Exploring the Potential of Humoral Immune Response to Commensal Bifidobacterium as a Biomarker for Human Health, including Both Malignant and Non-Malignant Diseases: A Perspective on Detection Strategies and Future Directions
Abstract
1. Introduction
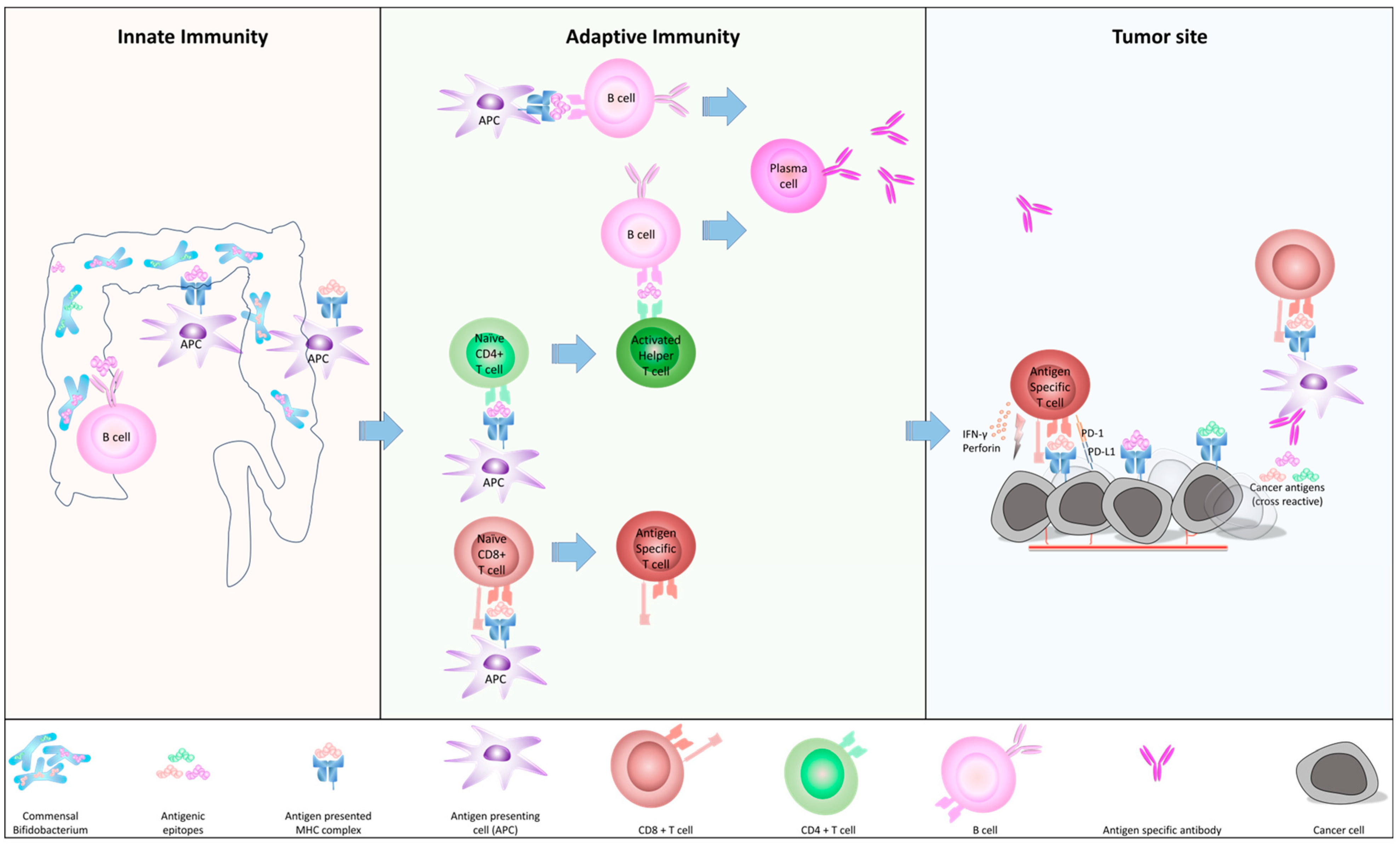
2. Microbiome and Immunosurveillance in Human
2.1. How c-BIF Interact with Human Immune System
2.2. The Homology of c-BIF with Tumor Antigens and Their Biological Role
2.3. The Role of c-BIF for Anti-Tumor Immunity
3. The Importance of c-BIF in Infectious and Inflammatory Diseases
3.1. c-BIF Plays Pivotal Role in Managing Symptoms of COVID
3.2. Preventive Therapeutic Effect of c-BIF on HPV
3.3. The Therapeutic Effects of c-BIF as Probiotics for Inflammatory Diseases
3.4. Microbial Signatures May Play a Critical Role for Immune-Related Adverse Events from Immunotherapy
4. The Potential Use of c-BIF as Biomarkers and Treatment for Cancer Immunotherapies
4.1. Using c-BIF as Predictive and Prognostic Biomarkers for Colorectra Cancer (CRC)
4.2. Humoral Immune Responses to Cytotoxic T lymphocyte (CTL) Epitope Peptides Shares Its Motifs with c-BIF Which Correlates to Overall Survival of Cancer
4.3. The Composition of the Gut Microbiome Is Recognized as a Key Factor in Influencing Effective Immuno-Oncology
4.4. Exploring Potential Obstacles and Limitations in Immunotherapies Utilizing Microbiome Composition
- (a)
- Safety Concerns:
- (b)
- Regulatory Challenges:
- (c)
- Limited Understanding of Microbiome:
- (d)
- Treatment Specificity:
- (e)
- Ethical and Psychological Factors:
- (f)
- Standardization and Accessibility:
- (g)
- Potential Long-Term Risks:
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ruiz, L.; Bottacini, F.; Boinett, C.J.; Cain, A.K.; O’connell-Motherway, M.; Lawley, T.D.; van Sinderen, D. The essential genomic landscape of the commensal Bifidobacterium breve UCC2003. Sci. Rep. 2017, 7, 5648. [Google Scholar] [CrossRef] [PubMed]
- Collen, A. 10% Human: How Your Body’s Microbes Hold the Key to Health and Happiness; Harper Paperbacks: New York, NY, USA, 2015. [Google Scholar]
- Aghamajidi, A.; Vareki, S.M. The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers 2022, 14, 3563. [Google Scholar] [CrossRef] [PubMed]
- Reboldi, A.; Cyster, J.G. Peyer’s patches: Organizing B-cell responses at the intestinal frontier. Immunol. Rev. 2016, 271, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Shale, M.; Schiering, C.; Powrie, F. CD4+ T-cell subsets in intestinal inflammation. Immunol. Rev. 2013, 252, 164–182. [Google Scholar] [CrossRef] [PubMed]
