Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Prediction of Dyspnea after 12 Months of an Acute Episode of COVID-19
Abstract
:1. Introduction
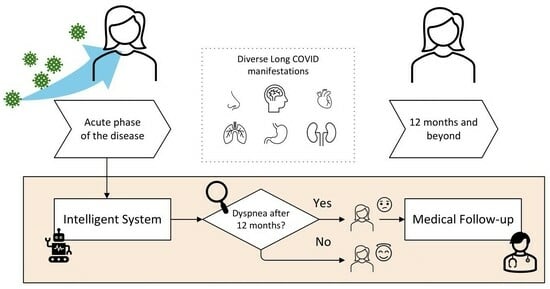
- To introduce at a conceptual level the architecture of a new intelligent clinical decision support system applied to the prediction of dyspnea after 12 months of an acute episode of COVID-19.
- Starting from the SeguiCovid database, select a reduced set of variables that can be used for the prediction of dyspnea after 12 months of a COVID-19 episode.
- Develop a new architecture, supported by a cascade of expert systems, whose knowledge bases are automatically generated using the Wang-Mendel automatic rule generation algorithm [52].
- Implement the intelligent clinical decision support system through a software artefact and demonstrate its utility through a case study.
2. Materials and Methods
2.1. System Definition
2.1.1. Database Usage
2.1.2. Conceptual Design
Stage 1: Data Collection
Stage 2: Data Processing
- Stage 2.1—Selection of variables: Before building the knowledge bases of the expert systems, it is essential to determine the input variables for each of them. To identify the variables with a higher predictive power, it is proposed to use a CART-type classification tree [56,57]. The use of decision trees as a feature selection tool is a common practice and is supported by the scientific literature [58,59,60]. After this process, it is observed that, among the variables present in Table 1, those with the greatest predictive power are sex, age, lung diseases, smoking, presence of dyspnea in the third month and DLNO.
- Stage 2.2—Definition of the expert system cascade: Once the variables with the highest predictive power have been identified, their treatment by the system is addressed. For this purpose, a cascade of expert systems is used, all of them using fuzzy inference engines of the Mamdani type [61,62,63,64]. The cascade has two levels, as shown in Figure 1. In the first level of the cascade, Expert System 1 is fed with sex, age and dyspnea at month 3, while Expert System 2 is fed with lung diseases, smoking and DLNO, obtaining at its output the risks R1 and R2, respectively. Then, at the second level of the cascade, these risks are fed to Expert System 3, which obtains the Dyspnea Risk at 12 months. In this way, the use of the cascade reduces uncertainty and facilitates the creation of more accurate knowledge bases [38,65], with simpler rules due to the smaller number of antecedents. In general, one of the most complex tasks in the development of an expert system is the creation of the knowledge base. Although there have been various studies and proposals aimed at identifying the risk factors associated with the development of long COVID, there is currently no explicit knowledge or experience of how a patient might develop 12 months after the episode and whether he or she might present with dyspnea. Statistical approaches could be used to solve this problem, with the associated loss of explanatory power; however, in this work we opt to use the Wang-Mendel automatic rule generation algorithm [52]. In this way, starting from the dataset, it is possible to generate a knowledge base based on fuzzy rules for each of the expert systems.
- Stage 2.3—Inference: Once the expert systems are defined and given a new patient’s data, their treatment is approached and the Dyspnea Risk at 12 months is obtained.
Stage 3: Alert Generation and Decision-Making
2.2. System Implementation
2.2.1. Data Collection
2.2.2. Data Processing
Variable Selection
Definition of the Expert System Cascade
- Step prior to applying the method: First, the shape of the membership functions is defined. Triangular membership functions are chosen for both antecedents and consequents, as in the original Wang-Mendel paper [52]. In addition, the value of a parameter N is defined, which is related to the number of sections that the membership function of each variable will have (the number of sections of the membership function is equal to 2N + 1, to ensure that there is a central section). In this case, a value of N = 1 is set for both the antecedents and the consequents, giving a total of 3 sections per variable. In the case of the categorical input variables (dyspnea at 3 months, sex, lung diseases and smoking) and the output variable (the label), although there are three sections, only the extreme ones are used (this is because there are two unique values: 0 and 1).
