The Wnt/β-catenin/TCF/Sp5/Zic4 Gene Network That Regulates Head Organizer Activity in Hydra Is Differentially Regulated in Epidermis and Gastrodermis
Abstract
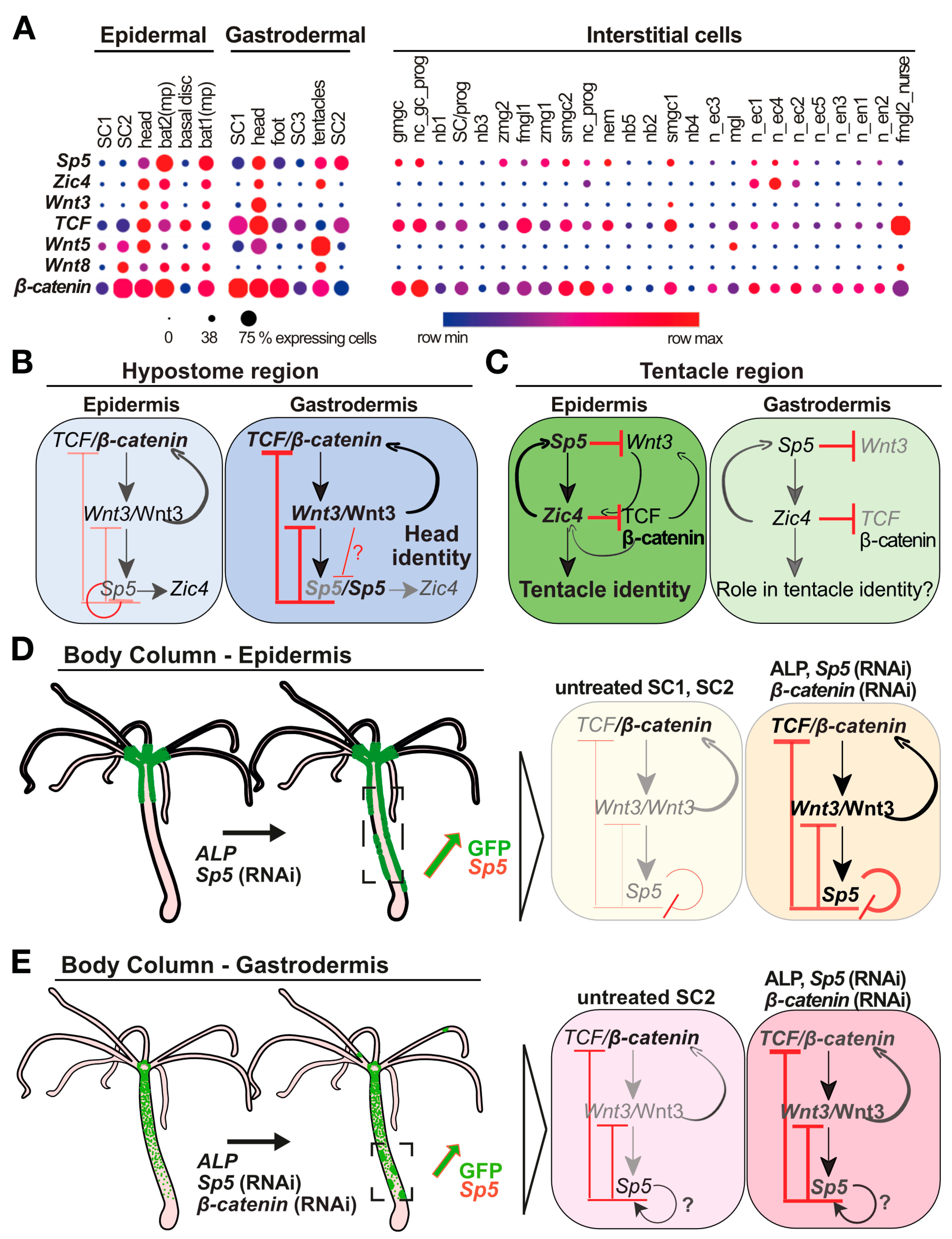
:1. Introduction
2. Materials and Methods
2.1. Animal Culture and Drug Treatment
2.2. Mapping of the Transcriptional Start Sites (TSS)
2.3. Reporter Constructs Expressed in Hydra or in HEK293T Cells
2.4. Generation of the Hydra Transgenic Lines
2.5. RNA Interference
2.6. Quantitative RT-PCR
2.7. Whole-Mount In Situ Hybridization (WM-ISH)
2.8. Immunofluorescence
2.9. Nuclear Extracts (NEs) and Electro-Mobility Shift Assay (EMSA)
2.10. Production of Anti-Sp5 Antibodies
2.11. Cell Culture and Whole Cell Extracts (WCEs) and Western Blotting
2.12. Chromatin Immuno-Precipitation and Quantitative PCR (ChIP-qPCR)
2.13. Imaging
2.14. Statistical Analyses
3. Results
3.1. Differential Sp5 Regulation in the Epidermal and Gastrodermal Layers along the Body Axis
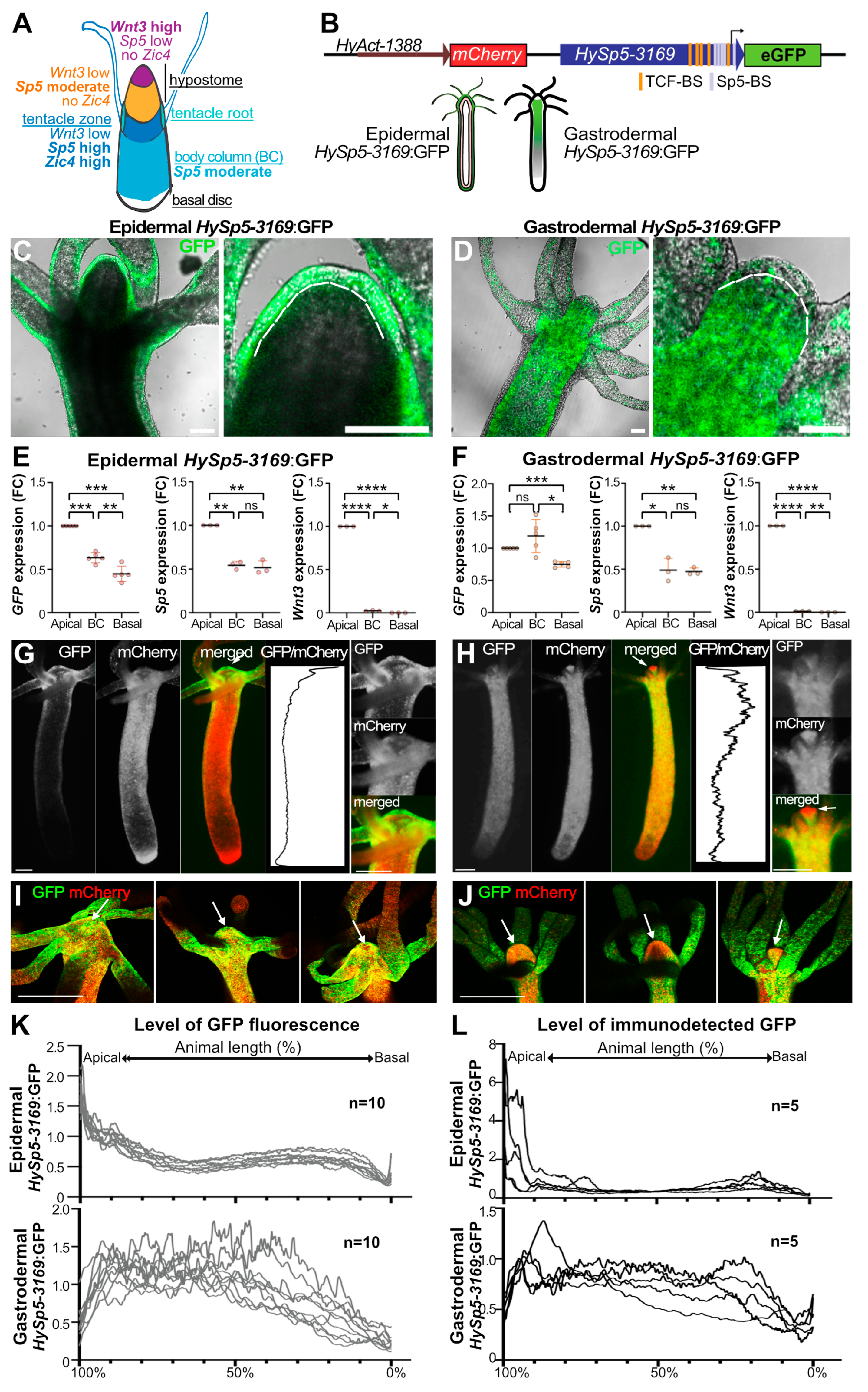
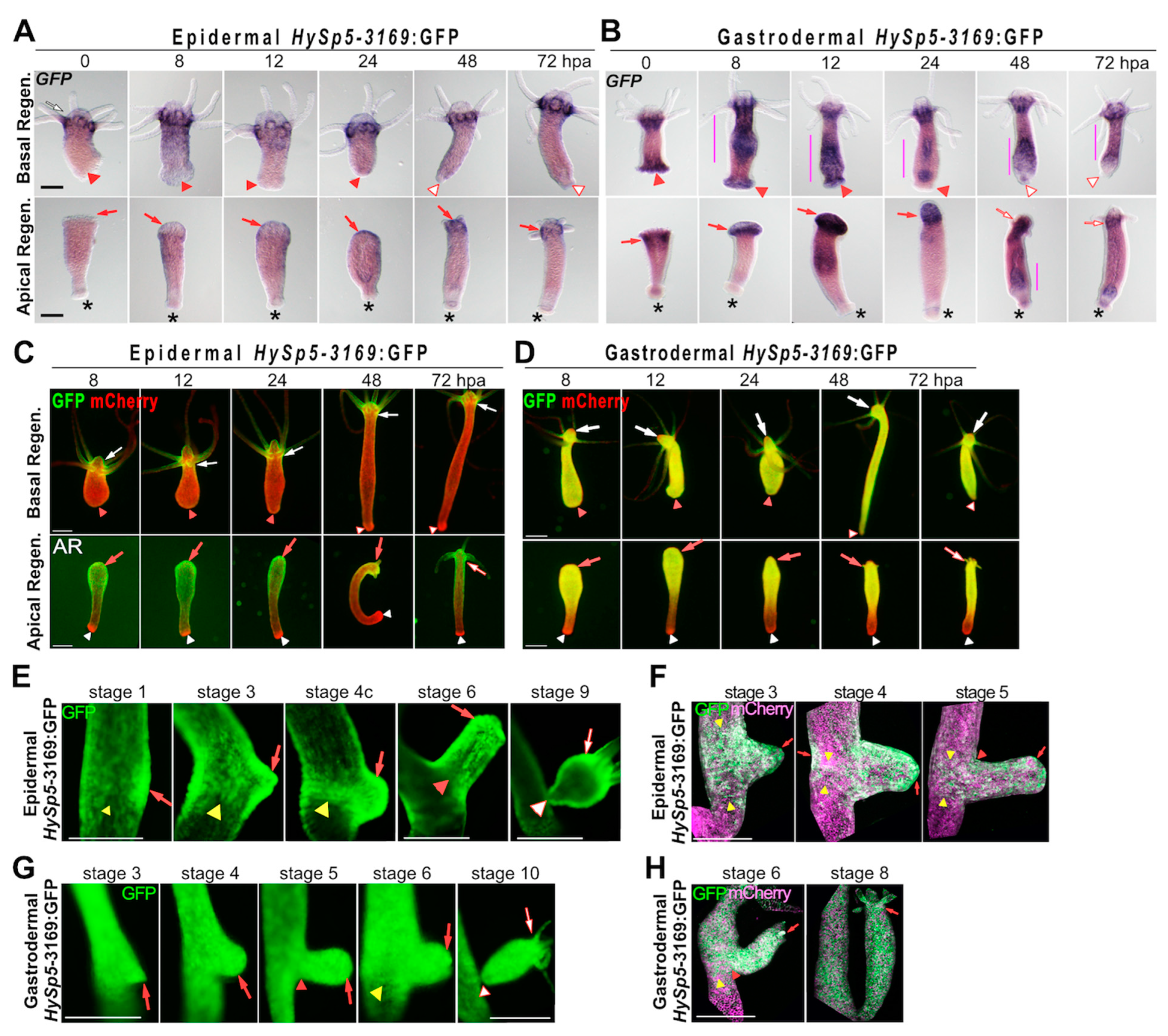
3.2. Sp5 Regulation after Bisection Is Systemic in Gastrodermis but Localized in Epidermis
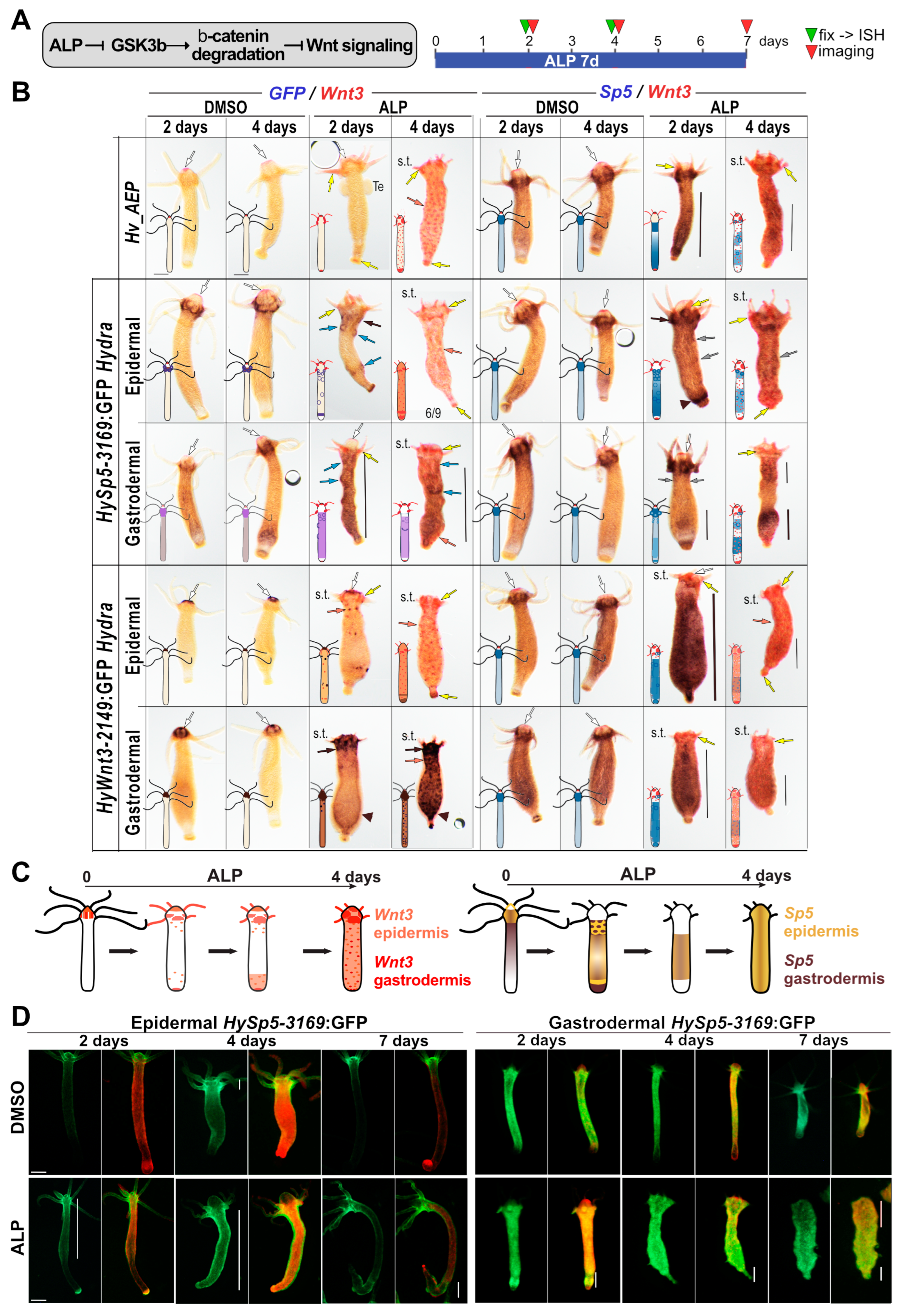
3.3. Layer-Specific Modulations of Sp5 Expression upon Alsterpaullone (ALP) Treatment