- Konjar, Š.; Ferreira, C.; Blankenhaus, B.; Veldhoen, M. Intestinal Barrier Interactions with Specialized CD8 T Cells. Front. Immunol. 2017, 8, 1281. [Google Scholar] [CrossRef] [PubMed]
- Van Der Bruggen, P.; Traversari, C.; Chomez, P.; Lurquin, C.; De Plaen, E.; Van Den Eynde, B.; Knuth, A.; Boon, T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991, 254, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Tureci, O.; Schmitt, H.; Cochlovius, B.; Johannes, T.; Schmits, R.; Stenner, F.; Luo, G.; Schobert, I.; Pfreundschuh, M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl. Acad. Sci. USA 1995, 92, 11810–11813. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Scanlan, M.J.; Sahin, U.; Türeci, Ö.; Gure, A.O.; Tsang, S.; Williamson, B.; Stockert, E.; Pfreundschuh, M.; Old, L.J. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. USA 1997, 94, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
- Matsueda, S.; Komatsu, N.; Kusumoto, K.; Koga, S.; Kuromatsu, R.; Yamada, S.; Seki, R.; Yutani, S.; Shichijo, S.; Mine, T.; et al. Humoral immune responses to CTL epitope peptides from tumor-associated antigens are widely detectable in humans: A new biomarker for overall survival of patients with malignant diseases. Dev. Comp. Immunol. 2013, 41, 68–76. [Google Scholar] [CrossRef]
- Suekane, S.; Yutani, S.; Yamada, A.; Sasada, T.; Matsueda, S.; Takamori, S.; Toh, U.; Kawano, K.; Yoshiyama, K.; Sakamoto, S.; et al. Identification of biomarkers for personalized peptide vaccination in 2,588 cancer patients. Int. J. Oncol. 2020, 56, 1479–1489. [Google Scholar] [CrossRef]
- Itoh, K.; Schichijo, S.; Suekane, S. Sequence similarity between commensal Bifidobacterium and cytotoxic T lymphocyte epitope peptides against human tumor associated antigens. 2023; under submission. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Kawakami, Y.; Eliyahu, S.; Delgado, C.H.; Robbins, P.F.; Rivoltini, L.; Topalian, S.L.; Miki, T.; Rosenberg, S.A. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc. Natl. Acad. Sci. 1994, 91, 3515–3519. [Google Scholar] [CrossRef]
- Shichijo, S.; Nakao, M.; Imai, Y.; Takasu, H.; Kawamoto, M.; Niiya, F.; Yang, D.; Toh, Y.; Yamana, H.; Itoh, K. A Gene Encoding Antigenic Peptides of Human Squamous Cell Carcinoma Recognized by Cytotoxic T Lymphocytes. J. Exp. Med. 1998, 187, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Suekane, S.; Yutani, S.; Toh, U.; Yoshiyama, K.; Itoh, K. Immune responses of patients without cancer recurrence after a cancer vaccine over a long term. Mol. Clin. Oncol. 2022, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Bessell, C.A.; Isser, A.; Havel, J.J.; Lee, S.; Bell, D.R.; Hickey, J.W.; Chaisawangwong, W.; Bieler, J.G.; Srivastava, R.; Kuo, F.; et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. J. Clin. Investig. 2020, 5, e135597. [Google Scholar] [CrossRef] [PubMed]
