Plasma Circular RNAs as Biomarkers for Breast Cancer
Abstract
:1. Breast Cancer
2. circRNAs
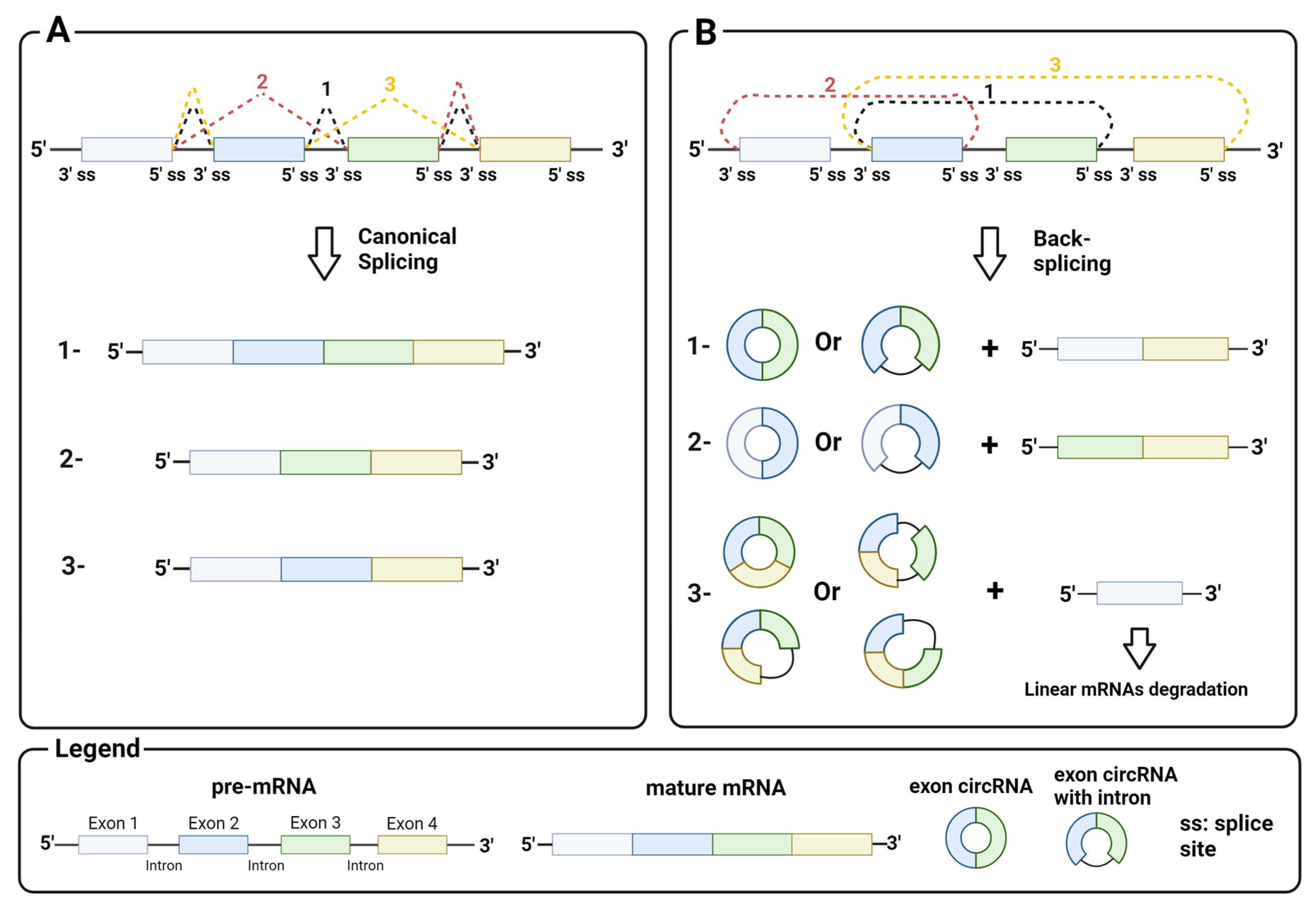
2.1. Biosynthesis
2.2. Biological Function
3. Liquid Biopsy and Circulating circRNAs
- Circulating tumor cells (CTCs): As the name suggests, CTCs are cells that have detached from a tumor and migrated into the bloodstream. They have a low concentration (about 10 cells/mL blood), but can help to preserve the biological, molecular, and histological characteristics of the primary tumor [52,53].
- Cell-free circulating nucleic acids (ccfNAs): These are essentially nucleic acids, DNA and RNA, that circulate freely in body fluids. Like CTCs, they have a low concentration and are highly fragmented. Nevertheless, the concentration, presence of mutations and integrity of ccfNAs have been shown to provide important diagnostic, prognostic and predictive information in cancer [54].
- Tumor-educated platelets (TEPs): These are platelets that have been altered by the tumor through the exchange of biomolecules to make them receptive to cancer signals. They play an active role in metastasis by covering the tumor cells in the bloodstream and protecting them from the immune system. Therefore, the concentration and molecular composition of TEPs can provide useful information about tumor progression [55].
- Exosomes: These are a class of extracellular vesicles that transport biomolecules from the cells of origin. Exosomes can be found in the body fluids of patients with various diseases and, therefore, have great potential as biomarkers. The characterization of their content is helpful to deepen the molecular profile of the pathological cells of origin [56].
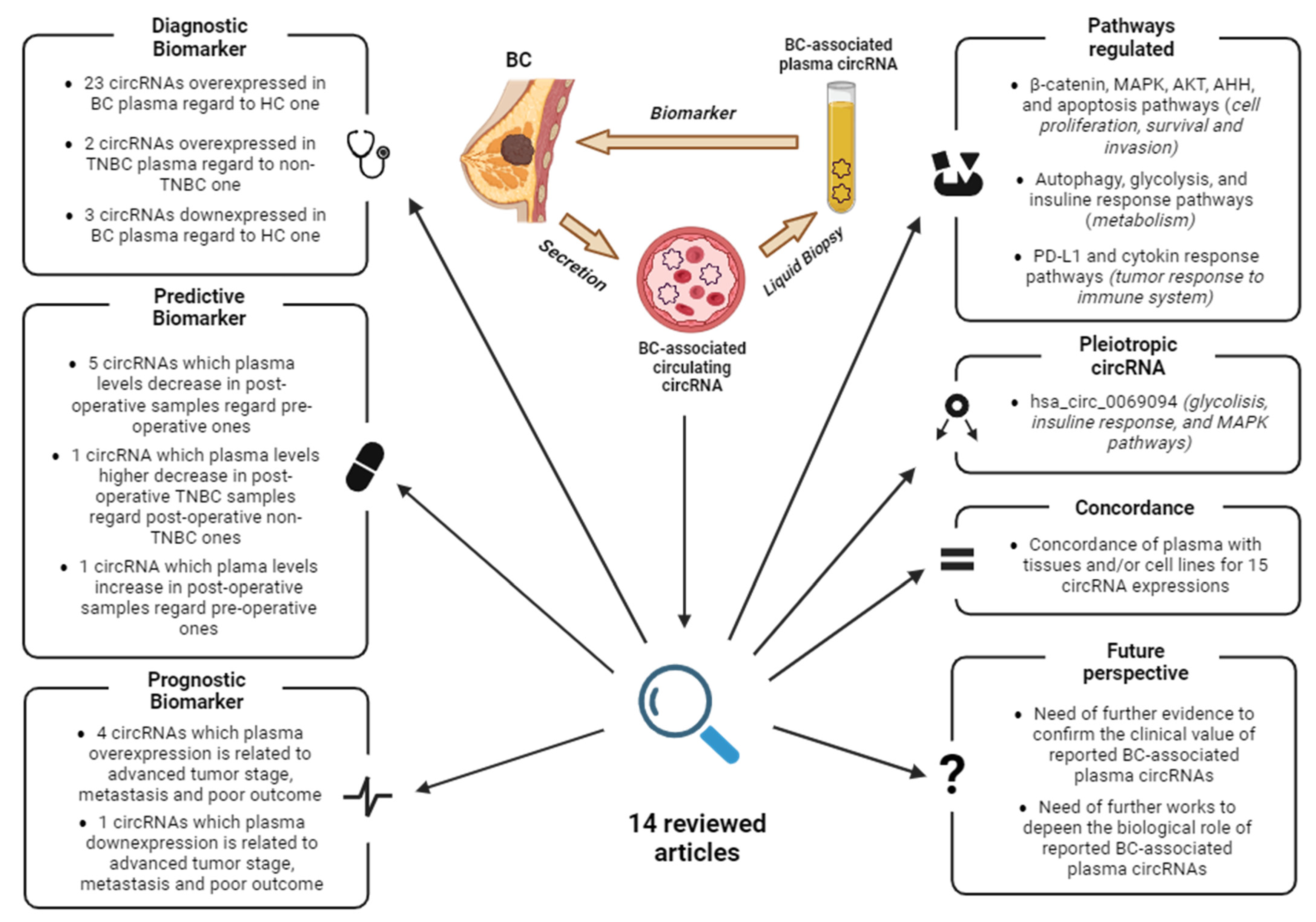
4. BC-Associated ccircRNAs
4.1. Identification of ccircRNAs by Expression Profiling of Plasma
4.2. Identification of ccircRNAs by Expression Profiling of Tissue
4.3. Identification of ccircRNAs by Literature and Database Screening
4.4. TNBC-Associated ccircRNAs
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| circRNA | Circular RNA |
| ccircRNA | Circulating circRNA |
| lncRNA | Long non-coding RNA |
| cRBP | circRNA binding protein |
| IRES | Internal ribosome entry site |
| M6A | N6-methyladenosine |
| CTC | Circulating tumor cells |
| ccfNA | Circulating cell-free nucleic acid |
| TEP | Tumor-educated platelets |
| EV | Extracellular vesicle |
| BBT | Benign breast tumor |
| ER | Estrogen receptor breast cancer |
| PR | Progesterone receptor breast cancer |
| OS | Overall survival |
| DSS | Disease-specific survival |
| TNBC | Triple-negative breast cancer |
| ALN | Axillary lymph node |
| BMBC | Brain metastasis breast cancer |
| BMFS | Brain metastasis-free survival |
| DFS | Disease-free survival |
| THP | Pirarubicin |
| NAC | Neo-adjuvant chemotherapy |
| HCC | Hepatocellular cancer |
| PC | Prostate cancer |
| OC | Ovarian cancer |
| CC | Colorectal cancer |
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.K.Y.; Tan, P.H. Liquid Biopsy in Breast Cancer: A Focused Review. Arch. Pathol. Lab. Med. 2020, 145, 678–686. [Google Scholar] [CrossRef]
- Tierno, D.; Grassi, G.; Zanconati, F.; Bortul, M.; Scaggiante, B. An Overview of Circulating Cell-Free Nucleic Acids in Diagnosis and Prognosis of Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2023, 24, 1799. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific Expression and Functions of Circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Mascrez, B.; Htuin, D.; Bailleul1, B. Mis-Splicing Yields Circular RNA Molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A Map of Human Circular RNAs in Clinically Relevant Tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Feng, J.; Lei, L.; Hu, J.; Xia, L.; Wang, J.; Xiang, Y.; Liu, L.; Zhong, S.; Han, L.; et al. Comprehensive Characterization of Tissue-Specific Circular RNAs in the Human and Mouse Genomes. Briefings Bioinform. 2016, 18, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, T.; Doss C, G.P. Non-Coding RNAs in Human Health and Disease: Potential Function as Biomarkers and Therapeutic Targets. Funct. Integr. Genom. 2023, 23, 33. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Yang, L. Regulation of CircRNA Biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Lovci, M.T.; Ghanem, D.; Marr, H.; Arnold, J.; Gee, S.; Parra, M.; Liang, T.Y.; Stark, T.J.; Gehman, L.T.; Hoon, S.; et al. Rbfox Proteins Regulate Alternative MRNA Splicing through Evolutionarily Conserved RNA Bridges. Nat. Struct. Mol. Biol. 2013, 20, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned from MicroRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Jesenko, T.; Brezar, S.K.; Cemazar, M.; Biasin, A.; Tierno, D.; Scaggiante, B.; Grassi, M.; Grassi, C.; Dapas, B.; Truong, N.H.; et al. Targeting Non-Coding RNAs for the Development of Novel Hepatocellular Carcinoma Therapeutic Approaches. Pharmaceutics 2023, 15, 1249. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the CeRNA Hypothesis with Quantitative Measurements of MiRNA and Target Abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, X.; Zhang, Z.; Lai, Y.; Lin, X.; Lin, B.; Ma, M.; Liang, X.; Li, X.; Lv, W.; et al. CircCD44 Plays Oncogenic Roles in Triple-Negative Breast Cancer by Modulating the MiR-502–5p/KRAS and IGF2BP2/Myc Axes. Mol. Cancer 2021, 20, 138. [Google Scholar] [CrossRef]
- Hall, I.F.; Climent, M.; Quintavalle, M.; Farina, F.M.; Schorn, T.; Zani, S.; Carullo, P.; Kunderfranco, P.; Civilini, E.; Condorelli, G.; et al. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating MiRNA-145 Function. Circ. Res. 2019, 124, 498–510. [Google Scholar] [CrossRef]
- Xu, B.; Yang, T.; Wang, Z.; Zhang, Y.; Liu, S.; Shen, M. CircRNA CDR1as/MiR-7 Signals Promote Tumor Growth of Osteosarcoma with a Potential Therapeutic and Diagnostic Value. Cancer Manag. Res. 2018, 10, 4871–4880. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ji, M.; He, G.; Yang, L.; Niu, Z.; Jian, M.; Wei, Y.; Ren, L.; Xu, J. Silencing CDR1as Inhibits Colorectal Cancer Progression through Regulating MicroRNA-7. OncoTargets Ther. 2017, 10, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, E.; Miyakawa, Y.; Kishikawa, T.; Seimiya, T.; Iwata, T.; Funato, K.; Odawara, N.; Sekiba, K.; Yamagami, M.; Suzuki, T.; et al. Expression of Circular RNA CDR1-AS in Colon Cancer Cells Increases Cell Surface PD-L1 Protein Levels. Oncol. Rep. 2019, 42, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, D.; Wei, Y. Overexpressed CDR1as Functions as an Oncogene to Promote the Tumor Progression via MiR-7 in Non-Small-Cell Lung Cancer. OncoTargets Ther. 2018, 11, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, F.-B.; Huang, M.; Xie, K.; Xie, Q.-S.; Liu, C.-H.; Shen, M.-J.; Huang, Q. Circular RNA CiRS-7 Promotes the Proliferation and Metastasis of Pancreatic Cancer by Regulating MiR-7-Mediated EGFR/STAT3 Signaling Pathway. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, F.; Zhu, Z.; Ding, A.; Luo, J. CircRNA CDR1as/MiR-1287/Raf1 Axis Modulates Hepatocellular Carcinoma Progression through MEK/ERK Pathway. Cancer Manag. Res. 2020, 12, 8951–8964. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Liu, S.; Ding, P.; Chang, S.; Sang, M. Circular RNA CiRS-7 Inhibits Autophagy of ESCC Cells by Functioning as MiR-1299 Sponge to Target EGFR Signaling. J. Cell. Biochem. 2019, 121, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, X.; Yuan, W.; Yang, C.; Zhang, X.; Han, J.; Wang, J.; Deng, X.; Yang, H.; Li, P.; et al. CircRNA-Cdr1as Exerts Anti-Oncogenic Functions in Bladder Cancer by Sponging MicroRNA-135a. Cell. Physiol. Biochem. 2018, 46, 1606–1616. [Google Scholar] [CrossRef]
- Lukiw, W.J. Circular RNA (CircRNA) in Alzheimer’s Disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of Tumor Apoptosis through a Circular RNA Enhancing Foxo3 Activity. Cell Death Differ. 2016, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes with P21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and Function of the CGAS-STING Pathway of Cytosolic DNA Sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-Coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Du, W.W.; Awan, F.M.; Dong, J.; Yang, B.B. The Circular RNA Circ-Ccnb1 Dissociates Ccnb1/Cdk1 Complex Suppressing Cell Invasion and Tumorigenesis. Cancer Lett. 2019, 459, 216–226. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A Peptide Encoded by Circular Form of LINC-PINT Suppresses Oncogenic Transcriptional Elongation in Glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Mao, Y.; Chen, X.; Xiao, J.; Qin, Y.; Zhao, L. The Functional Roles, Cross-Talk and Clinical Implications of M6a Modification and CircRNA in Hepatocellular Carcinoma. Int. J. Biol. Sci. 2021, 17, 3059–3079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A Novel Protein Encoded by the Circular Form of the SHPRH Gene Suppresses Glioma Tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Galardi, A.; Colletti, M.; Palma, A.; Di Giannatale, A. An Update on Circular RNA in Pediatric Cancers. Biomedicines 2022, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, D.; Pei, X.; Zhang, Y.; Zhang, Y.; Gu, Y.; Li, Y. Circska3 Modulates Foxm1 to Facilitate Cell Proliferation, Migration, and Invasion While Confine Apoptosis in Medul-loblastoma via Mir-383-5p. Cancer Manag. Res. 2020, 12, 13415–13426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Guan, J.; Luo, M. Circ-SKA3 Upregulates ID3 Expression by Decoying MiR-326 to Accelerate the Development of Medulloblastoma. J. Clin. Neurosci. 2021, 86, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-C.; Wang, F.-C.; Wang, J.-H.; Zhao, J.-Y.; Ye, S.-Y. The Circular RNA CircSKA3 Facilitates the Malignant Biological Behaviors of Medulloblastoma via MiR-520 h/CDK6 Pathway. Mol. Biotechnol. 2022, 64, 1022–1033. [Google Scholar] [CrossRef]
- Chen, N.; Chao, D.; Li, X. Circular RNA Has_circ_0000527 Participates in Proliferation, Invasion and Migration of Retinoblastoma Cells via MiR-646/BCL-2 Axis. Cell Biochem. Funct. 2020, 38, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yuan, H.-F.; Xu, D.; Chen, K.-J.; Tan, N.; Zheng, Q.-J. Circular RNA Circ_0000034 Upregulates STX17 Level to Promote Human Retinoblastoma Development via Inhibiting MiR-361-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12080–12092. [Google Scholar] [PubMed]
- Sun, Z.; Zhang, A.; Hou, M.; Jiang, T. Circular RNA Hsa_circ_0000034 Promotes the Progression of Retinoblastoma via Sponging MicroRNA-361-3p. Bioengineered 2020, 11, 949–957. [Google Scholar] [CrossRef]
- Ren, S.; Huang, M.; Bai, R.; Chen, L.; Yang, J.; Zhang, J.; Guo, W.; Ji, W.; Chen, Y. Efficient Modulation of Exon Skipping via Antisense Circular RNAs. Research 2023, 6, 0045. [Google Scholar] [CrossRef]
- Xie, J.; Ye, F.; Deng, X.; Tang, Y.; Liang, J.-Y.; Huang, X.; Sun, Y.; Tang, H.; Lei, J.; Zheng, S.; et al. Circular RNA: A Promising New Star of Vaccine. J. Transl. Intern. Med. 2022, 11, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bode, A.M.; Dong, Z. Circulating Tumor Cells: Moving Biological Insights into Detection. Theranostics 2017, 7, 2606–2619. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Bortul, M.; Giudici, F.; Tierno, D.; Generali, D.; Scomersi, S.; Grassi, G.; Bottin, C.; Cappelletti, M.R.; Zanconati, F.; Scaggiante, B. A Case–Control Study by DdPCR of ALU 260/111 and LINE-1 266/97 Copy Number Ratio in Circulating Cell-Free DNA in Plasma Revealed LINE-1 266/97 as a Potential Biomarker for Early Breast Cancer Detection. Int. J. Mol. Sci. 2023, 24, 8520. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.E.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for Exosome Isolation and Analysis: Enabling Liquid Biopsy for Personalized Medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal CircRNAs: Biogenesis, Effect and Application in Human Diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Fu, K.; Sun, H.; Rong, D.; Wang, H.; Cao, H. CircRNA Microarray Profiling Identifies a Novel Circulating Biomarker for Detection of Gastric Cancer. Mol. Cancer 2018, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Z.; Jiang, P.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Bi, H.; Liu, X.; Li, X. Circular RNA IARS (Circ-IARS) Secreted by Pancreatic Cancer Cells and Located within Exosomes Regulates Endothelial Monolayer Permeability to Promote Tumor Metastasis. J. Exp. Clin. Cancer Res. 2018, 37, 177. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.-B.; Yan, M.-G.; Fang, X.; Guo, J.-J.; Xiong, W.; Zhang, R.-P. Circulating Circular RNA Hsa_circ_0001785 Acts as a Diagnostic Biomarker for Breast Cancer Detection. Clin. Chim. Acta 2018, 487, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, J.; Lin, W.; Weng, J.; Hong, W.; Zou, J.; Zhang, T.; Ye, C.; Chen, Y. Circular RNA Hsa_circ_0001785 Inhibits the Proliferation, Migration and Invasion of Breast Cancer Cells in Vitro and in Vivo by Sponging MiR-942 to Upregulate SOCS3. Cell Cycle 2020, 19, 2811–2825. [Google Scholar] [CrossRef] [PubMed]
- Carow, B.; Rottenberg, M.E. SOCS3, a Major Regulator of Infection and Inflammation. Front. Immunol. 2014, 5, 58. [Google Scholar] [CrossRef]
- Yang, C.; Wang, M.; Huang, R.; Ou, L.; Li, M.; Wu, W.; Lei, R. Circ_0108942 Regulates the Progression of Breast Cancer by Regulating the MiR-1178-3p/TMED3 Axis. Clin. Breast Cancer 2022, 23, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, Y.; Li, Q. TMED3 Promotes Proliferation and Migration in Breast Cancer Cells by Activating Wnt/β-Catenin Signaling. OncoTargets Ther. 2020, 13, 5819–5830. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Lei, L.; Dong, S.; Liu, D. Circular RNA Hsa_circ_0068033 Acts as a Diagnostic Biomarker and Suppresses the Progression of Breast Cancer through Sponging MiR-659. OncoTargets Ther. 2020, 13, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Cai, G.-X.; Zhai, X.-M.; Yang, X.-X.; Li, M.; Li, K.; Zhou, C.-L.; Liu, T.-C.; Han, B.-W.; Liu, Z.-J.; et al. Plasma-Derived Extracellular Vesicles Circular RNAs Serve as Biomarkers for Breast Cancer Diagnosis. Front. Oncol. 2021, 11, 752651. [Google Scholar] [CrossRef]
- Hu, Y.; Song, Q.; Zhao, J.; Ruan, J.; He, F.; Yang, X.; Yu, X. Identification of Plasma Hsa_circ_0008673 Expression as a Potential Biomarker and Tumor Regulator of Breast Cancer. J. Clin. Lab. Anal. 2020, 34, 23393. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, S.; Wang, T.; Bi, S. Hsa_circ_0008673 Promotes Breast Cancer Progression by MiR-578/GINS4 Axis. Clin. Breast Cancer 2022, 23, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, Y.; Ma, J.; Luo, L.; Ma, B. Expression and Prognosis Analysis of GINS Subunits in Human Breast Cancer. Medicine 2021, 100, e24827. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, F.; Wu, L.; Zhang, X.; Tian, J.; Li, J.; Cao, J.; Ma, Y.; Zhang, L.; Wang, L. Identification of Hsa_circ_0104824 as a Potential Biomarkers for Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820960745. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Z.; Hu, G.; Zhang, Y.; Feng, Y.; Jiang, Y.; Wang, J. Profiling and Integrated Analysis of Differentially Expressed CircRNAs as Novel Biomarkers for Breast Cancer. J. Cell. Physiol. 2020, 235, 7945–7959. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Wang, X.; Liu, J.; Zhang, M.; Feng, K.; Wang, X. Hsa_circ_0069094 Accelerates Cell Malignancy and Glycolysis through Regulating the MiR-591/HK2 Axis in Breast Cancer. Cell. Signal. 2020, 79, 109878. [Google Scholar] [CrossRef] [PubMed]
- Tan, V.P.; Miyamoto, S. HK2/Hexokinase-II Integrates Glycolysis and Autophagy to Confer Cellular Protection. Autophagy 2015, 11, 963–964. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Qu, W.; Lian, Y.; Feng, C.; Zhan, Y. Hsa_circ_0069094 Knockdown Inhibits Cell Proliferation, Migration, Invasion and Glycolysis, While Induces Cell Apoptosis by MiR-661/HMGA1 Axis in Breast Cancer. Anti-Cancer Drugs 2021, 32, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Chiefari, E.; Nevolo, M.T.; Arcidiacono, B.; Maurizio, E.; Nocera, A.; Iiritano, S.; Sgarra, R.; Possidente, K.; Palmieri, C.; Paonessa, F.; et al. HMGA1 is a Novel Downstream Nuclear Target of the Insulin Receptor Signaling Pathway. Sci. Rep. 2012, 2, 251. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Han, Q.; Zhu, B.; Wan, L.; Feng, E. Circ_0069094 Regulates Malignant Phenotype and Paclitaxel Resistance in Breast Cancer Cells via Targeting the MiR-136-5p/YWHAZ Axis. Thorac. Cancer 2023, 14, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Neukamm, S.S.; Toth, R.; Morrice, N.; Campbell, D.G.; MacKintosh, C.; Lehmann, R.; Haering, H.-U.; Schleicher, E.D.; Weigert, C. Identification of the Amino Acids 300–600 of IRS-2 as 14-3-3 Binding Region with the Importance of IGF-1/Insulin-Regulated Phosphorylation of Ser-573. PLoS ONE 2012, 7, e43296. [Google Scholar] [CrossRef]
- Chen, S.; Synowsky, S.; Tinti, M.; MacKintosh, C. The Capture of Phosphoproteins by 14-3-3 Proteins Mediates Actions of Insulin. Trends Endocrinol. Metab. 2011, 22, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.; Li, J.; Lv, Q.; Feng, D. Hsa_circ_0069094 Positively Regulates the Expression of Oncogenic ZNF217 by Competitively Targeting MiR-758–3p to Promote the Development of Breast Cancer. Reprod. Biol. 2022, 22, 100708. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, K.G.; Verger, A.; Yaswen, P.; Crossley, M. Amplification of Zinc Finger Gene 217 (ZNF217) and Cancer: When Good Fingers Go Bad. Biochim. et Biophys. Acta (BBA) Rev. Cancer 2007, 1775, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zheng, W.; Ji, C.; Wang, X.; Chen, M.; Hua, K.; Deng, X.; Fang, L. Tumor-Derived CircRNAs as Circulating Biomarkers for Breast Cancer. Front. Pharmacol. 2022, 13, 811856. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.-F.; Chen, F.-F.; Fan, Y.-F.; Zhang, K.; Chen, H.-H. Circ-0000512 Inhibits PD-L1 Ubiquitination through Sponging MiR-622/CMTM6 Axis to Promote Triple-Negative Breast Cancer and Immune Escape. J. Immunother. Cancer 2023, 11, e005461. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 Maintains the Expression of PD-L1 and Regulates Anti-Tumour Immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Peng, X.; Liu, Y.; Hao, R.; Zhao, R.; Zhang, L.; Zhao, F.; Liu, Q.; Liu, Y.; Qi, Y. The Diagnostic Value of Serum Exosomal Has_circ_0000615 for Breast Cancer Patients. Int. J. Gen. Med. 2021, 14, 4545–4554. [Google Scholar] [CrossRef] [PubMed]
- Reed, N.P.; Henderson, M.A.; Oltz, E.M.; Aune, T.M. Reciprocal Regulation of Rag Expression in Thymocytes by the Zinc-Finger Proteins, Zfp608 and Zfp609. Genes Immun. 2012, 14, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.L.v.D.; Azzarelli, R.; Oishi, K.; Martynoga, B.; Urbán, N.; Dekkers, D.H.; Demmers, J.A.; Guillemot, F. Nipbl Interacts with Zfp609 and the Integrator Complex to Regulate Cortical Neuron Migration. Neuron 2016, 93, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Liu, Z.; Liang, M.; Pan, J.; Lin, M.; Lin, H.; Luo, Y.; Zhou, X.; Yao, W. Identification of CircRNA–MiRNA–MRNA Networks Contributes to Explore Underlying Pathogenesis and Therapy Strategy of Gastric Cancer. J. Transl. Med. 2021, 19, 226. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.S.; Zheng, R.N.; Guo, L.B.; Fu, X.J. Circular RNA Circ_0000615 Knockdown Suppresses the Development of Nasopharyngeal Cancer through Regulating the MiR-338-3p/FGF2 Axis. Neoplasma 2020, 67, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Zhou, M.; Du, D.; Wang, C.; Yao, J.; Li, H.; Luo, Y.; He, F.; He, J. EIF4A3-Mediated Circ_0042881 Activates the RAS Pathway via MiR-217/SOS1 Axis to Facilitate Breast Cancer Progression. Cell Death Dis. 2023, 14, 559. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jia, L.; Shao, J.; Liu, H.; Wu, Q.; Wu, X. Circular RNA CircNF1 SiRNA Silencing Inhibits Glioblastoma Cell Proliferation by Promoting the Maturation of MiR-340. Front. Neurol. 2021, 12, 658076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, K.; Pitts, S.; Cheng, Y.; Liu, X.; Ke, X.; Kovaka, S.; Ashktorab, H.; Smoot, D.T.; Schatz, M.; et al. Novel Circular RNA CircNF1 Acts as a Molecular Sponge, Promoting Gastric Cancer by Absorbing MiR-16. Endocr. Relat. Cancer 2019, 26, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Zhang, A.; Li, M.; Pan, L.; Tang, W.; An, M.; Liu, W.; Zhang, J. Circular RNA Profile of Breast Cancer Brain Metastasis: Identification of Potential Biomarkers and Therapeutic Targets. Epigenomics 2018, 10, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Liu, W.; Zhu, C.; Li, P.; Wang, L.; Pan, L.; Li, K.; Cai, P.; Meng, M.; Wang, Y.; et al. Circular Rna Circbcbm1 Promotes Breast Cancer Brain Metastasis by Modulating Mir-125a/Brd4 Axis. Int. J. Biol. Sci. 2021, 17, 3104–3117. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Hirosue, A.; Nakamoto, M.; Yoshida, R.; Sakata, J.; Matsuoka, Y.; Kawahara, K.; Nagao, Y.; Nagata, M.; Takahashi, N.; et al. BRD4 Promotes Metastatic Potential in Oral Squamous Cell Carcinoma through the Epigenetic Regulation of the MMP2 Gene. Br. J. Cancer 2020, 123, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Sui, X.-M.; Sui, Y.-N.; Zhu, Q.-W.; Yan, K.; Wang, L.-S.; Wang, F.; Zhou, J.-H. BRD4 Induces Cell Migration and Invasion in HCC Cells through MMP-2 and MMP-9 Activation Mediated by the Sonic hedgehog Signaling Pathway. Oncol. Lett. 2015, 10, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Borri, F.; Granaglia, A. Pathology of Triple Negative Breast Cancer. Semin. Cancer Biol. 2020, 72, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A Regulated by FUS Accelerates Triple-Negative Breast Cancer Progression by Modulating NFIB Expression and Translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef] [PubMed]
- Ponente, M.; Campanini, L.; Cuttano, R.; Piunti, A.; Delledonne, G.A.; Coltella, N.; Valsecchi, R.; Villa, A.; Cavallaro, U.; Pattini, L.; et al. PML Promotes Metastasis of Triple-Negative Breast Cancer through Transcriptional Regulation of HIF1A Target Genes. J. Clin. Investig. 2017, 2, e87380. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.A.; Hassan, K.; Naik, M.; Mohapatra, N.; Balan, P.; Korrapati, P.S.; Dixit, M. EEF1A2 Promotes HIF1A Mediated Breast Cancer Angiogenesis in Normoxia and Participates in a Positive Feedback Loop with HIF1A in Hypoxia. Br. J. Cancer 2023, 130, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.C.; Cech, T.R.; Parker, R.R. Biochemical Properties and Biological Functions of FET Proteins. Annu. Rev. Biochem. 2015, 84, 355–379. [Google Scholar] [CrossRef] [PubMed]
- Darbeheshti, F.; Mansoori, Y.; Azizi-Tabesh, G.; Zolfaghari, F.; Kadkhoda, S.; Rasti, A.; Rezaei, N.; Shakoori, A. Evaluation of Circ_0000977-Mediated Regulatory Network in Breast Cancer: A Potential Discriminative Biomarker for Triple-Negative Tumors. Biochem. Genet. 2023, 61, 1487–1508. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhao, Z.; Ma, L.; Zhao, W.; Hu, Y.; Song, Y. Novel Exosomal CircEGFR Facilitates Triple Negative Breast Cancer Autophagy via Promoting TFEB Nuclear Trafficking and Modulating MiR-224-5p/ATG13/ULK1 Feedback Loop. Oncogene 2024, 43, 821–836. [Google Scholar] [CrossRef] [PubMed]
- Cocco, S.; Leone, A.; Piezzo, M.; Caputo, R.; Di Lauro, V.; Di Rella, F.; Fusco, G.; Capozzi, M.; di Gioia, G.; Budillon, A.; et al. Targeting Autophagy in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 7836. [Google Scholar] [CrossRef] [PubMed]
- Alers, S.; Wesselborg, S.; Stork, B. ATG13: Just a Companion, or an Executor of the Autophagic Program? Autophagy 2014, 10, 944–956, Erratum in Autophagy 2014, 10, 1481. [Google Scholar] [CrossRef] [PubMed]
- Zachari, M.; Ganley, I.G. The Mammalian ULK1 Complex and Autophagy Initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef]
- Corà, D.; Bussolino, F.; Doronzo, G. Tfeb Signalling-Related Micrornas and Autophagy. Biomolecules 2021, 11, 985. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, F.; Chen, C.; Ji, J.; Huang, P.; Wei, D.; Zhang, Y.; Ren, L. Identification of Prognostic Genes Signature and Construction of CeRNA Network in Pirarubicin Treatment of Triple-Negative Breast Cancer. Breast Cancer 2023, 30, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, K.; Frye, D.; Newman, R.A.; Walters, R.; Theriault, R.; Fraschini, G.; Smith, T.; Buzdar, A.; Hortobagyi, G.N. Phase II Clinical and Pharmacological Study of Pirarubicin in Combination with 5-Fluorouracil and Cyclophosphamide in Metastatic Breast Cancer. Clin. Cancer Res. 1995, 1, 691–697. [Google Scholar] [PubMed]
- Ma, J.; Chen, C.; Fan, Z.; Zhang, Y.; Ji, J.; Wei, D.; Zhang, F.; Sun, B.; Huang, P.; Ren, L. CircEGFR Reduces the Sensitivity of Pirarubicin and Regulates the Malignant Progression of Triple-Negative Breast Cancer via the MiR-1299/EGFR Axis. Int. J. Biol. Macromol. 2023, 244, 125295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wei, D.; Xie, S.; Ren, L.; Qiao, S.; Li, L.; Ji, J.; Fan, Z. CircZCCHC2 Decreases Pirarubicin Sensitivity and Promotes Triple-Negative Breast Cancer Development via the MiR-1200/TPR Axis. iScience 2024, 27, 109057. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Pan, M.; Shi, H.; Wang, L.; Bai, Y.; Ge, Q. Cell-Free DNA Fragmentomics: The Novel Promising Biomarker. Int. J. Mol. Sci. 2023, 24, 1503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, P.; Liu, T.; Li, D.; Liu, X. Circ_0006404 Enhances Hepatocellular Carcinoma Progression by Regulating MiR-624. Environ. Sci. Pollut. Res. 2022, 29, 69980–69987. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Z.; Li, S.; Wang, L. Circ_0006404 Accelerates Prostate Cancer Progression through Regulating Mir-1299/Cfl2 Signaling. OncoTargets Ther. 2021, 14, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Wan, X.; Lu, Y.; Zhang, Y.; Huang, Y.; Xu, Y.; Liu, Y.; Zhao, P.; Xiang, X.; Li, L.; et al. Circular RNA CircFOXO3 Promotes Prostate Cancer Progression through Sponging MiR-29a-3p. J. Cell. Mol. Med. 2019, 24, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Tai, Y.-C. Hsa_circ_0006404 and Hsa_circ_0000735 Regulated Ovarian Cancer Response to Docetaxel Treatment via Regulating P-GP Expression. Biochem. Genet. 2021, 60, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.-Y.; Min, Y.-J.; Zhou, K.; Yang, Q.-S.; Peng, M.; Cui, Z.-R.; Zhu, X.-L.; Liu, H.; Wang, M.; Zhang, X.; et al. Identification of the Tumorsuppressive Role of Circular RNAFOXO3 in Colorectal Cancer via Regulation of MiR543/LATS1 Axis. Oncol. Rep. 2021, 46, 239. [Google Scholar] [CrossRef] [PubMed]
- Omid-Shafaat, R.; Moayeri, H.; Rahimi, K.; Menbari, M.; Vahabzadeh, Z.; Hakhamaneshi, M.; Nouri, B.; Ghaderi, B.; Abdi, M. Serum Circ-FAF1/Circ-ELP3: A Novel Potential Biomarker for Breast Cancer Diagnosis. J. Clin. Lab. Anal. 2021, 35, e24008. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, X.; Lin, Z.; Luo, L.; Wen, D. The Potential Value of Serum Chemerin in Patients with Breast Cancer. Sci. Rep. 2021, 11, 6564. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-K.; Zhai, S.-N.; Yang, L. Approaches and Challenges in Genome-Wide Circular RNA Identification and Quantification. Trends Genet. 2023, 39, 897–907. [Google Scholar] [CrossRef] [PubMed]



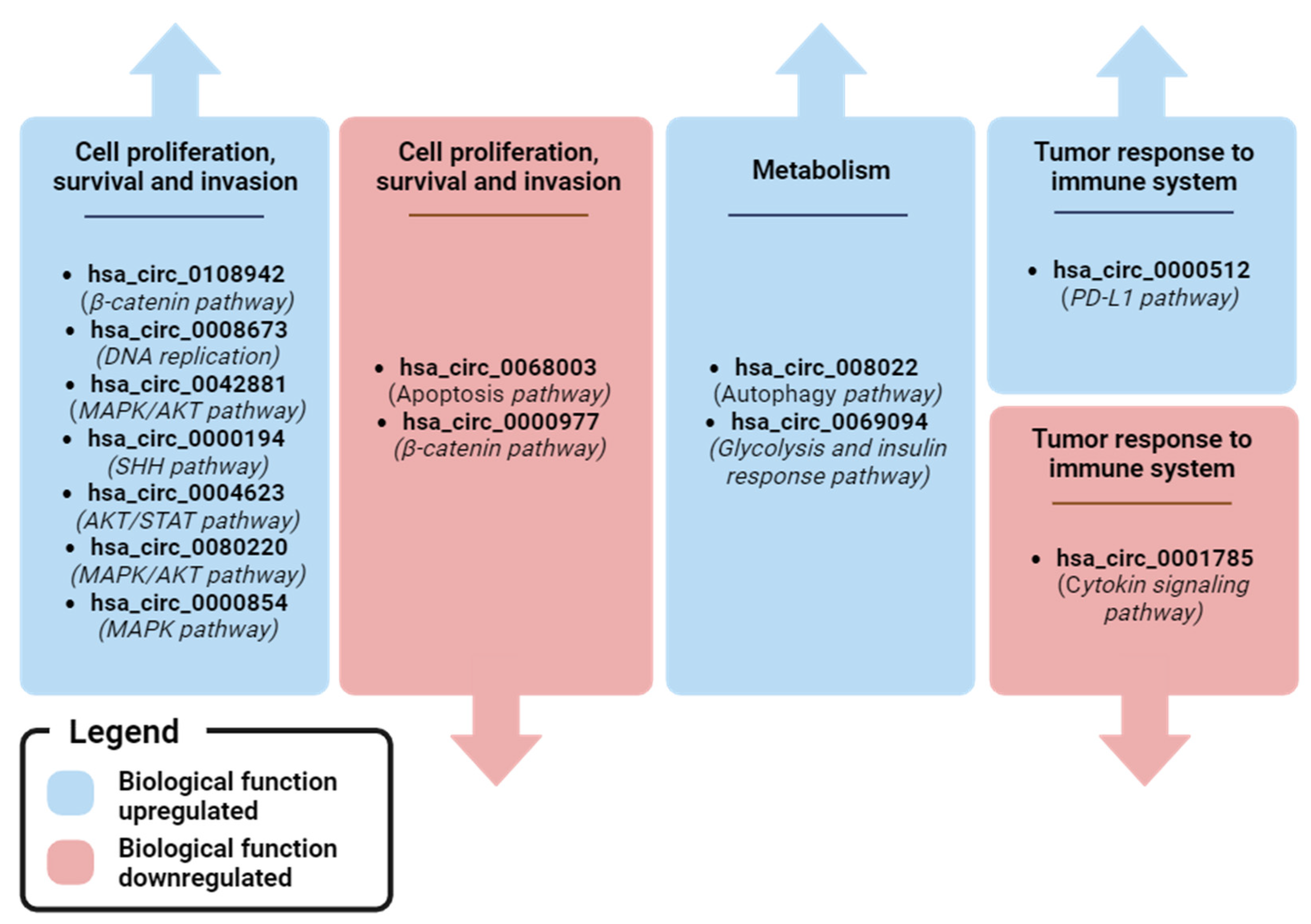
| Cit. | Sample (n) | circBase ID (Host Gene) | Clinical Value | AUC | Findings | Expression Match with Tissues and Cell Lines | Biological Function (Pathway Regulated) |
|---|---|---|---|---|---|---|---|
| hsa_circ_0001785 (ELP3) | Diagnostic, Prognostic, Predictive | 0.771 | Higher levels in BC patients than in healthy controls. High levels were associated with advanced tumor stages and metastasis. Levels decrease in post-operative samples compared to matched pre-operative ones | NR | miRNA sponging: miR-942/SOCS3 axis (cytokine signaling pathway) | ||
| Yin et al. [60] | Pre-operative BC (83): 57 matched post-operative; Healthy controls (25) | hsa_circ_0108942 (ANKRD12) | Diagnostic | 0.701 | Higher levels in BC patients than in healthy controls | NR | miRNA sponging: miR-1178-3p/TMED3 axis (β-catenin pathway) |
| hsa_circ_0068033 (NAALADL2) | Diagnostic | 0.619 | Lower levels in BC patients than in healthy controls. | NR | miRNA sponging: miR-659 (Apoptosis pathway) | ||
| Ju et al. [89] | Pre-operative BC (74); Healthy controls (46) | hsa_circ_0042881 (NF1) | Diagnostic | 0.802 | Higher levels in BC patients than in healthy controls | Yes, with tissues | miRNA sponging: miR-217/SOS1 axis (MAPK/AKT pathway) |
| Zehuan Li et al. [71] | Pre-operative BC (127); Healthy controls (50) | hsa_circ_0069094 (S100P) | Diagnostic | 0.680 | Higher levels in BC patients than in healthy controls | Yes, with tissues and cell lines | miRNA sponging: miR-591/HK2 axis (Glycolysis), miR-136-5p/YWHAZ axis (insulin response pathway), miR-758-3p/ZNF217 axis (MAPK/AKT pathway), miR-661/HMGA1 axis (glycolysis, insulin response pathways) |
| Chen et al. [97] | BC (24); Healthy controls (68) | hsa_circ_0004623 (HIF1A) | Diagnostic | 0.897 | Higher levels in BC patients than in healthy controls | Yes, with tissues and cell lines | miRNA sponging: miR-149-5p/NFIB axis (AKT/STAT pathway) |
| Darbeheshti et al. [101] | TNBC (20); Healthy controls (20) | hsa_circ_0000977 (NOL10) | Diagnostic | 0.960 | Lower levels in TNBC patients than in healthy controls | Yes, with tissues and cell lines | miRNA sponging: miR-135b-5p/APC/GATA3 axis (B-catenin pathway) |
| Song et al. [102] | BC (74); Healthy controls (24) | hsa_circ_0080222 (EGFR) | Diagnostic | NR | Higher levels in BC patients than in healthy controls | Yes, with tissues and cell lines | miRNA sponging: miR-224-5p/ATG13/ULK1 axis, protein sponging: ANXA2/TFEB axis (Autophagy pathway) |
| Jiulong et al. [109] | TNBC (49); non-TNBC (40); TNBC with NAC (10) | hsa_circ_0080220 (EGFR) | Diagnostic, Predictive | NR | Higher levels in TNBC patients than in non-TNBC ones. Lower levels in NAC-treated TNBC patients than in untreated TNBC ones | Yes, with tissues and cell lines | miRNA sponging: miR-1299/EGFR axis (MAPK/AKT pathway) |
| Fan Zhang et al. [110] | TNBC (42); non-TNBC (35) | hsa_circ_0000854 (ZCCHC2) | Diagnostic | NR | Higher levels in TNBC patients than in non-TNBC ones | Yes, with tissues and cell lines | miRNA sponging: miR-1200/TPR axis (MAPK pathway). |
| Bo Fu et al. [93] | BMBC (20); non-metastatic BC (20) | hsa_circ_0001944 (FIRRE) | Prognostic | NR | Higher levels in BMBC patients than in non-metastatic BC ones | Yes, with tissues and cell lines | miRNA sponging: miR-125a/BRD4 axis (SHH pathway) |
| Youting et al. [67] | Pre-operative BC plasma (384): 30 matched post-operative; Healthy controls (108) | hsa_circ_0008673 (BRCA1) | Diagnostic, Prognostic, Predictive | 0.833 | Higher levels in BC patients than in healthy controls. High levels are associated with large tumor size, metastasis, poor OS and DSS, and hormone receptor status. Levels decrease in post-operative samples compared to pre-operative ones | Yes, with cell lines | miRNA sponging: miR-578/GINS4 axis (DNA replication pathway) |
| Yuhne et al. [81] | Pre-operative BC (212): 102 matched post-operative; Healthy controls (212) | hsa_Circ_0000512 (RPPH1) | Diagnostic, Predictive | 0.704 | Higher levels in BC patients than in healthy controls. Levels decrease in post-operative samples compared to pre-operative ones. | Yes, with tissues | miRNA sponging: miR-622 (PD-L1 pathway) |
| Cit. | Sample (n) | circBase ID (Host Gene) | Clinical Value | AUC | Findings | Expression Match with Tissues and Cell Lines |
|---|---|---|---|---|---|---|
| Jiani Liu et al. [84] | BC (95); Healthy controls (95) | hsa_circ_0000615 (ZNF609) | Diagnostic, Prognostic, Predictive | 0.904 | Higher levels in BC patients than in healthy controls. High levels are associated with advanced tumor stage, metastasis, high recurrence risk after surgery and TNBC subtype | Yes, with tissues and cell lines |
| hsa_circ_0002190 (KLHDC10) | ||||||
| hsa_Circ_0007177 (CCZ1) | ||||||
| hsa_circ_0000642 (ZFAND6) | ||||||
| hsa_circ_0001439 (SCLT1) | ||||||
| Lin et al. [66] | BC (158); Healthy controls (43); BBT (88) | hsa_circ_0001417 (ANKRD17) | Diagnostic | 0.83 (for the panel) | Higher levels in BC patients than in healthy controls and BBT | NR |
| hsa_circ_0005552 (EHBP1) | ||||||
| hsa_circ_0001073 (ACVR2A) | ||||||
| hsa_circ_0000267 (FAM53B) | ||||||
| hsa_circ_0006404 (FOXO3) | ||||||
| Xiaohan Li et al. [70] | BC (83); Healthy controls (49) | hsa_circ_0104824 (NTRK3) | Diagnostic | 0.849 | Higher levels in BC patients than healthy controls | Yes, with tissues |
| hsa_circ_0079876 (ANLN) | 0.622 | Yes, with tissues | ||||
| Zehuan Li et al. [71] | BC (127); Healthy controls (50) | hsa_circ_0017650 (ITIH5) | Diagnostic | 0.758 | Higher levels in BC patients than in healthy controls | No, with tissues |
| hsa_circ_0017536 (AKR1C1) | 0.615 | No, with tissues | ||||
| Yuhne et al. [81] | Pre-operative BC (212): 102 matched postoperative; Healthy controls (212) | hsa_circ_0000091 (RPAP2) | Diagnostic, Prognostic, Predictive | 0.825 | Lower levels in BC patients than in healthy controls. Levels increase in post-operative samples compared to pre-operative ones. Low levels are associated with advanced TNM stages and ALN metastasis | Yes, with tissue |
| hsa_Circ_0067772 (SLC33A1) | Diagnostic, Predictive | 0.730 | Higher levels in BC patients than in healthy controls. Levels decrease in post-operative samples compared to pre-operative ones | Yes, with tissues |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tierno, D.; Grassi, G.; Zanconati, F.; Dapas, B.; Scaggiante, B. Plasma Circular RNAs as Biomarkers for Breast Cancer. Biomedicines 2024, 12, 875. https://doi.org/10.3390/biomedicines12040875
Tierno D, Grassi G, Zanconati F, Dapas B, Scaggiante B. Plasma Circular RNAs as Biomarkers for Breast Cancer. Biomedicines. 2024; 12(4):875. https://doi.org/10.3390/biomedicines12040875
Chicago/Turabian StyleTierno, Domenico, Gabriele Grassi, Fabrizio Zanconati, Barbara Dapas, and Bruna Scaggiante. 2024. "Plasma Circular RNAs as Biomarkers for Breast Cancer" Biomedicines 12, no. 4: 875. https://doi.org/10.3390/biomedicines12040875








