genes and Related Pathways in Cancers
3.3. SMA-Associated Upregulated Secretory Protein-Coding Genes and Their Predictive Values for Patient Survival
3.4. Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types and Genders
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aversa, Z.; Costelli, P.; Muscaritoli, M. Cancer-induced muscle wasting: Latest findings in prevention and treatment. Ther. Adv. Med. Oncol. 2017, 9, 369–382. [Google Scholar] [CrossRef]
- de Castro, G.S.; Simoes, E.; Lima, J.D.; Ortiz-Silva, M.; Festuccia, W.T.; Tokeshi, F.; Alcântara, P.S.; Otoch, J.P.; Coletti, D.; Seelaender, M. Human Cachexia Induces Changes in Mitochondria, Autophagy and Apoptosis in the Skeletal Muscle. Cancers 2019, 11, 1264. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.L.; Lu, J.; Song, Y.; Kwak, K.S.; Jiao, Q.; Rosenfeld, R.; Chen, Q.; Boone, T.; Simonet, W.S.; et al. Reversal of Cancer Cachexia and Muscle Wasting by ActRIIB Antagonism Leads to Prolonged Survival. Cell 2010, 142, 531–543. [Google Scholar] [CrossRef]
- García-Prat, L.; Martínez-Vicente, M.; Perdiguero, E.; Ortet, L.; Rodríguez-Ubreva, J.; Rebollo, E.; Ruiz-Bonilla, V.; Gutarra, S.; Ballestar, E.; Serrano, A.L.; et al. Autophagy maintains stemness by preventing senescence. Nature 2016, 529, 37–42. [Google Scholar] [CrossRef]
- Mulder, S.E.; Dasgupta, A.; King, R.J.; Abrego, J.; Attri, K.S.; Murthy, D.; Shukla, S.K.; Singh, P.K. JNK signaling contributes to skeletal muscle wasting and protein turnover in pancreatic cancer cachexia. Cancer Lett. 2020, 491, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-V.; Wu, W.-T.; Chen, Y.-H.; Chen, L.-R.; Hsu, W.-H.; Lin, Y.-L.; Han, D.-S. Enhanced serum levels of tumor necrosis factor-α, interleukin-1β, and -6 in sarcopenia: Alleviation through exercise and nutrition intervention. Aging 2023, 15, 13471–13485. [Google Scholar] [CrossRef]
- Lipshitz, M.; Visser, J.; Anderson, R.; Nel, D.G.; Smit, T.; Steel, H.C.; Rapoport, B. Emerging markers of cancer cachexia and their relationship to sarcopenia. J. Cancer Res. Clin. Oncol. 2010, 149, 17511–17527. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Semprun-Prieto, L.; Sukhanov, S.; Delafontaine, P.; Forrester, S.J.; Booz, G.W.; Sigmund, C.D.; Coffman, T.M.; Kawai, T.; Rizzo, V.; et al. IGF-1 prevents ANG II-induced skeletal muscle atrophy via Akt- and Foxo-dependent inhibition of the ubiquitin ligase atrogin-1 expression. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H1565–H1570. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Markov, S.D.; Attri, K.S.; Vernucci, E.; King, R.J.; Dasgupta, A.; Grandgenett, P.M.; Hollingsworth, M.A.; Singh, P.K.; Yu, F.; et al. Macrophages potentiate STAT3 signaling in skeletal muscles and regulate pancreatic cancer cachexia. Cancer Lett. 2020, 484, 29–39. [Google Scholar] [CrossRef]
- Coletti, D.; Moresi, V.; Adamo, S.; Molinaro, M.; Sassoon, D. Tumor necrosis factor-alpha gene transfer induces cachexia and inhibits muscle regeneration. Genesis 2005, 43, 120–128. [Google Scholar] [CrossRef]
- Talbert, E.E.; Lewis, H.L.; Farren, M.R.; Ramsey, M.L.; Chakedis, J.M.; Rajasekera, P.; Haverick, E.; Sarna, A.; Bloomston, M.; Pawlik, T.M.; et al. Circulating monocyte chemoattractant protein-1 (MCP-1) is associated with cachexia in treatment-naive pancreatic cancer patients. J. Cachexia Sarcopenia Muscle 2018, 9, 358–368. [Google Scholar] [CrossRef]
- Fukawa, T.; Yan-Jiang, B.C.; Min-Wen, J.C.; Jun-Hao, E.T.; Huang, D.; Qian, C.-N.; Ong, P.; Li, Z.; Chen, S.; Mak, S.Y. Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia. Nat. Med. 2016, 22, 666–671. [Google Scholar] [CrossRef]
- Armstrong, V.S.; Fitzgerald, L.W.; Bathe, O.F. Cancer-Associated Muscle Wasting-Candidate Mechanisms and Molecular Pathways. Int. J. Mol. Sci. 2020, 21, 9268. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Perez, S.L.; Haus, J.M.; Sheean, P.; Patel, B.; Mar, W.; Chaudhry, V.; McKeever, L.; Braunschweig, C. Measuring Abdominal Circumference and Skeletal Muscle from a Single Cross-Sectional Computed Tomography Image: A Step-by-Step Guide for Clinicians Using National Institutes of Health ImageJ. JPEN J. Parenter. Enteral. Nutr. 2016, 40, 308–318. [Google Scholar] [CrossRef]
- Derstine, B.A.; Holcombe, S.A.; Goulson, R.L.; Ross, B.E.; Wang, N.C.; Sullivan, J.A.; Su, G.L.; Wang, S.C. Quantifying Sarcopenia Reference Values Using Lumbar and Thoracic Muscle Areas in a Healthy Population. J. Nutr. Health Aging 2017, 21, 180–185. [Google Scholar] [CrossRef]
- Fuchs, G.; Chretien, Y.R.; Mario, J.; Do, S.; Eikermann, M.; Liu, B.; Yang, K.; Fintelmann, F.J. Quantifying the effect of slice thickness, intravenous contrast and tube current on muscle segmentation: Implications for body composition analysis. Eur. Radiol. 2018, 28, 2455–2463. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.M.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Derstine, B.A.; Holcombe, S.A.; Ross, B.E.; Wang, N.C.; Su, G.L.; Wang, S.C. Skeletal muscle cutoff values for sarcopenia diagnosis using T10 to L5 measurements in a healthy US population. Sci. Rep. 2018, 8, 11369. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Baeyens, J.P.; Bauer, J.M.; Boirie, Y.; Cederholm, T.; Landi, F.; Martin, F.C.; Michel, J.-P.; Rolland, Y.; Schneider, S.M.; et al. Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age Ageing 2010, 39, 412–423. [Google Scholar] [CrossRef]
- Martin, L.; Birdsell, L.; MacDonald, N.; Reiman, T.; Clandinin, M.T.; McCargar, L.J.; Murphy, R.; Ghosh, S.; Sawyer, M.B.; Baracos, V.E. Cancer Cachexia in the Age of Obesity: Skeletal Muscle Depletion Is a Powerful Prognostic Factor, Independent of Body Mass Index. J. Clin. Oncol. 2013, 31, 1539–1547. [Google Scholar] [CrossRef]
- Freire, P.P.; Fernandez, G.J.; de Moraes, D.; Cury, S.S.; Pai-Silva, M.D.; dos Reis, P.P.; Rogatto, S.R.; Carvalho, R.F. The expression landscape of cachexia-inducing factors in human cancers. J. Cachexia Sarcopenia Muscle 2020, 11, 947–961. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucl. Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucl. Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Srivastava, S.; Lindzen, M.; Sas-Chen, A.; Sheffer, M.; Lauriola, M.; Enuka, Y.; Noronha, A.; Mancini, M.; Lavi, S.; et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci. Signal. 2018, 11, eaan0949. [Google Scholar] [CrossRef] [PubMed]
- van Vlodrop, I.J.H.; Joosten, S.C.; De Meyer, T.; Smits, K.M.; Van Neste, L.; Melotte, V.; Baldewijns, M.M.L.L.; Schouten, L.J.; van den Brandt, P.A.; Jeschke, J.; et al. A Four-Gene Promoter Methylation Marker Panel Consisting of GREM1, NEURL, LAD1, and NEFH Predicts Survival of Clear Cell Renal Cell Cancer Patients. Clin. Cancer Res. 2017, 23, 2006–2018. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.; Yang, S.-J.; Park, S.M.; Lee, S.-H.; Song, K.S.; Jeong, E.-J.; Park, M.; Kim, J.-S.; Yeom, Y.I.; Kim, J.-A. LAD1 expression is associated with the metastatic potential of colorectal cancer cells. BMC Cancer 2020, 20, 1180. [Google Scholar] [CrossRef]
- Prokopchuk, O.; Grünwald, B.; Nitsche, U.; Jäger, C.; Prokopchuk, O.L.; Schubert, E.C.; Friess, H.; Martignoni, M.E.; Krüger, A. Elevated systemic levels of the matrix metalloproteinase inhibitor TIMP-1 correlate with clinical markers of cachexia in patients with chronic pancreatitis and pancreatic cancer. BMC Cancer 2018, 18, 128. [Google Scholar] [CrossRef]



| Cancer Type $ | Total # | SMA Data * | Clinical Data & | Transcriptomic Data ! |
|---|---|---|---|---|
| TCGA-UCEC | 65 | 52 | 52 | 3 |
| TCGA-STAD | 46 | 45 | 45 | 43 |
| TCGA-OV | 143 | 129 | 128 | 96 |
| TCGA-LUAD | 67 | 28 | 16 | 16 |
| TCGA-LIHC | 97 | 53 | 53 | 53 |
| TCGA-KIRP | 33 | 21 | 21 | 21 |
| TCGA-KIRC | 267 | 199 | 199 | 196 |
| TCGA-ESCA | 16 | 15 | 15 | 14 |
| TCGA-COAD | 25 | 24 | 24 | 19 |
| TCGA-BLCA | 120 | 100 | 100 | 99 |
| Pan-cancer | 879 | 666 | 653 | 560 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, X.; Zeng, Y.; Huang, J.; Chen, X.; Kang, H.; Huang, S. Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types: A Pan-Cancer Analysis. Biomedicines 2024, 12, 329. https://doi.org/10.3390/biomedicines12020329
Gan X, Zeng Y, Huang J, Chen X, Kang H, Huang S. Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types: A Pan-Cancer Analysis. Biomedicines. 2024; 12(2):329. https://doi.org/10.3390/biomedicines12020329
Chicago/Turabian StyleGan, Xin, Yunqian Zeng, Jiaquan Huang, Xin Chen, Hao Kang, and Shuaiwen Huang. 2024. "Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types: A Pan-Cancer Analysis" Biomedicines 12, no. 2: 329. https://doi.org/10.3390/biomedicines12020329
APA StyleGan, X., Zeng, Y., Huang, J., Chen, X., Kang, H., & Huang, S. (2024). Tumor-Derived Sarcopenia Factors Are Diverse in Different Tumor Types: A Pan-Cancer Analysis. Biomedicines, 12(2), 329. https://doi.org/10.3390/biomedicines12020329