- Fluckiger, A.; Daillère, R.; Liu, P.; Loos, F.; Richard, C.; Rabu, C.; Alou, M.T.; Goubet, A.-G.; Lemaitre, F.; Ferrere, G.; et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 2020, 369, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Kappler, K.; Hennet, T. Emergence and significance of carbohydrate-specific antibodies. Genes Immun. 2020, 21, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Matsueda, S.; Itoh, K.; Shichijo, S. Antitumor activity of antibody against cytotoxic T lymphocyte epitope peptide of lymphocyte-specific protein tyrosine kinase. Cancer Sci. 2018, 109, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Genaro, S.C.; Reis, L.S.L.d.S.; Reis, S.K.; Socca, E.A.R.; Fávaro, W.J. Probiotic supplementation attenuates the aggressiveness of chemically induced colorectal tumor in rats. Life Sci. 2019, 237, 116895. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lau, H.C.-H.; Cheng, W.Y.; Yu, J. Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment. Genom. Proteom. Bioinform. 2023, 21, 84–96. [Google Scholar] [CrossRef]
- Khan, R.; Petersen, F.C.; Shekhar, S. Commensal bacteria: An emerging player in defense against respiratory pathogens. Front. Immunol. 2019, 10, 1203. [Google Scholar] [CrossRef]
- Tong, J.; Chen, Y.; He, M.; Wang, W.; Wang, Y.; Li, N.; Xia, Q. The triangle relationship between human genome, gut microbiome, and COVID-19: Opening of a Pandora’s box. Front. Microbiol. 2023, 14, 1190939. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J.; Barrows, B.D.; Quigley, E.M.; Borody, T.J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871. [Google Scholar] [CrossRef]
- Bozkurt, H.S.; Quigley, E.M. The probiotic Bifidobacterium in the management of Coronavirus: A theoretical basis. Int. J. Immunopathol. Pharmacol. 2020, 34, 2058738420961304. [Google Scholar] [CrossRef]
- Fujimoto, K.; Kimura, Y.; Allegretti, J.R.; Yamamoto, M.; Zhang, Y.-Z.; Katayama, K.; Tremmel, G.; Kawaguchi, Y.; Shimohigoshi, M.; Hayashi, T.; et al. Functional Restoration of Bacteriomes and Viromes by Fecal Microbiota Transplantation. Gastroenterology 2021, 160, 2089–2102.e12. [Google Scholar] [CrossRef] [PubMed]
- NIH (National Cancer Institute). HPV and Cancer. Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-and-cancer (accessed on 18 October 2023).