- Step 1—Division of the input and output spaces into fuzzy regions: After setting the algorithm configuration, the division of the initial spaces, both antecedent and consequent, is addressed using the parameters set in the previous stage. Figure 3 shows a generic case for N = 1 with three sections of the membership function (Low, Medium and High). In line with the proposal of the original Wang-Mendel paper, the overlapping of the triangles is considered so that, if the top vertex of the central triangle has a maximum membership degree, the vertices of the neighbouring triangles at the same point have minimum membership degrees. For each of the expert systems, Figure 4 shows the different associated membership functions. For categorical variables, given their binary nature, the membership functions are simplified by eliminating the intermediate section. This decision is based on the observation that, since these variables can only assume two states (e.g., “yes” or “no”), an intermediate section would not contribute to the automatic rule generation process, nor to the subsequent inference. On the other hand, for the antecedents of Expert System 3, which are obtained after the inference and defuzzification of Expert Systems 1 and 2, we have kept three sections in the membership functions. This is because the values of R1 and R2 are continuous values obtained after the defuzzification process in Expert System 1 and 2, respectively, reflecting the risk of dyspnea.
- Step 2—Generation of fuzzy rules: After the division of the starting spaces, we move on to the generation of fuzzy rules. For this purpose, the degree of membership associated with each of the sections of the membership functions for each line of the dataset is determined. After that, in each of the lines of the dataset, each variable is assigned to the section with the maximum degree of membership, determining a rule for each line. In this case, as there are 130 patients reserved for the construction of the model, 130 rules are initially obtained in each knowledge base. It is important to clarify that the knowledge base of Expert System 3 is constructed once those of Expert Systems 1 and 2 have been determined, as it is necessary to construct a derived dataset with the values of R1, R2 and the label from the initial data.
- Step 3—Assigning a degree to each rule to resolve conflicts: After generating rules in Step 2, it may happen that there are rules with the same antecedents but different consequents. To solve this problem, in the original proposal by Wang-Mendel [52], a coefficient is determined for each rule, which is the product of the degrees of membership of the observation that gave rise to it, and the rule that maximises this value in the case of conflict is selected while the rest are discarded. In this way, the initial set of rules is greatly reduced. In this particular case, the knowledge base of Expert System 1 goes from 130 to 12 rules, that of Expert System 2 from 130 to 10 rules and that of Expert System 3 from 130 to 8 rules.
- Step 4—Construction of the combined fuzzy knowledge base: after the automatic generation of the knowledge base, the need to integrate linguistic rules proposed by experts could be identified. The experts should propose the rule together with its degree of importance, so that, in conflict situations, as observed in the previous stage, the rule with the higher coefficient is given priority. In this case, no rules other than those generated in Step 3 were added.
- Step 5—Inference: Once the knowledge bases have been established, it is possible to analyze data from new patients and obtain the risk indicators at the output of each of the expert systems. These risk indicators initially vary between 0 and 1; however, to facilitate interpretation of the risk at the output of the cascade, the Dyspnea Risk at 12 months is scaled between 0 and 100.
Prof Test Results and Determination of Optimum Threshold Value
2.2.3. Alert Generation and Decision-Making
3. Case Study
3.1. Initial Data Collection
3.2. Data Processing