3.4. Layer-Specific Modulations of Wnt3 Expression Induced by ALP Treatment
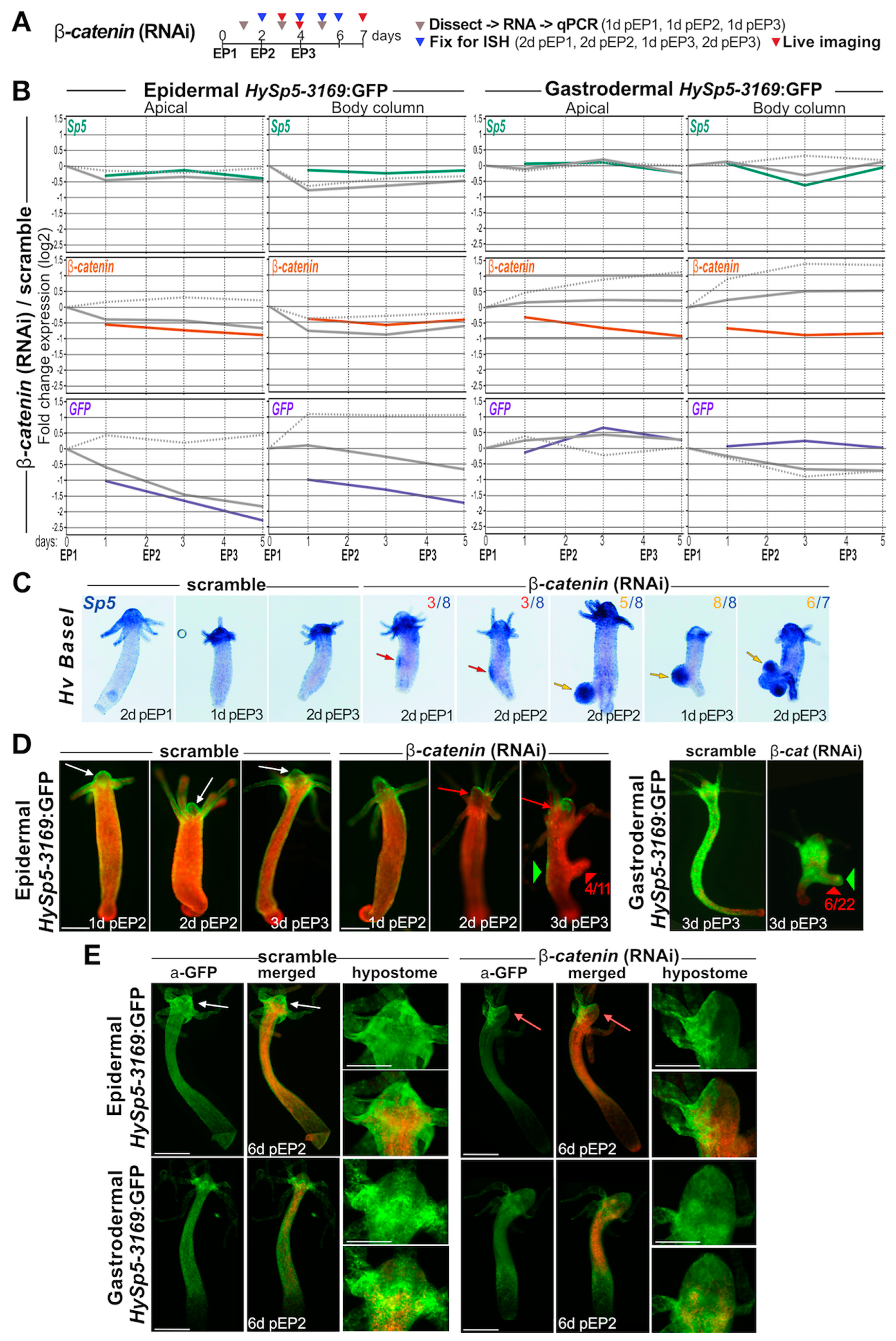
3.5. β-Catenin Knock-Down Differentially Impacts Sp5 Expression in Epidermis and Gastrodermis
3.6. β-catenin Knock-Down Leads to Formation of Bud-like Structures Expressing Gastrodermal Sp5
3.7. Negative Auto-Regulation of Sp5 in the Epidermis
3.8. Up-regulation of β-catenin after Sp5(RNAi) in Epidermis and Gastrodermis
3.9. Identification of Five Active Sp5-Binding Sites within the Proximal Hydra Sp5 Promoter
3.10. The Sp5 Proximal Promoter Is Involved in Sp5-Negative Autoregulation
3.11. The Zic4 Transcription Factor Positively Regulates Sp5 Expression
4. Discussion
4.1. Epithelial Layer-Specific Regulations of Sp5 in Intact Animals
4.2. Three Architectures of the Wnt3/β-Catenin/TCF/Sp5/Zic4 GRN in Intact Hydra
4.3. Sp5-Negative Autoregulation in the Epidermis
4.4. Distinct Configurations of the Wnt3/β-Catenin/Sp5 GRN in the Homeostatic and Developmental Head Organizers
4.5. Phenotypic Changes Induced by Dysregulations of the Wnt3/β-Catenin/TCF/Sp5/Zic4 GRN
4.6. Variability of Head Organizer Inhibitor Strength across Hydra Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Experimental Conditions | Phenotypes Tested Genes | Epidermal GFP HySp5-3169:GFP Animals | Gastrodermal GFP HySp5-3169:GFP Animals | Figures Supplement. |