- Cha, M.-K.; Lee, D.-K.; An, H.-M.; Lee, S.-W.; Shin, S.-H.; Kwon, J.-H.; Kim, K.-J.; Ha, N.-J. Antiviral activity of Bifidobacterium adolescentisSPM1005-A on human papillomavirus type 16. BMC Med. 2012, 10, 72. [Google Scholar] [CrossRef]
- Abdolalipour, E.; Mahooti, M.; Salehzadeh, A.; Torabi, A.; Mohebbi, S.R.; Gorji, A.; Ghaemi, A. Evaluation of the antitumor immune responses of probiotic Bifidobacterium bifidum in human papillomavirus-induced tumor model. Microb. Pathog. 2020, 145, 104207. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Hao, W.; Wang, X.; Zhou, R.; Lin, Q. Probiotics for the treatment of ulcerative colitis: A review of experimental research from 2018 to 2022. Front. Microbiol. 2023, 14, 1211271. [Google Scholar] [CrossRef]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; de Foy, J.-M.P.; Dequenne, I.; de Timary, P.; Cani, P.D. How Probiotics Affect the Microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef]
- Bozkurt, H.S.; Kara, B. A new treatment for ulcerative colitis: Intracolonic Bifidobacterium and xyloglucan application. Eur. J. Inflamm. 2020, 18, 1–7. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Jiang, J.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Identification of the key characteristics of Bifidobacterium longum strains for the alleviation of ulcerative colitis. Food Funct. 2021, 12, 3476–3492. [Google Scholar] [CrossRef]
- Halsey, T.M.; Thomas, A.S.; Hayase, T.; Ma, W.; Abu-Sbeih, H.; Sun, B.; Parra, E.R.; Jiang, Z.-D.; DuPont, H.L.; Sanchez, C.; et al. Microbiome alteration via fecal microbiota transplantation is effective for refractory immune checkpoint inhibitor–induced colitis. Sci. Transl. Med. 2023, 15, eabq4006. [Google Scholar] [CrossRef] [PubMed]
- Groenewegen, B.; Terveer, E.M.; Joosse, A.; Barnhoorn, M.C.; Zwittink, R.D. Fecal Microbiota Transplantation for Immune Checkpoint Inhibitor-Induced Colitis Is Safe and Contributes to Recovery: Two Case Reports. J. Immunother. 2023, 46, 216–220. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Suehiro, Y.; Hashimoto, S.; Hoshida, T.; Fujimoto, M.; Watanabe, M.; Imanaga, D.; Sakai, K.; Matsumoto, T.; Nishioka, M.; et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J. Gastroenterol. 2018, 53, 517–524. [Google Scholar] [CrossRef]
- Temraz, S.; Nassar, F.; Nasr, R.; Charafeddine, M.; Mukherji, D.; Shamseddine, A. Gut Microbiome: A Promising Biomarker for Immunotherapy in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4155. [Google Scholar] [CrossRef] [PubMed]
- Bupathi, M.; Wu, C. Biomarkers for immune therapy in colorectal cancer: Mismatch-repair deficiency and others. J. Gastrointest. Oncol. 2016, 7, 713–720. [Google Scholar] [CrossRef]
- Oh, H.J.; Kim, J.H.; Bae, J.M.; Kim, H.J.; Cho, N.-Y.; Kang, G.H. Prognostic Impact of Fusobacterium nucleatum Depends on Combined Tumor Location and Microsatellite Instability Status in Stage II/III Colorectal Cancers Treated with Adjuvant Chemotherapy. J. Pathol. Transl. Med. 2019, 53, 40–49. [Google Scholar] [CrossRef]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Yi, C.; Zhu, H. Predictive biomarkers of colon cancer immunotherapy: Present and future. Front. Immunol. 2022, 13, 1032314. [Google Scholar] [CrossRef] [PubMed]
- Viale, G.; Trapani, D.; Curigliano, G. Mismatch Repair Deficiency as a Predictive Biomarker for Immunotherapy Efficacy. BioMed Res. Int. 2017, 2017, 4719194. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Chang, M.; Chang, H.M.; Chang, F. Microsatellite Instability: A Predictive Biomarker for Cancer Immunotherapy. Appl. Immunohistochem. Mol. Morphol. 2018, 26, e15–e21. [Google Scholar] [CrossRef] [PubMed]
- Rezasoltani, S.; Aghdaei, H.A.; Jasemi, S.; Gazouli, M.; Dovrolis, N.; Sadeghi, A.; Schlüter, H.; Zali, M.R.; Sechi, L.A.; Feizabadi, M.M. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers 2022, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Nutrition Division. Probiotics in Food; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2006; p. 56. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Torres-Maravilla, E.; Boucard, A.-S.; Mohseni, A.H.; Taghinezhad-S, S.; Cortes-Perez, N.G.; Bermúdez-Humarán, L.G. Role of Gut Microbiota and Probiotics in Colorectal Cancer: Onset and Progression. Microorganisms 2021, 9, 1021. [Google Scholar] [CrossRef]
- Kvakova, M.; Kamlarova, A.; Stofilova, J.; Benetinova, V.; Bertkova, I. Probiotics and postbiotics in colorectal cancer: Prevention and complementary therapy. World J. Gastroenterol. 2022, 28, 3370–3382. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Q.; Yu, B.; Zhou, J.; Zhang, J.; Zhang, R.; Xie, J.; Wang, Q.; Zhao, S. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci. Rep. 2019, 9, 20128. [Google Scholar] [CrossRef]
- Uccello, M.; Malaguarnera, G.; Basile, F.; D’agata, V.; Malaguarnera, M.; Bertino, G.; Vacante, M.; Drago, F.; Biondi, A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012, 12 (Suppl. S1), S35. [Google Scholar] [CrossRef] [PubMed]
- Cruz, B.C.S.; Sarandy, M.M.; Messias, A.C.; Gonçalves, R.V.; Ferreira, C.L.L.F.; Peluzio, M.C.G. Preclinical and clinical relevance of probiotics and synbiotics in colorectal carcinogenesis: A systematic review. Nutr. Rev. 2020, 78, 667–687. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-Y.; Li, S.-C.; Lin, H.-P.; Shih, C.-K. Germinated brown rice combined with Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis inhibits colorectal carcinogenesis in rats. Food Sci. Nutr. 2018, 7, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Kakizaki, N.; Okuno, A.; Maekawa, T.; Tsuji, N.M. Lactococcus lactis subsp. Cremoris C60 restores T Cell Population in Small Intestinal Lamina Propria in Aged Interleukin-18 Deficient Mice. Nutrients 2020, 12, 3287. [Google Scholar] [CrossRef]
- Hradicka, P.; Beal, J.; Kassayova, M.; Foey, A.; Demeckova, V. A Novel Lactic Acid Bacteria Mixture: Macrophage-Targeted Prophylactic Intervention in Colorectal Cancer Management. Microorganisms 2020, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Boesmans, L.; Valles-Colomer, M.; Wang, J.; Eeckhaut, V.; Falony, G.; Ducatelle, R.; Van Immerseel, F.; Raes, J.; Verbeke, K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus pullicaecorum to Healthy Volunteers. mSystems 2018, 3, e00094-18. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-De-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Fahmy, C.A.; Gamal-Eldeen, A.M.; El-Hussieny, E.A.; Raafat, B.M.; Mehanna, N.S.; Talaat, R.M.; Shaaban, M.T. Bifidobacterium longum Suppresses Murine Colorectal Cancer through the Modulation of oncomiRs and Tumor Suppressor miRNAs. Nutr. Cancer 2019, 71, 688–700. [Google Scholar] [CrossRef]
- Shirahama, T.; Muroya, D.; Matsueda, S.; Yamada, A.; Shichijo, S.; Naito, M.; Yamashita, T.; Sakamoto, S.; Okuda, K.; Itoh, K.; et al. A randomized phase II trial of personalized peptide vaccine with low dose cyclophosphamide in biliary tract cancer. Cancer Sci. 2017, 108, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Yanagimoto, H.; Shiomi, H.; Satoi, S.; Mine, T.; Toyokawa, H.; Yamamoto, T.; Tani, T.; Yamada, A.; Kwon, A.-H.; Komatsu, N.; et al. A phase II study of personalized peptide vaccination combined with gemcitabine for non-resectable pancreatic cancer patients. Oncol. Rep. 2010, 24, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Borcoman, E.; Kanjanapan, Y.; Champiat, S.; Kato, S.; Servois, V.; Kurzrock, R.; Goel, S.; Bedard, P.; Le Tourneau, C. Novel patterns of response under immunotherapy. Ann. Oncol. 2019, 30, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.-M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.; Allen, L.; Zain, N.M.M.; Forbes, B.; Shawcross, D.L.; Goldenberg, S.D. Regulation, risk and safety of Faecal Microbiota Transplant. Infect. Prev. Pract. 2020, 2, 100069. [Google Scholar] [CrossRef] [PubMed]
- DeFilipp, Z.; Bloom, P.P.; Soto, M.T.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- FDA (Food and Drug Administration). Information Pertaining to Additional Safety Protections Regarding Use of Fecal Microbiota for Transplantation—Screening and Testing of Stool Donors for Multi-Drug Resistant Organisms. Available online: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/information-pertaining-additional-safety-protections-regarding-use-fecal-microbiota-transplantation (accessed on 18 June 2019).
- Khoruts, A.; Hoffmann, D.E.; Palumbo, F.B. The Impact of Regulatory Policies on the Future of Fecal Microbiota Transplantation. J. Law Med. Ethic 2019, 47, 482–504. [Google Scholar] [CrossRef]
- Bunnik, E.M.; Aarts, N.; Chen, L.A. Physicians Must Discuss Potential Long-Term Risks of Fecal Microbiota Transplantation to Ensure Informed Consent. Am. J. Bioeth. 2017, 17, 61–63. [Google Scholar] [CrossRef][Green Version]
| Peptide Name | Original Protein | Position | Sequence | HLA-IA Restriction | Positive/Negative | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy (a) (n = 66) | HCV (b) (n = 20) | Flu (c) (n = 20) | RA (d) (n = 20) | IgA Nephropathy (e) (n = 20) | Leukemia (f) (n = 26) | Liver Cancer (g) (n = 41) | Solid Cancer (h) (n = 2588) | |||||
| CypB-129 | Cyclophilin B | 129–138 | KLKHYGPGWV | A2/A3 supertype | 37/29 | 15/5 | 16/4 | 18/2 | 13/7 | 12/14 | 36/5 | 1888/700 |
| Lck-246 | p56 lck | 246–254 | KLVERLGAA | A2 | 46/20 | 16/4 | 17/3 | 17/3 | 16/4 | 11/15 | 35/6 | 1815/773 |
| Lck-422 | p56 lck | 422–430 | DVWSFGILL | A2/A3 supertype | 19/47 | 6/14 | 11/9 | 8/12 | 7/13 | 5/21 | 13/28 | 316/2272 |
| ppMAPkkk-432 | ppMAPkkk | 432–440 | DLLSHAFFA | A2/A26 | 27/39 | 10/10 | 10/10 | 10/10 | 5/15 | 9/17 | 26/15 | 1308/1280 |
| WHSC2-103 | WHSC2 | 103–111 | ASLDSDPWV | A2/A26/A3 supertype | 64/2 | 20/0 | 20/0 | 20/0 | 19/1 | 24/2 | 41/0 | 1673/915 |
| HNRPL-501 | HNRPL | 501–510 | NVLHFFNAPL | A2/A26 | 15/51 | 13/7 | 10/10 | 14/6 | 4/16 | 5/21 | 27/14 | 1189/1399 |
| UBE2V-43 | UBE2V | 43–51 | RLQEWCSVI | A2 | 57/9 | 16/4 | 20/0 | 20/0 | 20/0 | 19/7 | 39/2 | 1238/1350 |
| UBE2V-85 | UBE2V | 85–93 | LIADFLSGL | A2 | 7/59 | 1/19 | 3/17 | 5/15 | 1/19 | 3/23 | 11/30 | 362/2226 |
| WHSC2-141 | WHSC2 | 141–149 | ILGELREKV | A2 | 53/13 | 17/3 | 18/2 | 18/2 | 16/4 | 8/18 | 37/4 | 1908/680 |
| HNRPL-140 | HNRPL | 140–148 | ALVEFEDVL | A2 | 52/14 | 17/3 | 18/2 | 19/1 | 16/4 | 10/16 | 39/2 | 1295/1293 |
| SART3-302 | SART3 | 302–310 | LLQAEAPRL | A2 | 45/21 | 16/4 | 18/2 | 14/6 | 14/6 | 12/14 | 24/17 | 1640/948 |
| SART3-309 | SART3 | 309–317 | RLAEYQAYI | A2 | 48/18 | 16/4 | 18/2 | 19/1 | 15/5 | 15/11 | 41/0 | 1921/667 |
| SART2-93 | SART2 | 93–101 | DYSARWNEI | A24 | 64/2 | 20/0 | 20/0 | 20/0 | 20/0 | 25/1 | 41/0 | 2248/340 |
| SART3-109 | SART3 | 109–118 | VYDYNCHVDL | A24/A26/A3 supertype | 15/51 | 6/14 | 7/13 | 7/13 | 3/17 | 6/20 | 33/8 | 1206/1382 |
| Lck-208 | p56 lck | 208–216 | HYTNASDGL | A24 | 31/35 | 11/9 | 13/7 | 9/11 | 7/13 | 4/22 | 22/19 | 560/2028 |
| PAP-213 | PAP | 213–221 | LYCESVHNF | A24 | 54/12 | 16/4 | 17/3 | 19/1 | 18/2 | 22/4 | 39/2 | 1422/1166 |
| PSA-248 | PSA | 248–257 | HYRKWIKDTI | A24 | 51/15 | 15/5 | 19/1 | 20/0 | 18/2 | 21/5 | 40/1 | 1286/1302 |
| EGFR-800 | EGF-R | 800–809 | DYVREHKDNI | A24 | 59/7 | 18/2 | 19/1 | 20/0 | 18/2 | 19/7 | 40/1 | 1542/1046 |
| MRP3-503 | MRP3 | 503–511 | LYAWEPSFL | A24 | 3/63 | 1/19 | 2/18 | 2/18 | 1/19 | 2/24 | 4/37 | 467/2121 |
| MRP3-1293 | MRP3 | 1293–1302 | NYSVRYRPGL | A24 | 59/7 | 18/2 | 20/0 | 19/1 | 16/4 | 19/7 | 41/0 | 1558/1030 |
| SART2-161 | SART2 | 161–169 | AYDFLYNYL | A24 | 13/53 | 2/18 | 2/18 | 7/13 | 1/19 | 7/19 | 18/23 | 947/1641 |
| Lck-486 | p56 lck | 486–494 | TFDYLRSVL | A24 | 53/13 | 20/0 | 18/2 | 20/0 | 19/1 | 17/9 | 38/3 | 2184/404 |
| Lck-488 | p56 lck | 488–497 | DYLRSVLEDF | A24 | 64/2 | 20/0 | 20/0 | 20/0 | 20/0 | 25/1 | 41/0 | 2147/441 |
| PSMA-624 | PSMA | 624–632 | TYSVSFDSL | A24 | 41/25 | 20/0 | 14/6 | 17/3 | 8/12 | 11/15 | 38/3 | 950/1638 |
| EZH2-735 | EZH2 | 735–743 | KYVGIEREM | A24 | 15/51 | 7/13 | 7/13 | 9/11 | 3/17 | 3/23 | 17/24 | 558/2030 |
| PTHrP-102 | PTHrP | 102–111 | RYLTQETNKV | A24 | 35/31 | 14/6 | 17/3 | 15/5 | 16/4 | 5/21 | 32/9 | 886/1702 |
| SART3-511 | SART3 | 511–519 | WLEYYNLER | A3 supertype | 60/6 | 17/3 | 20/0 | 20/0 | 20/0 | 21/5 | 40/1 | 1967/621 |
| SART3-734 | SART3 | 734–742 | QIRPIFSNR | A3 supertype | 66/0 | 20/0 | 20/0 | 20/0 | 20/0 | 25/1 | 41/0 | 2041/547 |
| Lck-90 | p56 lck | 90–99 | ILEQSGEWWK | A3 supertype | 63/3 | 19/1 | 20/0 | 20/0 | 20/0 | 23/3 | 41/0 | 2087/501 |
| Lck-449 | p56 lck | 449–458 | VIQNLERGYR | A3 supertype | 52/14 | 19/1 | 18/2 | 20/0 | 18/2 | 14/12 | 39/2 | 1406/1182 |
| PAP-248 | PAP | 248–257 | GIHKQKEKSR | A3 supertype | 49/17 | 17/3 | 16/4 | 14/6 | 14/6 | 5/21 | 22/19 | 2035/553 |
| Query | Subject | Position | Sequence | Identities (%) |
|---|---|---|---|---|
| Lck-90 | 90–99 | ILEQSGEWWK | - | |
| Bifidobacterium animalis | 497–504 | *LQQSDEWW* | 6/8 (75%) | |
| Bifidobacterium longum | 480–487 | IFEQNGEW** | 6/8 (75%) | |
| 924–932 | *LEQSGDDEW | 7/9 (78%) | ||
| 953–961 | *LEQSGDDEW | 7/9 (78%) | ||
| Bifidobacterium sp. | 2625–2630 | ***EQSGEW | 6/6 (100%) | |
| Lck-208 | 208–216 | HYTNASDGL | - | |
| Bifidobacterium bifidum | 270–277 | HYTSATDG* | 6/8 (75%) | |
| Bifidobacterium castoris | 242–248 | **TNASDGL | 7/7 (100%) | |
| 207–213 | **TNASDGL | 7/7 (100%) | ||
| Bifidobacterium dentium | 607–615 | HYSDVSDGL | 6/9 (67%) | |
| Bifidobacterium italicum | 301–308 | *YQNASDGL | 7/8 (88%) | |
| Lck-246 | 246–254 | KLVERLGAA | - | |
| Bifidobacterium avesanii | 363–369 | KLVERLG** | 7/7 (100%) | |
| Bifidobacterium asteroides | 181–188 | MVERLGAA | 7/8 (88%) | |
| Bifidobacterium breve | 471–478 | KLVERFGA* | 7/8 (88%) | |
| Bifidobacterium longum | 300–307 | *LVERLDAA | 7/8 (88%) | |
| 292–299 | *LVERLDAA | 7/8 (88%) | ||
| Lck-422 | 422–430 | DVWSFGILL | - | |
| Bifidobacterium crudilactis | 312–318 | DVWSFAI** | 6/7 (86%) | |
| Bifidobacterium longum | 350–356 | **WSIGILL | 6/7 (86%) | |
| 144–150 | DVWNYGI | 5/7 (71%) | ||
| Bifidobacterium breve | 178–184 | DVWNYGI | 5/7 (71%) | |
| Bifidobacterium lemurum | 79–87 | *VWKFGLILL | 7/9 (78%) | |
| Lck-449 | 449–458 | VIQNLERGYR | ||
| Bifidobacterium felsineum | 1076–1082 | *IQNLERG** | 7/7 (100%) | |
| Bifidobacterium breve | 54–61 | VIENL**GYR | 7/10 (70%) | |
| Bifidobacterium cebidarum | 741–747 | *IQNLERG** | 7/7 (100%) | |
| Bifidobacterium pseudocatenulatum | 134–141 | *INNLERGY | 7/8 (88%) | |
| Bifidobacterium coryneforme | 437–444 | *QALDRGYR | 6/8 (75%) | |
| Lck-486 | 486–494 | TFDYLRSVL | - | |
| Bifidobacterium pullorum | 149–156 | TFDYIRSV* | 7/8 (88%) | |
| Bifidobacterium longum | 318–323 | *FDYLRS** | 6/6 (100%) | |
| 372–378 | *FDYLQSV* | 6/7 (80%) | ||
| Bifidobacterium breve | 12–18 | **DYLRSVL | 7/7 (100%) | |
| Bifidobacterium animalis | 120–125 | ***YLRSVL | 6/6 (100%) | |
| Lck-488 | 488–497 | DYLRSVLEDF | - | |
| Bifidobacterium breve | 12–18 | DYLRSVL*** | 7/7 (100%) | |
| Bifidobacterium pseudolongum | 322–327 | DYLRSV**** | 6/6 (100%) | |
| Bifidobacterium longum | 132–139 | **LRSILNDF | 6/8 (75%) | |
| Bifidobacterium animalis | 120–125 | *YLRSVL*** | 6/6 (100%) | |
| Bifidobacterium adolescentis | 76–84 | *YLRSIPENF | 6/9 (67%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Itoh, K.; Matsueda, S. Exploring the Potential of Humoral Immune Response to Commensal Bifidobacterium as a Biomarker for Human Health, including Both Malignant and Non-Malignant Diseases: A Perspective on Detection Strategies and Future Directions. Biomedicines 2024, 12, 803. https://doi.org/10.3390/biomedicines12040803
Itoh K, Matsueda S. Exploring the Potential of Humoral Immune Response to Commensal Bifidobacterium as a Biomarker for Human Health, including Both Malignant and Non-Malignant Diseases: A Perspective on Detection Strategies and Future Directions. Biomedicines. 2024; 12(4):803. https://doi.org/10.3390/biomedicines12040803
Chicago/Turabian StyleItoh, Kyogo, and Satoko Matsueda. 2024. "Exploring the Potential of Humoral Immune Response to Commensal Bifidobacterium as a Biomarker for Human Health, including Both Malignant and Non-Malignant Diseases: A Perspective on Detection Strategies and Future Directions" Biomedicines 12, no. 4: 803. https://doi.org/10.3390/biomedicines12040803
APA StyleItoh, K., & Matsueda, S. (2024). Exploring the Potential of Humoral Immune Response to Commensal Bifidobacterium as a Biomarker for Human Health, including Both Malignant and Non-Malignant Diseases: A Perspective on Detection Strategies and Future Directions. Biomedicines, 12(4), 803. https://doi.org/10.3390/biomedicines12040803