3.3. Alert Generation and Decision-Making
4. Discussion
Relevance of the Proposal
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raveendran, A.V.; Jayadevan, R.; Sashidharan, S. Long COVID: An Overview. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major Findings, Mechanisms and Recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, C.; Collins, L.F.; Malani, P. Long-Term Health Consequences of COVID-19. JAMA 2020, 324, 1723–1724. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, E.; Janvier, P.; Kherabi, Y.; Le Bot, A.; Hamon, A.; Gouze, H.; Doucet, L.; Berkani, S.; Oliosi, E.; Mallart, E.; et al. Post-Discharge Persistent Symptoms and Health-Related Quality of Life after Hospitalization for COVID-19. J. Infect. 2020, 81, e4–e6. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Nirantharakumar, K.; Hughes, S.; Myles, P.; Williams, T.; Gokhale, K.M.; Taverner, T.; Chandan, J.S.; Brown, K.; Simms-Williams, N.; et al. Symptoms and Risk Factors for Long COVID in Non-Hospitalized Adults. Nat. Med. 2022, 28, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, J.; Tang, H.T.; Wong, H.K.; Lyu, A.; Cheung, C.H.; Bian, Z. Prevalence and Risk Factors of Long COVID 6–12 Months after Infection with the Omicron Variant among Nonhospitalized Patients in Hong Kong. J. Med. Virol. 2023, 95, e28862. [Google Scholar] [CrossRef] [PubMed]
- Abu Hamdh, B.; Nazzal, Z. A Prospective Cohort Study Assessing the Relationship between Long-COVID Symptom Incidence in COVID-19 Patients and COVID-19 Vaccination. Sci. Rep. 2023, 13, 4896. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Huang, J.; Wong, Y.Y.; Wong, G.L.H.; Yip, T.C.F.; Chan, R.N.Y.; Chau, S.W.H.; Ng, S.C.; Wing, Y.K.; Chan, F.K.L. Epidemiology, Symptomatology, and Risk Factors for Long COVID Symptoms: Population-Based, Multicenter Study. JMIR Public Health Surveill. 2023, 9, e42315. [Google Scholar] [CrossRef] [PubMed]
- Notarte, K.I.; de Oliveira, M.H.S.; Peligro, P.J.; Velasco, J.V.; Macaranas, I.; Ver, A.T.; Pangilinan, F.C.; Pastrana, A.; Goldrich, N.; Kavteladze, D.; et al. Age, Sex and Previous Comorbidities as Risk Factors Not Associated with SARS-CoV-2 Infection for Long COVID-19: A Systematic Review and Meta-Analysis. J. Clin. Med. 2022, 11, 7314. [Google Scholar] [CrossRef]
- Stewart, S.; Newson, L.; Briggs, T.A.; Grammatopoulos, D.; Young, L.; Gill, P. Long COVID Risk—A Signal to Address Sex Hormones and Women’s Health. Lancet Reg. Health 2021, 11, 100242. [Google Scholar] [CrossRef]
- Yong, S.J. Long COVID or Post-COVID-19 Syndrome: Putative Pathophysiology, Risk Factors, and Treatments. Infect. Dis. 2021, 53, 737–754. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.J.; Williams, D.M.; Walker, A.J.; Mitchell, R.E.; Niedzwiedz, C.L.; Yang, T.C.; Huggins, C.F.; Kwong, A.S.F.; Silverwood, R.J.; Di Gessa, G.; et al. Long COVID Burden and Risk Factors in 10 UK Longitudinal Studies and Electronic Health Records. Nat. Commun. 2022, 13, 3528. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.R.; Souza Sampaio, V.; Cintho Ozahata, M.; Lopes Paixão da Silva, R.; Brito, A.; Bragatte, M.; Kalil, J.; Luiz Miraglia, J.; Tavares Malheiros, D.; Guozhang, Y.; et al. Risk Factors for Long COVID among Healthcare Workers, Brazil, 2020–2022. medRxiv 2023. [Google Scholar] [CrossRef]
- Cazé, A.B.; Cerqueira-Silva, T.; Bomfim, A.P.; de Souza, G.L.; Azevedo, A.C.A.; Brasil, M.Q.A.; Santos, N.R.; Khouri, R.; Dan, J.; Bandeira, A.C.; et al. Prevalence and Risk Factors for Long COVID after Mild Disease: A Cohort Study with a Symptomatic Control Group. J. Glob. Health 2023, 13, 06015. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.; Nair, C.V.; Kulirankal, K.G.; Corley, E.M.; Edathadathil, F.; Gutjahr, G.; Moni, M.; Sathyapalan, D.T. Characterization and Predictive Risk Scoring of Long COVID in a South Indian Cohort after Breakthrough COVID Infection; a Prospective Single Centre Study. BMC Infect. Dis. 2023, 23, 670. [Google Scholar] [CrossRef] [PubMed]
- Vimercati, L.; De Maria, L.; Quarato, M.; Caputi, A.; Gesualdo, L.; Migliore, G.; Cavone, D.; Sponselli, S.; Pipoli, A.; Inchingolo, F.; et al. Association between Long COVID and Overweight/Obesity. J. Clin. Med. 2021, 10, 4143. [Google Scholar] [CrossRef] [PubMed]
- Heubner, L.; Petrick, P.L.; Güldner, A.; Bartels, L.; Ragaller, M.; Mirus, M.; Rand, A.; Tiebel, O.; Beyer-Westendorf, J.; Rößler, M.; et al. Extreme Obesity Is a Strong Predictor for In-Hospital Mortality and the Prevalence of Long-COVID in Severe COVID-19 Patients with Acute Respiratory Distress Syndrome. Sci. Rep. 2022, 12, 18418. [Google Scholar] [CrossRef] [PubMed]
- Notarte, K.I.; Catahay, J.A.; Velasco, J.V.; Pastrana, A.; Ver, A.T.; Pangilinan, F.C.; Peligro, P.J.; Casimiro, M.; Guerrero, J.J.; Gellaco, M.M.L.; et al. Impact of COVID-19 Vaccination on the Risk of Developing Long-COVID and on Existing Long-COVID Symptoms: A Systematic Review. EClinicalMedicine 2022, 53, 101624. [Google Scholar] [CrossRef] [PubMed]
- Strain, W.D.; Sherwood, O.; Banerjee, A.; Van der Togt, V.; Hishmeh, L.; Rossman, J. The Impact of COVID Vaccination on Symptoms of Long COVID: An International Survey of People with Lived Experience of Long COVID. Vaccines 2022, 10, 652. [Google Scholar] [CrossRef]
- Ayoubkhani, D.; Bermingham, C.; Pouwels, K.B.; Glickman, M.; Nafilyan, V.; Zaccardi, F.; Khunti, K.; Alwan, N.A.; Walker, A.S. Trajectory of Long Covid Symptoms after Covid-19 Vaccination: Community Based Cohort Study. BMJ 2022, 377, e069676. [Google Scholar] [CrossRef]
- Durstenfeld, M.S.; Peluso, M.J.; Peyser, N.D.; Lin, F.; Knight, S.J.; Djibo, A.; Khatib, R.; Kitzman, H.; O’Brien, E.; Williams, N.; et al. Factors Associated With Long COVID Symptoms in an Online Cohort Study. Open Forum Infect. Dis. 2023, 10, ofad047. [Google Scholar] [CrossRef] [PubMed]
- Fernández-de-las-Peñas, C.; Palacios-Ceña, D.; Gómez-Mayordomo, V.; Florencio, L.L.; Cuadrado, M.L.; Plaza-Manzano, G.; Navarro-Santana, M. Prevalence of Post-COVID-19 Symptoms in Hospitalized and Non-Hospitalized COVID-19 Survivors: A Systematic Review and Meta-Analysis. Eur. J. Intern. Med. 2021, 92, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.N.; Hoang, V.T.; Dao, T.L.; Meddeb, L.; Lagier, J.C.; Million, M.; Raoult, D.; Gautret, P. Long-Term Persistence of Symptoms of Dyspnoea in COVID-19 Patients. Int. J. Infect. Dis. 2022, 115, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Fernández-De-las-Peñas, C.; Palacios-Ceña, D.; Gómez-Mayordomo, V.; Palacios-Ceña, M.; Rodríguez-Jiménez, J.; de-La-Llave-Rincón, A.I.; Velasco-Arribas, M.; Fuensalida-Novo, S.; Ambite-Quesada, S.; Guijarro, C.; et al. Fatigue and Dyspnoea as Main Persistent Post-COVID-19 Symptoms in Previously Hospitalized Patients: Related Functional Limitations and Disability. Respiration 2022, 101, 132–141. [Google Scholar] [CrossRef]
- Fernandes, A.S.; Alves, P.; Jarman, I.H.; Etchells, T.A.; Fonseca, J.M.; Lisboa, P.J.G. A Clinical Decision Support System for Breast Cancer Patients. IFIP Adv. Inf. Commun. Technol. 2010, 314, 122–129. [Google Scholar] [CrossRef]
- Hermsen, E.D.; VanSchooneveld, T.C.; Sayles, H.; Rupp, M.E. Implementation of a Clinical Decision Support System for Antimicrobial Stewardship. Infect. Control Hosp. Epidemiol. 2012, 33, 412–415. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, R.S.; Angstman, K.B.; Kesman, R.; Stroebel, R.J.; Bernard, M.E.; Scheitel, S.M.; Hunt, V.L.; Rahman, A.S.; Chaudhry, R. Use of a Clinical Decision Support System to Increase Osteoporosis Screening. J. Eval. Clin. Pract. 2012, 18, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Alaa, A.M.; Moon, K.H.; Hsu, W.; Van Der Schaar, M. ConfidentCare: A Clinical Decision Support System for Personalized Breast Cancer Screening. IEEE Trans. Multimed. 2016, 18, 1942–1955. [Google Scholar] [CrossRef]
- Wagholikar, K.B.; MacLaughlin, K.L.; Henry, M.R.; Greenes, R.A.; Hankey, R.A.; Liu, H.; Chaudhry, R.C. Clinical Decision Support with Automated Text Processing for Cervical Cancer Screening. J. Am. Med. Inform. Assoc. 2012, 19, 833–839. [Google Scholar] [CrossRef]
- Courbis, A.L.; Murray, R.B.; Arnavielhe, S.; Caimmi, D.; Bedbrook, A.; Van Eerd, M.; De Vries, G.; Dray, G.; Agache, I.; Morais-Almeida, M.; et al. Electronic Clinical Decision Support System for Allergic Rhinitis Management: MASK e-CDSS. Clin. Exp. Allergy 2018, 48, 1640–1653. [Google Scholar] [CrossRef]
- Sherimon, P.C.; Krishnan, R.; Sherimon, B.P.C.; In, R.C. OntoDiabetic: An Ontology-Based Clinical Decision Support System for Diabetic Patients. Arab. J. Sci. Eng. 2015, 41, 1145–1160. [Google Scholar] [CrossRef]
- Hashi, E.K.; Uz Zaman, M.S.; Hasan, M.R. An Expert Clinical Decision Support System to Predict Disease Using Classification Techniques. In Proceedings of the ECCE 2017—International Conference on Electrical, Computer and Communication Engineering, Cox’s Bazar, Bangladesh, 16–18 February 2017; pp. 396–400. [Google Scholar] [CrossRef]
- Sesen, M.B.; Peake, M.D.; Banares-Alcantara, R.; Tse, D.; Kadir, T.; Stanley, R.; Gleeson, F.; Brady, M. Lung Cancer Assistant: A Hybrid Clinical Decision Support Application for Lung Cancer Care. J. R Soc. Interface 2014, 11, 20140534. [Google Scholar] [CrossRef]
- Durieux, P.; Nizard, R.; Ravaud, P.; Mounier, N.; Lepage, E. Clinical Decision Support System for Prevention of Venous Thromboembolism: Effect on Physician Behavior. J. Am. Med. Assoc. 2000, 283, 2816–2821. [Google Scholar] [CrossRef]
- Hamedan, F.; Orooji, A.; Sanadgol, H.; Sheikhtaheri, A. Clinical Decision Support System to Predict Chronic Kidney Disease: A Fuzzy Expert System Approach. Int. J. Med. Inform. 2020, 138, 104134. [Google Scholar] [CrossRef]
- Casal-Guisande, M.; Comesaña-Campos, A.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B. Design and Development of a Methodology Based on Expert Systems, Applied to the Treatment of Pressure Ulcers. Diagnostics 2020, 10, 614. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wells, A.; Brufsky, A.; Neapolitan, R. A Clinical Decision Support System Learned from Data to Personalize Treatment Recommendations towards Preventing Breast Cancer Metastasis. PLoS ONE 2019, 14, e0213292. [Google Scholar] [CrossRef] [PubMed]
- Casal-Guisande, M.; Comesaña-Campos, A.; Dutra, I.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B. Design and Development of an Intelligent Clinical Decision Support System Applied to the Evaluation of Breast Cancer Risk. J. Pers. Med. 2022, 12, 169. [Google Scholar] [CrossRef]
- Uzoka, F.M.E.; Osuji, J.; Obot, O. Clinical Decision Support System (DSS) in the Diagnosis of Malaria: A Case Comparison of Two Soft Computing Methodologies. Expert. Syst. Appl. 2011, 38, 1537–1553. [Google Scholar] [CrossRef]
- Casal-Guisande, M.; Álvarez-Pazó, A.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B.; Peláez-Lourido, G.; Comesaña-Campos, A. Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Screening and Early Diagnosis of Breast Cancer. Cancers 2023, 15, 1711. [Google Scholar] [CrossRef]
- Casal-Guisande, M.; Torres-Durán, M.; Mosteiro-Añón, M.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B.; Fernández-Villar, A.; Comesaña-Campos, A. Design and Conceptual Proposal of an Intelligent Clinical Decision Support System for the Diagnosis of Suspicious Obstructive Sleep Apnea Patients from Health Profile. Int. J. Environ. Res. Public Health 2023, 20, 3627. [Google Scholar] [CrossRef]
- Casal-Guisande, M.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B.; Comesaña-Campos, A. Integration of the Wang & Mendel Algorithm into the Application of Fuzzy Expert Systems to Intelligent Clinical Decision Support Systems. Mathematics 2023, 11, 2469. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Chadaga, R.; Umakanth, S.; Bhat, D.; Shashi Kumar, G.S. Explainable Artificial Intelligence Approaches for COVID-19 Prognosis Prediction Using Clinical Markers. Sci. Rep. 2024, 14, 1783. [Google Scholar] [CrossRef] [PubMed]
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Prabhu, S.; Rajagopala Chadaga, P. A Machine Learning and Explainable Artificial Intelligence Triage-Prediction System for COVID-19. Decis. Anal. J. 2023, 7, 100246. [Google Scholar] [CrossRef]
- Ahmad, I.; Amelio, A.; Merla, A.; Scozzari, F. A Survey on the Role of Artificial Intelligence in Managing Long COVID. Front. Artif. Intell. 2023, 6, 1292466. [Google Scholar] [CrossRef] [PubMed]
- Haendel, M.A.; Chute, C.G.; Bennett, T.D.; Eichmann, D.A.; Guinney, J.; Kibbe, W.A.; Payne, P.R.O.; Pfaff, E.R.; Robinson, P.N.; Saltz, J.H.; et al. The National COVID Cohort Collaborative (N3C): Rationale, Design, Infrastructure, and Deployment. J. Am. Med. Inform. Assoc. 2021, 28, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, E.R.; Girvin, A.T.; Bennett, T.D.; Bhatia, A.; Brooks, I.M.; Deer, R.R.; Dekermanjian, J.P.; Jolley, S.E.; Kahn, M.G.; Kostka, K.; et al. Identifying Who Has Long COVID in the USA: A Machine Learning Approach Using N3C Data. Lancet Digit. Health 2022, 4, e532–e541. [Google Scholar] [CrossRef] [PubMed]
- Antony, B.; Blau, H.; Casiraghi, E.; Loomba, J.J.; Callahan, T.J.; Laraway, B.J.; Wilkins, K.J.; Antonescu, C.C.; Valentini, G.; Williams, A.E.; et al. Predictive Models of Long COVID. EBioMedicine 2023, 96, 104777. [Google Scholar] [CrossRef] [PubMed]
- Reme, B.A.; Gjesvik, J.; Magnusson, K. Predictors of the Post-COVID Condition Following Mild SARS-CoV-2 Infection. Nat. Commun. 2023, 14, 5839. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Jain, V.; Singh, A. Stacking Ensemble-Based Intelligent Machine Learning Model for Predicting Post-COVID-19 Complications. New Gener. Comput. 2022, 40, 987–1007. [Google Scholar] [CrossRef]
- Patel, M.A.; Knauer, M.J.; Nicholson, M.; Daley, M.; Van Nynatten, L.R.; Cepinskas, G.; Fraser, D.D. Organ and Cell-Specific Biomarkers of Long-COVID Identified with Targeted Proteomics and Machine Learning. Mol. Med. 2023, 29, 26. [Google Scholar] [CrossRef]
- Wang, L.X.; Mendel, J.M. Generating Fuzzy Rules by Learning from Examples. IEEE Trans. Syst. Man. Cybern. 1992, 22, 1414–1427. [Google Scholar] [CrossRef]
- Pérez-González, A.; Araújo-Ameijeiras, A.; Fernández-Villar, A.; Crespo, M.; Poveda, E.; Cabrera, J.J.; del Campo, V.; de Araujo, B.G.; Gómez, C.; Leiro, V.; et al. Long COVID in Hospitalized and Non-Hospitalized Patients in a Large Cohort in Northwest Spain, a Prospective Cohort Study. Sci. Rep. 2022, 12, 3369. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Fernández, M.; Ramos-Hernández, C.; García-Río, F.; Torres-Durán, M.; Nodar-Germiñas, A.; Tilve-Gómez, A.; Rodríguez-Fernández, P.; Valverde-Pérez, D.; Ruano-Raviña, A.; Fernández-Villar, A. Alterations in Respiratory Function Test Three Months after Hospitalisation for COVID-19 Pneumonia: Value of Determining Nitric Oxide Diffusion. J. Clin. Med. 2021, 10, 2119. [Google Scholar] [CrossRef]
- Núñez-Fernández, M.; Ramos-Hernández, C.; García-Río, F.; Pérez-González, A.; Tilve-Gómez, A.; Rodríguez-Fernández, P.; Nodar-Germiñas, A.; Fernández-García, A.; Ruano-Raviña, A.; Fernández-Villar, A. Evolution and Long-term Respiratory Sequelae after Severe COVID-19 Pneumonia: Nitric Oxide Diffusion Measurement Value. Respir. Res. 2023, 24, 48. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman & Hall/CRC: Boca Raton, FL, USA, 1984; ISBN 9781351460491. [Google Scholar]
- Lewis, R.J. An Introduction to Classification and Regression Tree (CART) Analysis Introduction to CART. In Proceedings of the 2000 Annual Meeting of the Society for Academic Emergency Medicine, San Francisco, CA, USA, 22–25 May 2000. [Google Scholar]
- Tran, V.T.; Yang, B.S.; Oh, M.S.; Tan, A.C.C. Fault Diagnosis of Induction Motor Based on Decision Trees and Adaptive Neuro-Fuzzy Inference. Expert. Syst. Appl. 2009, 36, 1840–1849. [Google Scholar] [CrossRef]
- Questier, F.; Put, R.; Coomans, D.; Walczak, B.; Heyden, Y. Vander the Use of CART and Multivariate Regression Trees for Supervised and Unsupervised Feature Selection. Chemom. Intell. Lab. Syst. 2005, 76, 45–54. [Google Scholar] [CrossRef]
- Dong, N.; die Zhai, M.; Zhao, L.; Wu, C.H. Cervical Cell Classification Based on the CART Feature Selection Algorithm. J. Ambient. Intell. Humaniz Comput. 2021, 12, 1837–1849. [Google Scholar] [CrossRef]
- Mamdani, E.H.; Assilian, S. An Experiment in Linguistic Synthesis with a Fuzzy Logic Controller. Int. J. Man Mach. Stud. 1975, 7, 1–13. [Google Scholar] [CrossRef]
- Mamdani, E.H. Advances in the Linguistic Synthesis of Fuzzy Controllers. Int. J. Man Mach. Stud. 1976, 8, 669–678. [Google Scholar] [CrossRef]
- Mamdani, E.H. Application of Fuzzy Logic to Approximate Reasoning Using Linguistic Synthesis. IEEE Trans. Comput. 1977, 26, 1182–1191. [Google Scholar] [CrossRef]
- Ross, T.J. Fuzzy Logic with Engineering Applications, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2010; ISBN 9781119994374. [Google Scholar]
- Casal-Guisande, M.; Ceide-Sandoval, L.; Mosteiro-Añón, M.; Torres-Durán, M.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B.; Fernández-Villar, A.; Comesaña-Campos, A. Design of an Intelligent Decision Support System Applied to the Diagnosis of Obstructive Sleep Apnea. Diagnostics 2023, 13, 1854. [Google Scholar] [CrossRef] [PubMed]
- Statistics and Machine Learning Toolbox—MATLAB. Available online: https://es.mathworks.com/products/statistics.html (accessed on 13 December 2023).
- Fuzzy Logic Toolbox–MATLAB. Available online: https://www.mathworks.com/products/fuzzy-logic.html (accessed on 1 November 2022).
- App Designer. Available online: https://www.mathworks.com/products/matlab/app-designer.html (accessed on 18 October 2022).
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal Classifier for Imbalanced Data Using Matthews Correlation Coefficient Metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Guilford, J.P. Psychometric Methods; McGraw-Hill: New York, NY, USA, 1954. [Google Scholar]
- Akinola, O.O.; Ezugwu, A.E.; Agushaka, J.O.; Zitar, R.A.; Abualigah, L. Multiclass Feature Selection with Metaheuristic Optimization Algorithms: A Review. Neural Comput. Appl. 2022, 34, 19751–19790. [Google Scholar] [CrossRef]
- Zaitseva, E.; Rabcan, J.; Levashenko, V.; Kvassay, M. Importance Analysis of Decision Making Factors Based on Fuzzy Decision Trees. Appl. Soft. Comput. 2023, 134, 109988. [Google Scholar] [CrossRef]







| Group | Variable | Type | Comment | |
|---|---|---|---|---|
| Independent variables | General and anthropometric data | Sex | Categorical | Man (1)/woman (0) |
| Age | Numerical | - | ||
| Body mass index (BMI) | Numerical | - | ||
| Toxic habits | Smoking | Categorical | Yes(1)/no(0) | |
| Previous comorbidities | Lung diseases | Categorical | Yes(1)/no(0) | |
| Heart diseases | Categorical | Yes(1)/no(0) | ||
| Diabetes | Categorical | Yes(1)/no(0) | ||
| Arterial hypertension | Categorical | Yes(1)/no(0) | ||
| Severity of the disease | Severity of pneumonia | Categorical | Refers to the unit to which the patient was admitted during hospitalisation for COVID-19-associated pneumonia: ICU(1)/IRCU(0). | |
| Situation after three months | Dyspnea after three months | Categorical | The Modified British Medical Research Council (mMRC) scale is used to assess dyspnea. If the score is zero, the patient has no dyspnea; if the score is greater than zero, the patient has symptoms of dyspnea. Yes(1)/No(0) | |
| Chest X-ray | Categorical | Affected X-ray (1)/No affect X-ray (0) | ||
| Alteration of pulmonary diffusion: DLNO | Numerical | Reference percentage of the diffusion capacity in relation to the theoretical value | ||
| Dependent variable | Situation after 12 months | Dyspnea after twelve months | Categorical | The Modified British Medical Research Council (mMRC) scale is used to assess dyspnea. If the score is zero, the patient has no dyspnea; if the score is greater than zero, the patient has symptoms of dyspnea. Yes (1)/No (0) |
| Expert System 1 | |
| Input | Dyspnea at 3 months, Sex and Age |
| Output | R1 |
| Expert System 2 | |
| Input | Lung diseases, Smoking and DLNO |
| Output | R2 |
| Expert System 3 | |
| Input | R1 and R2 |
| Output | Dyspnea Risk at 12 months |
| Variable | Value |
|---|---|
| Sex | Man |
| Age | 45 |
| Dyspnea at third month | Yes |
| Lung diseases | No |
| Smoking | No |
| DLNO | 70 |
| Expert System | Rules |
|---|---|
| Expert System 1 | IF (Dyspnea at third month is Yes) AND (Age is Low) AND (Sex is Man) THEN (R1 is Dyspnea) IF (Dyspnea at third month is Yes) AND (Age is Medium) AND (Sex is Man) THEN (R1 is Dyspnea) |
| Expert System 2 | IF (Lung diseases is No) AND (Smoking is No) AND (DLNO es medium) THEN (R2 is No Dyspnea) IF (Lung diseases is No) AND (Smoking is No) AND (DLNO es High) THEN (R2 is No Dyspnea) |
| Expert System 3 | IF (R1 is Medium) AND (R2 is Low) THEN (R3 is Dyspnea) IF (R1 is High) AND (R2 is Low) THEN (R3 is Dyspnea) |
| Reasoning | Scalability | Efficiency | Data Dependence | |
|---|---|---|---|---|
| Plaff et al. [47] | The system relies on statistical inference approaches. | The system is not scalable. | The authors employed XGBoost model for their analysis. They used an implicit approach to manage uncertainty based on probabilities. | This is a fully data-dependent approach, as it employs supervised learning approaches. |
| - | - | = | = | |
| Blessy Antony et al. [48] | The system relies on statistical inference approaches. | The system is not scalable. | The authors employed logistic regression and random forest for their analysis. They used an implicit approach to manage uncertainty based on probabilities. | This is a fully data-dependent approach, as it employs supervised learning approaches. |
| - | - | = | = | |
| Reme et al. [49] | The system relies on statistical inference approaches. | The system is not scalable. | The authors employed the LASSO and random forest models to analyse the data. They also utilised an implicit approach to manage uncertainty based on probabilities. | This is a fully data-dependent approach, as it employs supervised learning approaches. |
| - | - | = | = | |
| Gupta et al. [50] | The system relies on statistical inference approaches. | The system is not scalable. | The authors used a stacking ensemble model supported by Deep Neural Networks to analyse the data. They also utilised an implicit approach to manage uncertainty based on probabilities. | This is a fully data-dependent approach, as it employs supervised learning approaches. |
| - | - | = | = | |
| Patel et al. [51] | The system relies on statistical inference approaches. | The system is not scalable. | The authors employ a Random Forest model to analyse the data. They also utilised an implicit approach to manage uncertainty based on probabilities. | This is a fully data-dependent approach, as it employs supervised learning approaches. |
| - | - | = | = | |
| Our proposal | The proposed system relies on the use of symbolic inference approaches. | The proposed system is scalable, since it is possible to modify the inference engines. | The system uses fuzzy inference engines, which allow uncertainty management from a non-probabilistic point of view. | Data dependence is present, since they are necessary to define the knowledge bases of the expert systems. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casal-Guisande, M.; Comesaña-Campos, A.; Núñez-Fernández, M.; Torres-Durán, M.; Fernández-Villar, A. Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Prediction of Dyspnea after 12 Months of an Acute Episode of COVID-19. Biomedicines 2024, 12, 854. https://doi.org/10.3390/biomedicines12040854
Casal-Guisande M, Comesaña-Campos A, Núñez-Fernández M, Torres-Durán M, Fernández-Villar A. Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Prediction of Dyspnea after 12 Months of an Acute Episode of COVID-19. Biomedicines. 2024; 12(4):854. https://doi.org/10.3390/biomedicines12040854
Chicago/Turabian StyleCasal-Guisande, Manuel, Alberto Comesaña-Campos, Marta Núñez-Fernández, María Torres-Durán, and Alberto Fernández-Villar. 2024. "Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Prediction of Dyspnea after 12 Months of an Acute Episode of COVID-19" Biomedicines 12, no. 4: 854. https://doi.org/10.3390/biomedicines12040854
APA StyleCasal-Guisande, M., Comesaña-Campos, A., Núñez-Fernández, M., Torres-Durán, M., & Fernández-Villar, A. (2024). Proposal and Definition of an Intelligent Clinical Decision Support System Applied to the Prediction of Dyspnea after 12 Months of an Acute Episode of COVID-19. Biomedicines, 12(4), 854. https://doi.org/10.3390/biomedicines12040854