|---|---|---|---|---|
| INTACT ANIMALS | GFP, Sp5, Wnt3 transcripts GFP fluorescence GFP protein | Apical: tentacle ring, tentacles Hypostome: whole, tentacle ring + BC: upper only | Apical: tentacle ring, tentacles low at head tip, tentacle ring ++ BC: high from 90% to 40% | 1E, 1F 1C, 1D, 1G-1L S3, S4 |
| REGENERATING ANIMALS | GFP transcripts GFP fluorescence | Apical Reg.: GFP(+) from 24 hpa GFP(+) in upper AR half Basal Reg.: no GFP, no GFP | Apical Reg.: GFP(+) from 8 hpa GFP(+) in upper AR half Basal Reg: GFP(+) 8-12 hpa, then GFP(-) diffuse GFP along BR halves | 2A, 2B, S5 2C, 2D, S6 |
| BUDDING ANIMALS | GFP fluorescence | GFP(+) patch (st.1) GFP+ budding belt (st.3,4) GFP(+) bud (st. 5,6) GFP(+) head (st.9) | diffuse GFP(+) polyp (st.3, 4) diffuse GFP(+) bud (st.5, 6) low boundary GFP(+) belt (st. 6) diffuse GFP(+) bud (st.9) | 2E-2H |
| Alsterpaullone (ALP) treatment | Morphological phenotype | shortening of tentacles (4d) small and transient ectopic structures along the BC (2d, 4d) basal disc refined after 4d, apical-like: Wnt3+, Sp5+ | 3A, 3B, 3D, S7, S8, S9 | |
| Sp5 pattern | Loss of apical Sp5 (4d) Sp5 diffuse along BC Sp5 spots in upper BC (2d) Sp5 ring above basal disc (2d) | Loss of apical Sp5 (4d) Sp5 diffuse along BC (2d) Sp5 diffuse is higher in lower BC (4d) | 3A-3C, S7A, S8A, S8C S9A, S9C | |
| Wnt3 pattern | Wnt3 lost at the head tip Wnt3+ in tentacle roots (2d) Wnt3+ dots along BC (4d) High Wnt3 at basal pole (4d) | Wnt3 lost at the head tip Wnt3+ in tentacle roots (2d) Wnt3+ dots along BC (4d) High Wnt3 at basal pole (4d) | 3A-3C, S7, S8, S9 | |
| GFP pattern | Loss of GFP in tentacle ring GFP(+) spots in upper BC (2d) GFP(+) ring above basal disc | Loss of GFP in tentacle ring GFP diffuse along BC + rings GFP diffuse along lower BC | 3A-3C, S7B, S8B, S8D S9B, S9D | |
| GFP fluorescence | GFP diffuse along BC (4d) high GFP in peduncle (7d) | Loss of apical GFP fluo (4d) GFP fluo in lower half (7d) | 3D | |
| β-catenin(RNAi) | Morphological phenotype | Bud-like structures (GFP-) Size reduction | Bud-like structures (GFP+) Size reduction | 4A, 4C, 4D, 4E S11, S12, S13 |
| β-catenin transcripts | Apical: β-cat 2x decrease BC: no change | Apical: β-cat 2x decrease BC: β-cat 2x decrease | 4A, 4B, S10 | |
| Sp5 transcripts Sp5 pattern (HvBasel) | No Sp5 specific response | BC: Sp5 transient decrease | 4A, 4B, S10 4C, S11 | |
| GFP transcripts | Apical: GFP ~4x decrease BC: GFP ~4x decrease | Apical: GFP transient increase BC: no specific response | 4A, 4B, S10 | |
| GFP fluorescence GFP protein | Loss of apical fluorescence Ectopic patches along BC | Progressive reduction in GFP fluorescence along the lower body column | 4D, S12 4E, S13 | |
| Phenotype | NO MORPHOLOGICAL CHANGE | NO MORPHOLOGICAL CHANGE | 4D, S11, S12, S13, S15, S16 | |
| scramble RNAs | β-catenin transcripts | MINOR MODULATIONS | Apical: steady increase up to 2x BC: steady increase > 2x | S10 (4A, 4B) |
| Sp5 transcripts | BC: transiently decreased BC: transiently decreased ~2x | NO STRONG MODULATION BC: transiently decreased < -2x | S10 (4A, 4B), S11 S14A, S14B (5A) | |
| GFP transcripts | BC: increased ~2x BC: decreased ~2x | BC: decreased ~2x Ap, BC: Limited modulation | S10 (4A, 4B) S14A, S14B (5A) | |
| GFP fluorescence | NO STRONG MODULATION | BC: Progressive decrease along BC | S12, S13 | |
| Wnt3 transcripts | BC: increase > 2x | BC: increase > 2x | S14A, S14B (5A) | |
| Sp5(RNAi) | Phenotype | No multiheaded phenotype up to 2d pEP2 in the Hv_AEP2 transgenic animals | 5B,5C,5E, S15-S17 | |
| Sp5 transcripts | BC: increase ~ 2x 1d pEP2 | BC: increase < 2x pEP2 | 5A, S14 | |
| Wnt3 transcripts | BC: transient increase 16h pEP2 | BC: transient moderate increase 1d pEP2 | 5A, S14 | |
| GFP transcripts | Apical: transient increase 2x – 4x; BC: transient increase > 4x | No significant modulation | 5A, 5D, S14 | |
| GFP pattern GFP fluorescence | BC: Extended areas of GFP/GFP overexpression along the BC | Ectopic GFP/GFP spots 2d pEP2 in lower BC and in tentacles | 5B, 5C, 5E, S15, S16, S17 | |
| β-catenin | BC: increased ~1.5x 2d pEP2 | BC: increased ~1.5x 2d pEP2 | 5D | |
References
- Galliot, B. The Hydra Model System. Int. J. Dev. Biol. 2012, 56, 407–409. [Google Scholar] [CrossRef]
- Browne, E.N. The Production of New Hydranths in Hydra by the Insertion of Small Grafts. J. Exp. Zool. 1909, 7, 1–37. [Google Scholar] [CrossRef]
- Spemann, H.; Mangold, H. Über Die Induktion von Embryonalanlagen Durch Implantation Artfremder Organisatoren. Wilhem Roux Arch. Entw. Mech. 1924, 100, 599–638. [Google Scholar]
- Yao, T. Studies on the Organizer Problem in Pelmatohydra Oligactis. I. The Induction Potency of the Implants and the Nature of the Induced Hydranth. J. Exp. Biol. 1945, 21, 145–150. [Google Scholar] [CrossRef]
- MacWilliams, H.K. Hydra Transplantation Phenomena and the Mechanism of Hydra Head Regeneration. II. Properties of the Head Activation. Dev. Biol. 1983, 96, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Broun, M.; Bode, H.R. Characterization of the Head Organizer in Hydra. Development 2002, 129, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H. Transplantation Analysis of Developmental Mechanisms in Hydra. Int. J. Dev. Biol. 2012, 56, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Vogg, M.C.; Wenger, Y.; Galliot, B. How Somatic Adult Tissues Develop Organizer Activity. Curr. Top. Dev. Biol. 2016, 96, 391–414. [Google Scholar] [CrossRef] [PubMed]
- Vogg, M.C.; Galliot, B.; Tsiairis, C.D. Model Systems for Regeneration: Hydra. Development 2019, 146, dev177212. [Google Scholar] [CrossRef]
- De Robertis, E.M.; Wessely, O.; Oelgeschlager, M.; Brizuela, B.; Pera, E.; Larrain, J.; Abreu, J.; Bachiller, D. Molecular Mechanisms of Cell-Cell Signaling by the Spemann-Mangold Organizer. Int. J. Dev. Biol. 2001, 45, 189–197. [Google Scholar]
- Tickle, C.; Summerbell, D.; Wolpert, L. Positional Signalling and Specification of Digits in Chick Limb Morphogenesis. Nature 1975, 254, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Mallart, R.-M.; Martinez, S.; Lance-Jones, C.C. Pluripotentiality of the 2-Day-Old Avian Germinative Neuroepithelium. Dev. Biol. 1990, 139, 75–88. [Google Scholar] [CrossRef]
- Nakamura, H. Do CNS Anlagen Have Plasticity in Differentiation? Analysis in Quail-Chick Chimera. Brain Res. 1990, 511, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Joubin, K.; Stern, C.D. Formation and Maintenance of the Organizer among the Vertebrates. Int. J. Dev. Biol. 2001, 45, 165–175. [Google Scholar] [PubMed]
- Anderson, C.; Stern, C.D. Organizers in Development. Curr. Top. Dev. Biol. 2016, 117, 435–454. [Google Scholar] [CrossRef] [PubMed]
- Nye, H.L.; Cameron, J.A.; Chernoff, E.A.; Stocum, D.L. Regeneration of the Urodele Limb: A Review. Dev. Dyn. 2003, 226, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Brockes, J.P.; Kumar, A. Comparative Aspects of Animal Regeneration. Annu. Rev. Cell Dev. Biol. 2008, 24, 525–549. [Google Scholar] [CrossRef] [PubMed]
- Kragl, M.; Knapp, D.; Nacu, E.; Khattak, S.; Maden, M.; Epperlein, H.H.; Tanaka, E.M. Cells Keep a Memory of Their Tissue Origin during Axolotl Limb Regeneration. Nature 2009, 460, 60–65. [Google Scholar] [CrossRef]
- Rand, H.W.; Bovard, J.F.; Minnich, D.E. Localization of Formative Agencies in Hydra. Proc. Natl. Acad. Sci. USA 1926, 12, 565–570. [Google Scholar] [CrossRef]
- MacWilliams, H.K. Hydra Transplantation Phenomena and the Mechanism of Hydra Head Regeneration. I. Properties of the Head Inhibition. Dev. Biol. 1983, 96, 217–238. [Google Scholar] [CrossRef]
- Gierer, A.; Meinhardt, H. A Theory of Biological Pattern Formation. Kybernetik 1972, 12, 30–39. [Google Scholar] [CrossRef]
- Hobmayer, B.; Rentzsch, F.; Kuhn, K.; