Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine
Abstract
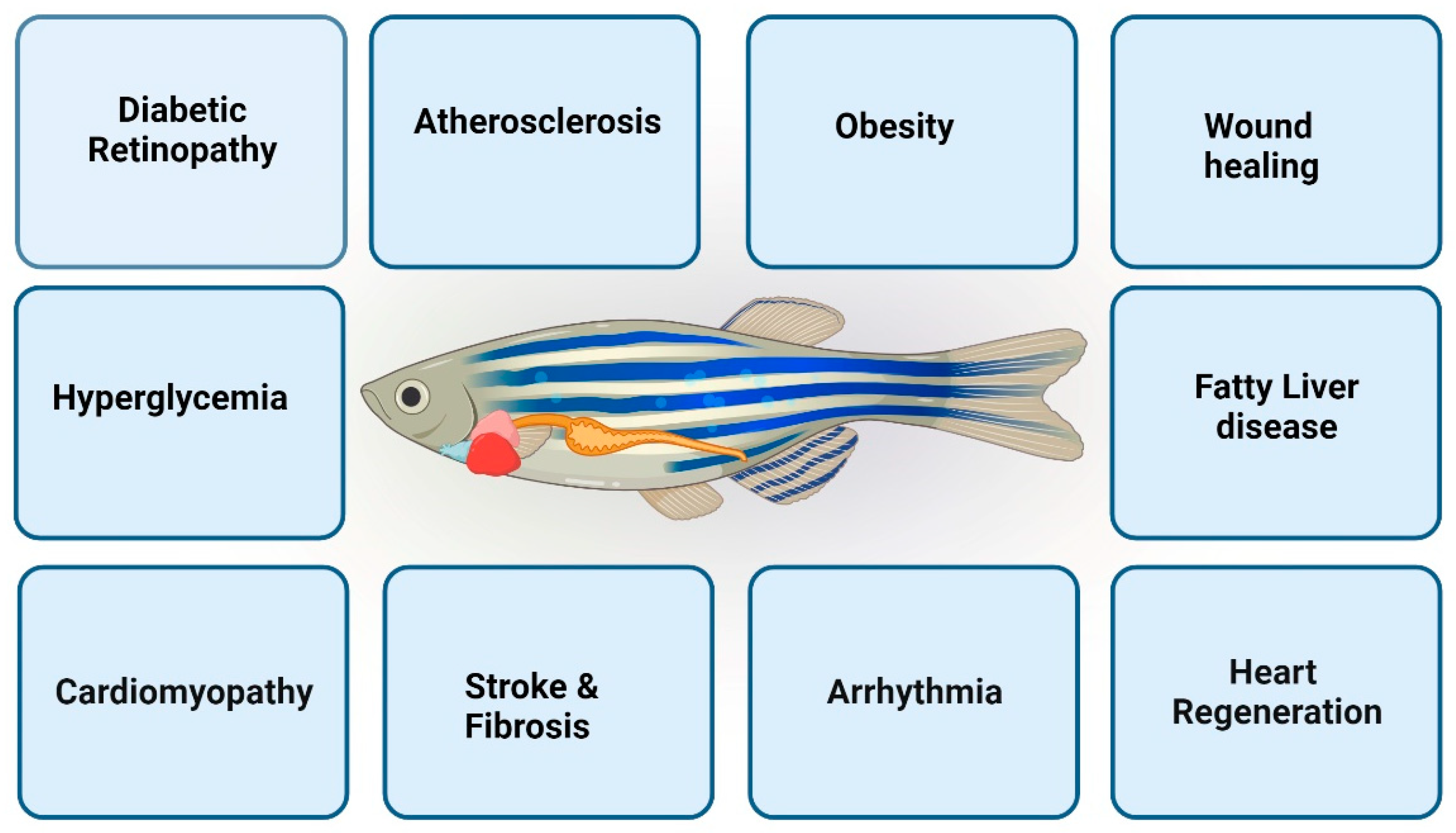
:1. Introduction
2. Genetic Studies Using Zebrafish
3. Methods
- Inclusion criteria:
- A.1.
- Research articles that report on genetic or non-genetic models of cardiovascular and metabolic disease.
- A.2.
- Research articles that involve the keywords heart failure, cardiomyopathy, hypertrophy, arrhythmia, regeneration, drug screen, obesity, diabetes, and atherosclerosis.
- A.3.
- Research articles that report the genetic tools used in zebrafish to investigate cardiovascular and metabolic diseases.
- The exclusion criteria were set to exclude the following articles:
- B.1.
- Articles that report on other animal models apart from zebrafish.
- B.2.
- Articles that report on zebrafish models of other vascular diseases or other diseases.
- B.3.
- Articles that report on aging-related disorders.
- B.4.
- Article published in languages other than English.
4. Zebrafish as Cardiovascular Disease Models
4.1. Heart Regeneration
4.2. Zebrafish Cardiomyopathy Models
4.3. Arteriogenesis
4.4. Thrombosis
4.5. Modeling Cardiac Rhythm Disorders in Zebrafish
4.6. Limitations of Zebrafish as a Cardiovascular Disease Model
4.6.1. Simplified Cardiovascular Anatomy
4.6.2. Limited Behavioral Studies
4.6.3. Temperature Sensitivity
5. Summary
6. Metabolic Disease Models
6.1. Obesity
6.2. Non-Alcoholic Fatty Liver Disease
6.3. Atherosclerosis
6.4. Diabetes
6.5. Limitations of Zebrafish as a Metabolic Disease Model
6.6. Mammalian-Specific Aspects
6.7. Limited Tissue Complexity
6.8. Limited Behavioral Studies
7. Summary
8. Conclusions
9. Future Directions
9.1. Precision Medicine and Personalized Therapies
9.2. Functional Genomics and Systems Biology
9.3. Single-Cell Analysis
9.4. Live Imaging and 3D Models
9.5. Integrative Models for Disease Complexities
9.6. Non-Coding RNA Biology
9.7. Environmental Influences and Nutritional Studies
9.8. Drug Discovery and Screening
9.9. Functional Validation in Mammalian Models
9.10. Innovations in Genome Editing
9.11. Genomics in Cardiovascular Disease
9.12. Transcriptomics in Cardiovascular Disease
9.13. Proteomics in Cardiovascular Disease
9.14. Metabolomics in Cardiovascular Disease
9.15. Epigenomics in Cardiovascular Disease
9.16. Integrated OMICS Approaches
Funding
Conflicts of Interest
References
- Streisinger, G.; Walker, C.; Dower, N.; Knauber, D.; Singer, F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 1981, 291, 293–296. [Google Scholar] [CrossRef]
- Grunwald, D.J.; Streisinger, G. Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet. Res. 1992, 59, 103–116. [Google Scholar] [CrossRef]
- Kimmel, C.B. Genetics and early development of zebrafish. Trends Genet. 1989, 5, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Haffter, P.; Granato, M.; Brand, M.; Mullins, M.C.; Hammerschmidt, M.; Kane, D.A.; Odenthal, J.; van Eeden, F.J.; Jiang, Y.J.; Heisenberg, C.P.; et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 1996, 123, 1–36. [Google Scholar] [CrossRef]
- Kim, C.H.; Oda, T.; Itoh, M.; Jiang, D.; Artinger, K.B.; Chandrasekharappa, S.C.; Driever, W.; Chitnis, A.B. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 2000, 407, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef]
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef]
- Bedell, V.M.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.G.; Krug, R.G., 2nd; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.; et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Sood, R.; Burgess, S.M. Understanding and Editing the Zebrafish Genome. Adv. Genet. 2015, 92, 1–52. [Google Scholar] [CrossRef]
- Sung, Y.H.; Kim, J.M.; Kim, H.T.; Lee, J.; Jeon, J.; Jin, Y.; Choi, J.H.; Ban, Y.H.; Ha, S.J.; Kim, C.H.; et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014, 24, 125–131. [Google Scholar] [CrossRef]
- Postlethwait, J.H.; Yan, Y.L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.; et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Barbazuk, W.B.; Korf, I.; Kadavi, C.; Heyen, J.; Tate, S.; Wun, E.; Bedell, J.A.; McPherson, J.D.; Johnson, S.L. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000, 10, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.T.; Fishman, M.C. From Zebrafish to human: Modular medical models. Annu. Rev. Genom. Hum. Genet. 2002, 3, 311–340. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Halpern, M.E.; Rhee, J.; Goll, M.G.; Akitake, C.M.; Parsons, M.; Leach, S.D. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 2008, 5, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Langenau, D.M.; Feng, H.; Berghmans, S.; Kanki, J.P.; Kutok, J.L.; Look, A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2005, 102, 6068–6073. [Google Scholar] [CrossRef]
- Neuhauss, S.C.; Solnica-Krezel, L.; Schier, A.F.; Zwartkruis, F.; Stemple, D.L.; Malicki, J.; Abdelilah, S.; Stainier, D.Y.; Driever, W. Mutations affecting craniofacial development in zebrafish. Development 1996, 123, 357–367. [Google Scholar] [CrossRef]
- Chen, J.-N.; Haffter, P.; Odenthal, J.; Vogelsang, E.; Brand, M.; Eeden, F.J.M.v.; Furutani-Seiki, M.; Granato, M.; Hammerschmidt, M.; Heisenberg, C.-P.; et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development 1996, 123, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Stainier, D.Y.R.; Fouquet, B.; Chen, J.-N.; Warren, K.S.; Weinstein, B.M.; Meiler, S.E.; Mohideen, M.-A.P.K.; Neuhauss, S.C.F.; Solnica-Krezel, L.; Schier, A.F.; et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 1996, 123, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.W.; Herzog, W.; Santoro, M.M.; Mitchell, T.S.; Frantsve, J.; Jungblut, B.; Beis, D.; Scott, I.C.; D’Amico, L.A.; Ober, E.A.; et al. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev. Biol. 2007, 307, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011, 91, 279–288. [Google Scholar] [CrossRef]
- González-Rosa, J.M. Zebrafish Models of Cardiac Disease: From Fortuitous Mutants to Precision Medicine. Circ. Res. 2022, 130, 1803–1826. [Google Scholar] [CrossRef]
- Ding, Y.; Bu, H.; Xu, X. Modeling Inherited Cardiomyopathies in Adult Zebrafish for Precision Medicine. Front. Physiol. 2020, 11, 599244. [Google Scholar] [CrossRef]
- Beis, D.; Stainier, D.Y. In vivo cell biology: Following the zebrafish trend. Trends Cell Biol. 2006, 16, 105–112. [Google Scholar] [CrossRef]
- Bradford, Y.M.; Toro, S.; Ramachandran, S.; Ruzicka, L.; Howe, D.G.; Eagle, A.; Kalita, P.; Martin, R.; Taylor Moxon, S.A.; Schaper, K.; et al. Zebrafish Models of Human Disease: Gaining Insight into Human Disease at ZFIN. ILAR J. 2017, 58, 4–16. [Google Scholar] [CrossRef]
- Kopp, R.; Schwerte, T.; Pelster, B. Cardiac performance in the zebrafish breakdance mutant. J. Exp. Biol. 2005, 208, 2123–2134. [Google Scholar] [CrossRef]
- Hassel, D.; Scholz, E.P.; Trano, N.; Friedrich, O.; Just, S.; Meder, B.; Weiss, D.L.; Zitron, E.; Marquart, S.; Vogel, B.; et al. Deficient zebrafish ether-à-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation 2008, 117, 866–875. [Google Scholar] [CrossRef]
- Verkerk, A.O.; Remme, C.A. Zebrafish: A novel research tool for cardiac (patho)electrophysiology and ion channel disorders. Front. Physiol. 2012, 3, 255. [Google Scholar] [CrossRef] [PubMed]
- Sarantis, P.; Gaitanaki, C.; Beis, D. Ventricular remodeling of single-chambered myh6−/− adult zebrafish hearts occurs via a hyperplastic response and is accompanied by elastin deposition in the atrium. Cell Tissue Res. 2019, 378, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S.S.; Dóró, E.; Magyary, I.; Egginton, S.; Sík, A.; Müller, F. Optimisation of embryonic and larval ECG measurement in zebrafish for quantifying the effect of QT prolonging drugs. PLoS ONE 2013, 8, e60552. [Google Scholar] [CrossRef]
- Milan, D.J.; Jones, I.L.; Ellinor, P.T.; MacRae, C.A. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am. J. Physiol.-Heart Circ. Physiol. 2006, 291, H269–H273. [Google Scholar] [CrossRef] [PubMed]
- Huttner, I.G.; Trivedi, G.; Jacoby, A.; Mann, S.A.; Vandenberg, J.I.; Fatkin, D. A transgenic zebrafish model of a human cardiac sodium channel mutation exhibits bradycardia, conduction-system abnormalities and early death. J. Mol. Cell. Cardiol. 2013, 61, 123–132. [Google Scholar] [CrossRef]
- Ding, Y.; Dvornikov, A.V.; Ma, X.; Zhang, H.; Wang, Y.; Lowerison, M.; Packard, R.R.; Wang, L.; Chen, J.; Zhang, Y.; et al. Haploinsufficiency of mechanistic target of rapamycin ameliorates bag3 cardiomyopathy in adult zebrafish. Dis. Models Mech. 2019, 12, dmm040154. [Google Scholar] [CrossRef]
- Sun, X.; Hoage, T.; Bai, P.; Ding, Y.; Chen, Z.; Zhang, R.; Huang, W.; Jahangir, A.; Paw, B.; Li, Y.-G.; et al. Cardiac Hypertrophy Involves Both Myocyte Hypertrophy and Hyperplasia in Anemic Zebrafish. PLoS ONE 2009, 4, e6596. [Google Scholar] [CrossRef]
- Orr, N.; Arnaout, R.; Gula, L.J.; Spears, D.A.; Leong-Sit, P.; Li, Q.; Tarhuni, W.; Reischauer, S.; Chauhan, V.S.; Borkovich, M.; et al. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat. Commun. 2016, 7, 11303. [Google Scholar] [CrossRef]
- Scheid, L.-M.; Mosqueira, M.; Hein, S.; Kossack, M.; Juergensen, L.; Mueller, M.; Meder, B.; Fink, R.H.A.; Katus, H.A.; Hassel, D. Essential light chain S195 phosphorylation is required for cardiac adaptation under physical stress. Cardiovasc. Res. 2016, 111, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Bu, H.; Ding, Y.; Li, J.; Zhu, P.; Shih, Y.-H.; Wang, M.; Zhang, Y.; Lin, X.; Xu, X. Inhibition of mTOR or MAPK ameliorates vmhcl/myh7 cardiomyopathy in zebrafish. JCI Insight 2021, 6, e154215. [Google Scholar] [CrossRef]
- Chi, N.C.; Bussen, M.; Brand-Arzamendi, K.; Ding, C.; Olgin, J.E.; Shaw, R.M.; Martin, G.R.; Stainier, D.Y.R. Cardiac conduction is required to preserve cardiac chamber morphology. Proc. Natl. Acad. Sci. USA 2010, 107, 14662–14667. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Z.; Chen, C.; Yao, S.; Yang, Q.; Li, F.; He, X.; Ai, C.; Wang, M.; Guan, M.-X. Deletion of Gtpbp3 in zebrafish revealed the hypertrophic cardiomyopathy manifested by aberrant mitochondrial tRNA metabolism. Nucleic Acids Res. 2019, 47, 5341–5355. [Google Scholar] [CrossRef]
- Cheng, F.; Miao, L.; Wu, Q.; Gong, X.; Xiong, J.; Zhang, J. Vinculin b deficiency causes epicardial hyperplasia and coronary vessel disorganization in zebrafish. Development 2016, 143, 3522–3531. [Google Scholar] [CrossRef]
- Asimaki, A.; Kapoor, S.; Plovie, E.; Karin Arndt, A.; Adams, E.; Liu, Z.; James, C.A.; Judge, D.P.; Calkins, H.; Churko, J.; et al. Identification of a New Modulator of the Intercalated Disc in a Zebrafish Model of Arrhythmogenic Cardiomyopathy. Sci. Transl. Med. 2014, 6, 240ra274. [Google Scholar] [CrossRef]
- Rottbauer, W.; Just, S.; Wessels, G.; Trano, N.; Most, P.; Katus, H.A.; Fishman, M.C. VEGF–PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 2005, 19, 1624–1634. [Google Scholar] [CrossRef]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart Regeneration in Zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, A.J.; Huq, A.; Weinstein, B.M.; Walker, C.; Fishman, M.; Stainier, D.Y.R. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002, 31, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Zebrowski, D.C.; Becker, R.; Engel, F.B. Towards regenerating the mammalian heart: Challenges in evaluating experimentally induced adult mammalian cardiomyocyte proliferation. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H1045–H1054. [Google Scholar] [CrossRef] [PubMed]
- Vihtelic, T.S.; Hyde, D.R. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J. Neurobiol. 2000, 44, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Kroehne, V.; Freudenreich, D.; Hans, S.; Kaslin, J.; Brand, M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 2011, 138, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Wullimann, M.F.; Becker, C.G.; Bernhardt, R.R.; Schachner, M. Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 1997, 377, 577–595. [Google Scholar] [CrossRef]
- Bise, T.; Sallin, P.; Pfefferli, C.; Jaźwińska, A. Multiple cryoinjuries modulate the efficiency of zebrafish heart regeneration. Sci. Rep. 2020, 10, 11551. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Karra, R.; Dickson, A.L.; Poss, K.D. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013, 382, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Engel, F.B.; Hsieh, P.C.H.; Lee, R.T.; Keating, M.T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. USA 2006, 103, 15546–15551. [Google Scholar] [CrossRef] [PubMed]
- Sander, V.; Suñe, G.; Jopling, C.; Morera, C.; Izpisua Belmonte, J.C. Isolation and in vitro culture of primary cardiomyocytes from adult zebrafish hearts. Nat. Protoc. 2013, 8, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Suñé, G.; Faucherre, A.; Fabregat, C.; Izpisua Belmonte, J.C. Hypoxia induces myocardial regeneration in zebrafish. Circulation 2012, 126, 3017–3027. [Google Scholar] [CrossRef]
- Parente, V.; Balasso, S.; Pompilio, G.; Verduci, L.; Colombo, G.I.; Milano, G.; Guerrini, U.; Squadroni, L.; Cotelli, F.; Pozzoli, O.; et al. Hypoxia/reoxygenation cardiac injury and regeneration in zebrafish adult heart. PLoS ONE 2013, 8, e53748. [Google Scholar] [CrossRef]
- Lam, N.T.; Currie, P.D.; Lieschke, G.J.; Rosenthal, N.A.; Kaye, D.M. Nerve growth factor stimulates cardiac regeneration via cardiomyocyte proliferation in experimental heart failure. PLoS ONE 2012, 7, e53210. [Google Scholar] [CrossRef]
- Chablais, F.; Veit, J.; Rainer, G.; Jaźwińska, A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev. Biol. 2011, 11, 21. [Google Scholar] [CrossRef]
- González-Rosa, J.M.; Martín, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef]
- González-Rosa, J.M.; Burns, C.E.; Burns, C.G. Zebrafish heart regeneration: 15 years of discoveries. Regeneration 2017, 4, 105–123. [Google Scholar] [CrossRef]
- González-Rosa, J.M.; Peralta, M.; Mercader, N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012, 370, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Schindler, Y.L.; Garske, K.M.; Wang, J.; Firulli, B.A.; Firulli, A.B.; Poss, K.D.; Yelon, D. Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 2014, 141, 3112–3122. [Google Scholar] [CrossRef] [PubMed]
- Vaparanta, K.; Jokilammi, A.; Paatero, I.; Merilahti, J.A.; Heliste, J.; Hemanthakumar, K.A.; Kivelä, R.; Alitalo, K.; Taimen, P.; Elenius, K. STAT5b is a key effector of NRG-1/ERBB4-mediated myocardial growth. EMBO Rep. 2023, 24, e56689. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, Z.; Yang, Y.; Feng, D.; Dong, Y.; Garbutt, T.A.; Hu, Z.; Wang, L.; Luan, C.; Cooper, C.D.; et al. Functional coordination of non-myocytes plays a key role in adult zebrafish heart regeneration. EMBO Rep. 2021, 22, e52901. [Google Scholar] [CrossRef]
- Hoage, T.; Sun, X.; Xu, X. Functions of the Wnt/β-catenin pathway in an anemia-induced zebrafish model of cardiomyopathy are location dependent. Biochem. Biophys. Res. Commun. 2011, 415, 490–496. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, X.; Redfield, M.; Kushwaha, S.; Xu, X. Target of rapamcyin (TOR)-based therapeutics for cardiomyopathy. Cell Cycle 2012, 11, 428–429. [Google Scholar] [CrossRef]
- Shih, Y.-H.; Zhang, Y.; Ding, Y.; Ross, C.A.; Li, H.; Olson, T.M.; Xu, X. Cardiac Transcriptome and Dilated Cardiomyopathy Genes in Zebrafish. Circ. Cardiovasc. Genet. 2015, 8, 261–269. [Google Scholar] [CrossRef]
- Gerull, B.; Gramlich, M.; Atherton, J.; McNabb, M.; Trombitás, K.; Sasse-Klaassen, S.; Seidman, J.G.; Seidman, C.; Granzier, H.; Labeit, S.; et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002, 30, 201–204. [Google Scholar] [CrossRef]
- Xu, X.; Meiler, S.E.; Zhong, T.P.; Mohideen, M.; Crossley, D.A.; Burggren, W.W.; Fishman, M.C. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 2002, 30, 205–209. [Google Scholar] [CrossRef]
- Seeley, M.; Huang, W.; Chen, Z.; Wolff, W.O.; Lin, X.; Xu, X. Depletion of Zebrafish Titin Reduces Cardiac Contractility by Disrupting the Assembly of Z-Discs and A-Bands. Circ. Res. 2007, 100, 238–245. [Google Scholar] [CrossRef]
- Steffen, L.S.; Guyon, J.R.; Vogel, E.D.; Howell, M.H.; Zhou, Y.; Weber, G.J.; Zon, L.I.; Kunkel, L.M. The zebrafish runzel muscular dystrophy is linked to the titin gene. Dev. Biol. 2007, 309, 180–192. [Google Scholar] [CrossRef]
- Knöll, R.; Postel, R.; Wang, J.; Krätzner, R.; Hennecke, G.; Vacaru, A.M.; Vakeel, P.; Schubert, C.; Murthy, K.; Rana, B.K.; et al. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation 2007, 116, 515–525. [Google Scholar] [CrossRef]
- Schönberger, J.; Levy, H.; Grünig, E.; Sangwatanaroj, S.; Fatkin, D.; MacRae, C.; Stäcker, H.; Halpin, C.; Eavey, R.; Philbin, E.F.; et al. Dilated cardiomyopathy and sensorineural hearing loss: A heritable syndrome that maps to 6q23-24. Circulation 2000, 101, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Schönberger, J.; Wang, L.; Shin, J.T.; Kim, S.D.; Depreux, F.F.; Zhu, H.; Zon, L.; Pizard, A.; Kim, J.B.; Macrae, C.A.; et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005, 37, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Hassel, D.; Dahme, T.; Erdmann, J.; Meder, B.; Huge, A.; Stoll, M.; Just, S.; Hess, A.; Ehlermann, P.; Weichenhan, D.; et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat. Med. 2009, 15, 1281–1288. [Google Scholar] [CrossRef]
- Ho, Y.L.; Lin, Y.H.; Tsai, W.Y.; Hsieh, F.J.; Tsai, H.J. Conditional antisense-knockdown of zebrafish cardiac troponin C as a new animal model for dilated cardiomyopathy. Circ. J. 2009, 73, 1691–1697. [Google Scholar] [CrossRef]
- Van’t Padje, S.; Chaudhry, B.; Severijnen, L.A.; van der Linde, H.C.; Mientjes, E.J.; Oostra, B.A.; Willemsen, R. Reduction in fragile X related 1 protein causes cardiomyopathy and muscular dystrophy in zebrafish. J. Exp. Biol. 2009, 212, 2564–2570. [Google Scholar] [CrossRef]
- Dhandapany, P.S.; Razzaque, M.A.; Muthusami, U.; Kunnoth, S.; Edwards, J.J.; Mulero-Navarro, S.; Riess, I.; Pardo, S.; Sheng, J.; Rani, D.S.; et al. RAF1 mutations in childhood-onset dilated cardiomyopathy. Nat. Genet. 2014, 46, 635–639. [Google Scholar] [CrossRef] [PubMed]
- González-Solá, M.; Al-Khayat, H.A.; Behra, M.; Kensler, R.W. Zebrafish cardiac muscle thick filaments: Isolation technique and three-dimensional structure. Biophys. J. 2014, 106, 1671–1680. [Google Scholar] [CrossRef]
- Paavola, J.; Schliffke, S.; Rossetti, S.; Kuo, I.Y.T.; Yuan, S.; Sun, Z.; Harris, P.C.; Torres, V.E.; Ehrlich, B.E. Polycystin-2 mutations lead to impaired calcium cycling in the heart and predispose to dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2013, 58, 199–208. [Google Scholar] [CrossRef]
- Lawson, N.D.; Weinstein, B.M. Arteries and veins: Making a difference with zebrafish. Nat. Rev. Genet. 2002, 3, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Gray, C.; Packham, I.M.; Wurmser, F.; Eastley, N.C.; Hellewell, P.G.; Ingham, P.W.; Crossman, D.C.; Chico, T.J.A. Ischemia is not required for arteriogenesis in zebrafish embryos. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Chiba, A.; Fukumoto, M.; Morooka, N.; Mochizuki, N. Zebrafish Vascular Development: General and Tissue-Specific Regulation. J. Lipid Atheroscler. 2021, 10, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.V.; Monzo, K.; Cha, Y.R.; Pan, W.; Weinstein, B.M. Vascular development in the zebrafish. Cold Spring Harb. Perspect. Med. 2012, 2, a006684. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.G.; Wang, L.; Jia, X.E.; Liu, Y.J.; Dong, Z.W.; Jin, Y.; Chen, Y.; Deng, M.; Zhou, Y.; Zhou, Y.; et al. Activated N-Ras signaling regulates arterial-venous specification in zebrafish. J. Hematol. Oncol. 2013, 6, 34. [Google Scholar] [CrossRef]
- Herold, J.; Pipp, F.; Fernandez, B.; Xing, Z.; Heil, M.; Tillmanns, H.; Braun-Dullaeus, R.C. Transplantation of monocytes: A novel strategy for in vivo augmentation of collateral vessel growth. Hum. Gene Ther. 2004, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Deng, Y.; Mukhopadhyay, A.; Lanahan, A.A.; Zhuang, Z.W.; Moodie, K.L.; Mulligan-Kehoe, M.J.; Byzova, T.V.; Peterson, R.T.; Simons, M. ERK1/2-Akt1 crosstalk regulates arteriogenesis in mice and zebrafish. J. Clin. Investig. 2010, 120, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.M.; Poole, A.W. Using zebrafish (Danio rerio) to assess gene function in thrombus formation. Methods Mol. Biol. 2012, 788, 305–319. [Google Scholar] [CrossRef]
- Jagadeeswaran, P.; Rao, P. Role of KIAA0472, a Novel Ser/Thr Kinase in zebrafish thrombosis. In Proceedings of the International Conference on Zebrafish Development and Genetics, Madison, WI, USA, 14–18 June 2006. [Google Scholar]
- Gregory, M.; Hanumanthaiah, R.; Jagadeeswaran, P. Genetic analysis of hemostasis and thrombosis using vascular occlusion. Blood Cells Mol. Dis. 2002, 29, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kretz, C.A.; Maeder, M.L.; Huarng, M.C.; Richter, C.E.; Vo, A.H.; Tsao, P.; Swaroop, A.; Joung, J.K.; Shavit, J.A. A Zebrafish Model Of Antithrombin III Deficiency Displays Bleeding and Thrombosis Secondary To Disseminated Intravascular Coagulation. Blood 2013, 122, 200. [Google Scholar] [CrossRef]
- Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [CrossRef] [PubMed]
- Soranzo, N.; Spector, T.D.; Mangino, M.; Kühnel, B.; Rendon, A.; Teumer, A.; Willenborg, C.; Wright, B.; Chen, L.; Li, M.; et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009, 41, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Tijssen, M.R.; Thijssen-Timmer, D.C.; Gusnanto, A.; Steward, M.; Burns, P.; Langford, C.F.; Ellis, P.D.; Dudbridge, F.; Zwaginga, J.-J.; et al. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood 2006, 109, 3260–3269. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, P.; Sheehan, J.P.; Craig, F.E.; Troyer, D. Identification and characterization of zebrafish thrombocytes. Br. J. Haematol. 1999, 107, 731–738. [Google Scholar] [CrossRef]
- Jagadeeswaran, P.; Gregory, M.; Day, K.; Cykowski, M.; Thattaliyath, B. Zebrafish: A genetic model for hemostasis and thrombosis. J. Thromb. Haemost. 2005, 3, 46–53. [Google Scholar] [CrossRef]
- Lin, H.F.; Traver, D.; Zhu, H.; Dooley, K.; Paw, B.H.; Zon, L.I.; Handin, R.I. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 2005, 106, 3803–3810. [Google Scholar] [CrossRef]
- Schurgers, E.; Moorlag, M.; Hemker, C.; Lindhout, T.; Kelchtermans, H.; de Laat, B. Thrombin Generation in Zebrafish Blood. PLoS ONE 2016, 11, e0149135. [Google Scholar] [CrossRef]
- Tournoij, E.; Weber, G.J.; Akkerman, J.W.N.; De Groot, P.G.; Zon, L.I.; Moll, F.L.; Schulte-Merker, S. Mlck1a is expressed in zebrafish thrombocytes and is an essential component of thrombus formation. J. Thromb. Haemost. 2010, 8, 588–595. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Liu, H.C.; Guo, S.Y.; Xia, B.; Song, R.S.; Lao, Q.C.; Xuan, Y.X.; Li, C.Q. A Zebrafish Thrombosis Model for Assessing Antithrombotic Drugs. Zebrafish 2016, 13, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Gut, P.; Reischauer, S.; Stainier, D.Y.R.; Arnaout, R. Little Fish, Big Data: Zebrafish as a Model for Cardiovascular and Metabolic Disease. Physiol. Rev. 2017, 97, 889–938. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lan, C.; Zhao, D.; Wang, N.; Du, D.; Luo, H.; Lu, H.; Peng, Z.; Wang, Y.; Qiao, Z.; et al. Wuliangye Baijiu but not ethanol reduces cardiovascular disease risks in a zebrafish thrombosis model. Npj Sci. Food 2022, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Gauvrit, S.; Bossaer, J.; Lee, J.; Collins, M.M. Modeling Human Cardiac Arrhythmias: Insights from Zebrafish. J. Cardiovasc. Dev. Dis. 2022, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Langheinrich, U. Zebrafish: A new model on the pharmaceutical catwalk. Bioessays 2003, 25, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Pott, A.; Rottbauer, W.; Just, S. Functional genomics in zebrafish as a tool to identify novel antiarrhythmic targets. Curr. Med. Chem. 2014, 21, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Warren, K.S.; Fishman, M.C. “Physiological genomics”: Mutant screens in zebrafish. Am. J. Physiol. 1998, 275, H1–H7. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.P. The zebrafish: A new model organism for integrative physiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R3–R9. [Google Scholar] [CrossRef]
- Jorgensen, E.M.; Mango, S.E. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 2002, 3, 356–369. [Google Scholar] [CrossRef]
- Créton, R.; Speksnijder, J.E.; Jaffe, L.F. Patterns of free calcium in zebrafish embryos. J. Cell Sci. 1998, 111 Pt 12, 1613–1622. [Google Scholar] [CrossRef]
- Vornanen, M.; Hassinen, M. Zebrafish heart as a model for human cardiac electrophysiology. Channels 2016, 10, 101–110. [Google Scholar] [CrossRef]
- Leong, I.U.; Skinner, J.R.; Shelling, A.N.; Love, D.R. Zebrafish as a model for long QT syndrome: The evidence and the means of manipulating zebrafish gene expression. Acta Physiol. 2010, 199, 257–276. [Google Scholar] [CrossRef]
- Nemtsas, P.; Wettwer, E.; Christ, T.; Weidinger, G.; Ravens, U. Adult zebrafish heart as a model for human heart? An electrophysiological study. J. Mol. Cell. Cardiol. 2010, 48, 161–171. [Google Scholar] [CrossRef]
- Baker, K.; Warren, K.S.; Yellen, G.; Fishman, M.C. Defective “pacemaker” current (Ih) in a zebrafish mutant with a slow heart rate. Proc. Natl. Acad. Sci. USA 1997, 94, 4554–4559. [Google Scholar] [CrossRef]
- Brette, F.; Luxan, G.; Cros, C.; Dixey, H.; Wilson, C.; Shiels, H.A. Characterization of isolated ventricular myocytes from adult zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 2008, 374, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Müller, I.I.; Melville, D.B.; Tanwar, V.; Rybski, W.M.; Mukherjee, A.; Shoemaker, M.B.; Wang, W.-D.; Schoenhard, J.A.; Roden, D.M.; Darbar, D.; et al. Functional modeling in zebrafish demonstrates that the atrial-fibrillation-associated gene GREM2 regulates cardiac laterality, cardiomyocyte differentiation and atrial rhythm. Dis. Models Mech. 2013, 6, 332–341. [Google Scholar] [CrossRef]
- Angom, R.S.; Joshi, A.; Patowary, A.; Sivadas, A.; Ramasamy, S.; Shamsudheen, K.V.; Kaushik, K.; Sabharwal, A.; Lalwani, M.K.; Subburaj, K.; et al. Forward genetic screen using a gene-breaking trap approach identifies a novel role of grin2bb-associated RNA transcript (grin2bbART) in zebrafish heart function. Front. Cell Dev. Biol. 2024. [Google Scholar] [CrossRef]
- Parker, T.; Libourel, P.A.; Hetheridge, M.J.; Cumming, R.I.; Sutcliffe, T.P.; Goonesinghe, A.C.; Ball, J.S.; Owen, S.F.; Chomis, Y.; Winter, M.J. A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. J. Pharmacol. Toxicol. Methods 2014, 69, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.M. Zebrafish as a model to explore cell metabolism. Trends Endocrinol. Metab. 2014, 25, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Hata, K.; Matsuoka, I.; Zang, L.; Kim, Y.; Chu, D.; Juneja, L.R.; Nishimura, N.; Shimada, Y. Anti-Obesity Natural Products Tested in Juvenile Zebrafish Obesogenic Tests and Mouse 3T3-L1 Adipogenesis Assays. Molecules 2020, 25, 5840. [Google Scholar] [CrossRef]
- Misselbeck, K.; Parolo, S.; Lorenzini, F.; Savoca, V.; Leonardelli, L.; Bora, P.; Morine, M.J.; Mione, M.C.; Domenici, E.; Priami, C. A network-based approach to identify deregulated pathways and drug effects in metabolic syndrome. Nat. Commun. 2019, 10, 5215. [Google Scholar] [CrossRef]
- Asaoka, Y.; Terai, S.; Sakaida, I.; Nishina, H. The expanding role of fish models in understanding non-alcoholic fatty liver disease. Dis. Model. Mech. 2013, 6, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Shimada, Y.; Zang, L.; Terasawa, M.; Nishiura, K.; Matsuda, K.; Toombs, C.; Langdon, C.; Nishimura, N. Novel Anti-Obesity Properties of Palmaria mollis in Zebrafish and Mouse Models. Nutrients 2018, 10, 1401. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.I. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010, 363, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Gieger, C.; Radhakrishnan, A.; Cvejic, A.; Tang, W.; Porcu, E.; Pistis, G.; Serbanovic-Canic, J.; Elling, U.; Goodall, A.H.; Labrune, Y.; et al. New gene functions in megakaryopoiesis and platelet formation. Nature 2011, 480, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, S.; Huang, J.; Liu, Y.; Wang, Q.; Chen, J.; Sun, L.; Tu, W. Mitochondrial dysfunction in metabolic disorders induced by per- and polyfluoroalkyl substance mixtures in zebrafish larvae. Environ. Int. 2023, 176, 107977. [Google Scholar] [CrossRef]
- Eisen, J.S.; Weston, J.A. Development of the Neural Crest in the Zebrafish. Dev. Biol. 1993, 159, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Gut, P.; Baeza-Raja, B.; Andersson, O.; Hasenkamp, L.; Hsiao, J.; Hesselson, D.; Akassoglou, K.; Verdin, E.; Hirschey, M.D.; Stainier, D.Y.R. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat. Chem. Biol. 2013, 9, 97–104. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, C.; Lu, G.; Chen, Z. Effect of adenosine monophosphate-activated protein kinase-p53-Krüppel-like factor 2a pathway in hyperglycemia-induced cardiac remodeling in adult zebrafish. J. Diabetes Investig. 2021, 12, 320–333. [Google Scholar] [CrossRef]
- Dorsemans, A.C.; Soulé, S.; Weger, M.; Bourdon, E.; Lefebvre d’Hellencourt, C.; Meilhac, O.; Diotel, N. Impaired constitutive and regenerative neurogenesis in adult hyperglycemic zebrafish. J. Comp. Neurol. 2017, 525, 442–458. [Google Scholar] [CrossRef]
- Dorsemans, A.C.; Lefebvre d’Hellencourt, C.; Ait-Arsa, I.; Jestin, E.; Meilhac, O.; Diotel, N. Acute and Chronic Models of Hyperglycemia in Zebrafish: A Method to Assess the Impact of Hyperglycemia on Neurogenesis and the Biodistribution of Radiolabeled Molecules. J. Vis. Exp. 2017, 124, e55203. [Google Scholar] [CrossRef]
- Couret, D.; Bourane, S.; Catan, A.; Nativel, B.; Planesse, C.; Dorsemans, A.C.; Ait-Arsa, I.; Cournot, M.; Rondeau, P.; Patche, J.; et al. A hemorrhagic transformation model of mechanical stroke therapy with acute hyperglycemia in mice. J. Comp. Neurol. 2018, 526, 1006–1016. [Google Scholar] [CrossRef]
- Moss, J.B.; Koustubhan, P.; Greenman, M.; Parsons, M.J.; Walter, I.; Moss, L.G. Regeneration of the pancreas in adult zebrafish. Diabetes 2009, 58, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Delaspre, F.; Beer, R.L.; Rovira, M.; Huang, W.; Wang, G.; Gee, S.; Vitery Mdel, C.; Wheelan, S.J.; Parsons, M.J. Centroacinar Cells Are Progenitors That Contribute to Endocrine Pancreas Regeneration. Diabetes 2015, 64, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.; Sarras, M.P., Jr.; Intine, R.V. Limb regeneration is impaired in an adult zebrafish model of diabetes mellitus. Wound Repair Regen. 2010, 18, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Benchoula, K.; Khatib, A.; Quzwain, F.M.C.; Che Mohamad, C.A.; Wan Sulaiman, W.M.A.; Abdul Wahab, R.; Ahmed, Q.U.; Abdul Ghaffar, M.; Saiman, M.Z.; Alajmi, M.F.; et al. Optimization of Hyperglycemic Induction in Zebrafish and Evaluation of Its Blood Glucose Level and Metabolite Fingerprint Treated with Psychotria malayana Jack Leaf Extract. Molecules 2019, 24, 1506. [Google Scholar] [CrossRef] [PubMed]
- Karanth, S.; Chaurasia, B.; Bowman, F.M.; Tippetts, T.S.; Holland, W.L.; Summers, S.A.; Schlegel, A. FOXN3 controls liver glucose metabolism by regulating gluconeogenic substrate selection. Physiol. Rep. 2019, 7, e14238. [Google Scholar] [CrossRef]
- Kimmel, R.A.; Dobler, S.; Schmitner, N.; Walsen, T.; Freudenblum, J.; Meyer, D. Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment. Sci. Rep. 2015, 5, 14241. [Google Scholar] [CrossRef]
- Maddison, L.A.; Joest, K.E.; Kammeyer, R.M.; Chen, W. Skeletal muscle insulin resistance in zebrafish induces alterations in β-cell number and glucose tolerance in an age- and diet-dependent manner. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E662–E669. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Nishimura, Y.; Zang, L.; Hirano, M.; Shimada, Y.; Wang, Z.; Umemoto, N.; Kuroyanagi, J.; Nishimura, N.; Tanaka, T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010, 10, 21. [Google Scholar] [CrossRef]
- Hasumura, T.; Shimada, Y.; Kuroyanagi, J.; Nishimura, Y.; Meguro, S.; Takema, Y.; Tanaka, T. Green tea extract suppresses adiposity and affects the expression of lipid metabolism genes in diet-induced obese zebrafish. Nutr. Metab. 2012, 9, 73. [Google Scholar] [CrossRef]
- Forn-Cuní, G.; Varela, M.; Fernández-Rodríguez, C.M.; Figueras, A.; Novoa, B. Liver immune responses to inflammatory stimuli in a diet-induced obesity model of zebrafish. J. Endocrinol. 2015, 224, 159–170. [Google Scholar] [CrossRef]
- Jais, A.; Einwallner, E.; Sharif, O.; Gossens, K.; Lu Tess, T.-H.; Soyal Selma, M.; Medgyesi, D.; Neureiter, D.; Paier-Pourani, J.; Dalgaard, K.; et al. Heme Oxygenase-1 Drives Metaflammation and Insulin Resistance in Mouse and Man. Cell 2014, 158, 25–40. [Google Scholar] [CrossRef]
- Lackey, D.E.; Burk, D.H.; Ali, M.R.; Mostaedi, R.; Smith, W.H.; Park, J.; Scherer, P.E.; Seay, S.A.; McCoin, C.S.; Bonaldo, P.; et al. Contributions of adipose tissue architectural and tensile properties toward defining healthy and unhealthy obesity. Am. J. Physiol. -Endocrinol. Metab. 2014, 306, E233–E246. [Google Scholar] [CrossRef]
- Wernstedt Asterholm, I.; Tao, C.; Morley, T.S.; Wang, Q.A.; Delgado-Lopez, F.; Wang, Z.V.; Scherer, P.E. Adipocyte Inflammation Is Essential for Healthy Adipose Tissue Expansion and Remodeling. Cell Metab. 2014, 20, 103–118. [Google Scholar] [CrossRef]
- Seth, A.; Stemple, D.L.; Barroso, I. The emerging use of zebrafish to model metabolic disease. Dis. Models Mech. 2013, 6, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A. Studying lipoprotein trafficking in zebrafish, the case of chylomicron retention disease. J. Mol. Med. 2015, 93, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.J.; Trent, C.M.; Rawls, J.F. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J. Lipid Res. 2009, 50, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Fraher, D.; Sanigorski, A.; Mellett, N.A.; Meikle, P.J.; Sinclair, A.J.; Gibert, Y. Zebrafish Embryonic Lipidomic Analysis Reveals that the Yolk Cell Is Metabolically Active in Processing Lipid. Cell Rep. 2016, 14, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Imrie, D.; Sadler, K.C. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev. Dyn. 2010, 239, 3013–3023. [Google Scholar] [CrossRef]
- Leibold, S.; Hammerschmidt, M. Long-Term Hyperphagia and Caloric Restriction Caused by Low- or High-Density Husbandry Have Differential Effects on Zebrafish Postembryonic Development, Somatic Growth, Fat Accumulation and Reproduction. PLoS ONE 2015, 10, e0120776. [Google Scholar] [CrossRef]
- McMenamin, S.K.; Minchin, J.E.N.; Gordon, T.N.; Rawls, J.F.; Parichy, D.M. Dwarfism and Increased Adiposity in the gh1 Mutant Zebrafish vizzini. Endocrinology 2013, 154, 1476–1487. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-Y.; Chen, C.-F.; Rajendran, R.S.; Shen, C.-N.; Chen, T.-H.; Yen, C.-C.; Chuang, C.-K.; Lin, D.-S.; Hsiao, C.-D. Overexpression of Akt1 Enhances Adipogenesis and Leads to Lipoma Formation in Zebrafish. PLoS ONE 2012, 7, e36474. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M.; Halaas, J.L. Leptin and the regulation of body weight in mammals. Nature 1998, 395, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.; Page-McCaw, P.S.; Chen, W.; Cone, R.D. Leptin signaling regulates glucose homeostasis, but not adipostasis, in the zebrafish. Proc. Natl. Acad. Sci. USA 2016, 113, 3084–3089. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.; Maddison, L.A.; Chen, W. Zebrafish as a Model for Obesity and Diabetes. Front. Cell Dev. Biol. 2018, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Smolińska, K.; Sobczyński, J.; Szopa, A.; Wnorowski, A.; Tomaszewska, E.; Muszyński, S.; Winiarska-Mieczan, A.; Czernecki, T.; Bielak, A.; Dobrowolska, K.; et al. Innovative high fat diet establishes a novel zebrafish model for the study of visceral obesity. Sci. Rep. 2024, 14, 3012. [Google Scholar] [CrossRef] [PubMed]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Minchin, J.E.N.; Dahlman, I.; Harvey, C.J.; Mejhert, N.; Singh, M.K.; Epstein, J.A.; Arner, P.; Torres-Vázquez, J.; Rawls, J.F. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc. Natl. Acad. Sci. USA 2015, 112, 4363–4368. [Google Scholar] [CrossRef]
- Minchin, J.E.N.; Rawls, J.F. Chapter 3—In vivo Analysis of White Adipose Tissue in Zebrafish. In Methods in Cell Biology; Detrich, H.W., Westerfield, M., Zon, L.I., Eds.; Academic Press: New York, NY, USA, 2011; Volume 105, pp. 63–86. [Google Scholar]
- Schlegel, A.; Gut, P. Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cell. Mol. Life Sci. 2015, 72, 2249–2260. [Google Scholar] [CrossRef]
- Farber, S.A.; Pack, M.; Ho, S.-Y.; Johnson, I.D.; Wagner, D.S.; Dosch, R.; Mullins, M.C.; Hendrickson, H.S.; Hendrickson, E.K.; Halpern, M.E. Genetic Analysis of Digestive Physiology Using Fluorescent Phospholipid Reporters. Science 2001, 292, 1385–1388. [Google Scholar] [CrossRef]
- Ho, S.-Y.; Lorent, K.; Pack, M.; Farber, S.A. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006, 3, 289–300. [Google Scholar] [CrossRef]
- Pérez-Victoria, F.J.; Schindler, C.; Magadán, J.G.; Mardones, G.A.; Delevoye, C.; Romao, M.; Raposo, G.; Bonifacino, J.S. Ang2/Fat-Free Is a Conserved Subunit of the Golgi-associated Retrograde Protein Complex. Mol. Biol. Cell 2010, 21, 3386–3395. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Mietus-Snyder, M.; Valente, A.; Schwarz, J.-M.; Lustig, R.H. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lorent, K.; Gong, W.; Koo, K.A.; Waisbourd-Zinman, O.; Karjoo, S.; Zhao, X.; Sealy, I.; Kettleborough, R.N.; Stemple, D.L.; Windsor, P.A.; et al. Identification of a plant isoflavonoid that causes biliary atresia. Sci. Transl. Med. 2015, 7, 286ra267. [Google Scholar] [CrossRef]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human Fatty Liver Disease: Old Questions and New Insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A. Studying non-alcoholic fatty liver disease with zebrafish: A confluence of optics, genetics, and physiology. Cell. Mol. Life Sci. 2012, 69, 3953–3961. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A.; Stainier, D.Y.R. Microsomal Triglyceride Transfer Protein Is Required for Yolk Lipid Utilization and Absorption of Dietary Lipids in Zebrafish Larvae. Biochemistry 2006, 45, 15179–15187. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Page-McCaw, P.; Chen, W. FGF1 Mediates Overnutrition-Induced Compensatory β-Cell Differentiation. Diabetes 2015, 65, 96–109. [Google Scholar] [CrossRef]
- Karanth, S.; Tran, V.M.; Kuberan, B.; Schlegel, A. Polyunsaturated fatty acyl-coenzyme As are inhibitors of cholesterol biosynthesis in zebrafish and mice. Dis. Models Mech. 2013, 6, 1365–1377. [Google Scholar] [CrossRef]
- Sakaguchi, T.F.; Sadler, K.C.; Crosnier, C.; Stainier, D.Y.R. Endothelial Signals Modulate Hepatocyte Apicobasal Polarization in Zebrafish. Curr. Biol. 2008, 18, 1565–1571. [Google Scholar] [CrossRef]
- Schaub, M.; Nussbaum, J.; Verkade, H.; Ober, E.A.; Stainier, D.Y.R.; Sakaguchi, T.F. Mutation of zebrafish Snapc4 is associated with loss of the intrahepatic biliary network. Dev. Biol. 2012, 363, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Hugo, S.E.; Cruz-Garcia, L.; Karanth, S.; Anderson, R.M.; Stainier, D.Y.R.; Schlegel, A. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 2012, 26, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Matthews, R.P.; Lorent, K.; Mañoral-Mobias, R.; Huang, Y.; Gong, W.; Murray, I.V.J.; Blair, I.A.; Pack, M. TNFα-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish s-adenosylhomocysteine hydrolase. Development 2009, 136, 865–875. [Google Scholar] [CrossRef]
- Nussbaum, J.M.; Liu, L.J.; Hasan, S.A.; Schaub, M.; McClendon, A.; Stainier, D.Y.R.; Sakaguchi, T.F. Homeostatic generation of reactive oxygen species protects the zebrafish liver from steatosis. Hepatology 2013, 58, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Wu, S.-Y.; Baek, J.-I.; Choi, S.Y.; Su, Y.; Flynn, C.R.; Gamse, J.T.; Ess, K.C.; Hardiman, G.; Lipschutz, J.H.; et al. A Post-Developmental Genetic Screen for Zebrafish Models of Inherited Liver Disease. PLoS ONE 2015, 10, e0125980. [Google Scholar] [CrossRef]
- Fang, L.; Harkewicz, R.; Hartvigsen, K.; Wiesner, P.; Choi, S.-H.; Almazan, F.; Pattison, J.; Deer, E.; Sayaphupha, T.; Dennis, E.A.; et al. Oxidized Cholesteryl Esters and Phospholipids in Zebrafish Larvae Fed a High Cholesterol Diet: Macrophage Binding and Activation. J. Biol. Chem. 2010, 285, 32343–32351. [Google Scholar] [CrossRef]
- Breslow, J.L. Mouse Models of Atherosclerosis. Science 1996, 272, 685–688. [Google Scholar] [CrossRef]
- Bradley, M.N.; Hong, C.; Chen, M.; Joseph, S.B.; Wilpitz, D.C.; Wang, X.; Lusis, A.J.; Collins, A.; Hseuh, W.A.; Collins, J.L.; et al. Ligand activation of LXRβ reverses atherosclerosis and cellular cholesterol overload in mice lacking LXRα and apoE. J. Clin. Investig. 2007, 117, 2337–2346. [Google Scholar] [CrossRef]
- Cruz-Garcia, L.; Schlegel, A. Lxr-driven enterocyte lipid droplet formation delays transport of ingested lipids. J. Lipid Res. 2014, 55, 1944–1958. [Google Scholar] [CrossRef]
- Ferreira, J. New zebrafish transgenic lines to further understand diabetes. Lab Anim. 2023, 52, 175. [Google Scholar] [CrossRef]
- Zang, L.; Shimada, Y.; Nishimura, N. Development of a Novel Zebrafish Model for Type 2 Diabetes Mellitus. Sci. Rep. 2017, 7, 1461. [Google Scholar] [CrossRef]
- Andersson, O.; Adams, B.A.; Yoo, D.; Ellis, G.C.; Gut, P.; Anderson, R.M.; German, M.S.; Stainier, D.Y.R. Adenosine Signaling Promotes Regeneration of Pancreatic β Cells In Vivo. Cell Metab. 2012, 15, 885–894. [Google Scholar] [CrossRef] [PubMed]
- D’Amour, K.A.; Bang, A.G.; Eliazer, S.; Kelly, O.G.; Agulnick, A.D.; Smart, N.G.; Moorman, M.A.; Kroon, E.; Carpenter, M.K.; Baetge, E.E. Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006, 24, 1392–1401. [Google Scholar] [CrossRef]
- Zhu, S.; Russ, H.A.; Wang, X.; Zhang, M.; Ma, T.; Xu, T.; Tang, S.; Hebrok, M.; Ding, S. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 2016, 7, 10080. [Google Scholar] [CrossRef] [PubMed]
- Dalgin, G.; Prince, V.E. Differential levels of Neurod establish zebrafish endocrine pancreas cell fates. Dev. Biol. 2015, 402, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.D.S.; Provost, E.; Leach, S.D.; Stainier, D.Y.R. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008, 22, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Field, H.A.; Dong, P.D.S.; Beis, D.; Stainier, D.Y.R. Formation of the digestive system in zebrafish. ii. pancreas morphogenesis. Dev. Biol. 2003, 261, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.J.; Pisharath, H.; Yusuff, S.; Moore, J.C.; Siekmann, A.F.; Lawson, N.; Leach, S.D. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 2009, 126, 898–912. [Google Scholar] [CrossRef]
- Sun, Z.; Hopkins, N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001, 15, 3217–3229. [Google Scholar] [CrossRef] [PubMed]
- Rovira, M.; Huang, W.; Yusuff, S.; Shim, J.S.; Ferrante, A.A.; Liu, J.O.; Parsons, M.J. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc. Natl. Acad. Sci. USA 2011, 108, 19264–19269. [Google Scholar] [CrossRef] [PubMed]
- Pack, M.; Solnica-Krezel, L.; Malicki, J.; Neuhauss, S.C.; Schier, A.F.; Stemple, D.L.; Driever, W.; Fishman, M.C. Mutations affecting development of zebrafish digestive organs. Development 1996, 123, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.S.; Andersson, O.; Row, R.; Kimelman, D.; Stainier, D.Y. Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.S.; Shin, C.H.; Stainier, D.Y. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev. Cell 2008, 15, 738–748. [Google Scholar] [CrossRef]
- Chung, W.S.; Stainier, D.Y. Intra-endodermal interactions are required for pancreatic beta cell induction. Dev. Cell 2008, 14, 582–593. [Google Scholar] [CrossRef]
- Dong, P.D.; Munson, C.A.; Norton, W.; Crosnier, C.; Pan, X.; Gong, Z.; Neumann, C.J.; Stainier, D.Y. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet. 2007, 39, 397–402. [Google Scholar] [CrossRef]
- Manfroid, I.; Delporte, F.; Baudhuin, A.; Motte, P.; Neumann, C.J.; Voz, M.L.; Martial, J.A.; Peers, B. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development 2007, 134, 4011–4021. [Google Scholar] [CrossRef] [PubMed]
- Stafford, D.; Prince, V.E. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 2002, 12, 1215–1220. [Google Scholar] [CrossRef] [PubMed]

| Genetic Conservation and Transparency: | References |
|---|---|
| Zebrafish display a high degree of genetic similarity with humans regarding cardiovascular development and function. Their transparency during early development allows for real-time visualization of the developing heart and blood vessels, providing unparalleled insights into the molecular and cellular processes involved in cardiovascular diseases. | [13,14,15] |
| High Reproductive Output: | [16,17] |
| The prolific reproduction of zebrafish facilitates large-scale genetic and drug-related screening studies. Their ability to generate many embryos enables researchers to investigate various genetic and environmental factors influencing cardiovascular health, promoting the identification of novel therapeutic targets. | |
| Conserved Cardiovascular Physiology: | [21,26,27,28] |
| Zebrafish possess a cardiovascular system with remarkable similarities to mammals. Their two-chambered heart, like the human heart during early development, allows for investigating basic cardiovascular processes, including heart development, contractility, and blood vessel formation, in a simplified yet biologically relevant context. | |
| Ease of Genetic Manipulation: | [6,7,11,12] |
| The advent of advanced genetic tools, such as ZFN, TALEN, CRISPR/Cas9, has made targeted genetic modifications in zebrafish relatively straightforward. This ease of genetic manipulation enables researchers to model specific cardiovascular diseases, study the effects of gene mutations, and identify potential therapeutic targets. | |
| Drug Discovery and Toxicity Screening: | [32,33,34,35] |
| Zebrafish’s permeability to small molecules makes them amenable to drug delivery. This characteristic, coupled with the ease of monitoring their cardiac function, allows for efficient drug discovery and toxicity screening. Zebrafish models contribute to the preclinical evaluation of drug candidates for cardiovascular diseases. |
| Zebrafish Gene | Human Orthologue | Model | Phenotype | Reference |
|---|---|---|---|---|
| amhc | MYH6 | Mutant line | Hypoplastic atrium/enlarged ventricle/CM hyperplasia | [32] |
| bag3 | BAG3 | Mutant line | Ventricle hypertrophy, Trabecular density, Myofibril degeneration, Apoptosis | [36] |
| scl4a1a | Mutant line | Cardiomyopathy, myofibril degeneration, apoptosis, CM hypertrophy and hyperplasia | [37] | |
| cmlc1 | MYL4 | Mutant line | Atrium enlargement, Disrupted sarcomeric structure | [38] |
| vmhcl | MYH7 | Mutant line | Disrupted sarcomeric structure, Trabecular density, Enlarged ventricle | [39,40] |
| gja3 | GJA3, CX46 | Mutant line | Pericardial edema | [41] |
| gtpbp3 | GTBP3 | Mutant line | CM hypertrophy, Abnormal mitochondrial morphology, Disrupted sarcomeric structure | [42] |
| vclb | VCL | Gene trap line | Disorganized coronary development, Fibrosis, Epicardial/myocardial hyperplasia | [43] |
| jupa | JUP | Mutant lines | Cardiomegaly, Thin atrial/ventricular walls, Disrupted sarcomeric structure | [44] |
| plcg1 | PLCG1 | Normal myocardial ultrastructure | [45] | |
| jag2b | JAG2 | Ablation NC-derived CMs | Enlarged ventricle, CM hypertrophy | [46] |
| Mutant line | Hypertrophic ventricle | |||
| tnnt2a | TNNT | Mutant line | Abnormal sarcomeric assembly | [47] |
| SL | Resemblances in Electrophysiology between Zebrafish and Humans | Reference |
|---|---|---|
| 1 | Similar long plateau phase | [112] |
| 2 | Presence of 0–4 phases | [112,113] |
| 3 | Ventricular AP has similar duration. The duration of zebrafish ventricular AP at 19 °C is like human ventricular AP at 37 °C | [114] |
| 4 | IK1 is present in atrial and ventricular myocytes | [112,114] |
| 5 | IK1 and IKr are the major repolarizing currents in both hearts | [115] |
| 6 | Similar fundamental current systems (INa, ICaL, and IK) are present in atrial and ventricular myocytes | [113,114] |
| 7 | Atrial and ventricular myocytes of the zebrafish heart have both T-type (ICaT) and L-type (ICaL) Ca2+ currents | [112,113,114] |
| 8 | The distinct QT intervals in ECG are similar in both | [112,114] |
| 9 | P, QRS, and T waves are clearly distinguishable in a zebrafish ECG | [112,114] |
| Disease | Method of Induction | References |
|---|---|---|
| Acute Hyperglycemia | Induced by intraperitoneal injection of D-glucose | [132,133] |
| Chronic Hyperglycemia | ||
| 1 | Induced by the destruction of pancreatic cells | [134,135,136] |
| 2 | The induction of chronic hyperglycemia by dissolving D-glucose in fish water | [137] |
| 3 | Genetic induction | [138,139,140] |
| Obesity | Overfeeding models | [141,142,143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angom, R.S.; Nakka, N.M.R. Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine. Biomedicines 2024, 12, 693. https://doi.org/10.3390/biomedicines12030693
Angom RS, Nakka NMR. Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine. Biomedicines. 2024; 12(3):693. https://doi.org/10.3390/biomedicines12030693
Chicago/Turabian StyleAngom, Ramcharan Singh, and Naga Malleswara Rao Nakka. 2024. "Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine" Biomedicines 12, no. 3: 693. https://doi.org/10.3390/biomedicines12030693
APA StyleAngom, R. S., & Nakka, N. M. R. (2024). Zebrafish as a Model for Cardiovascular and Metabolic Disease: The Future of Precision Medicine. Biomedicines, 12(3), 693. https://doi.org/10.3390/biomedicines12030693







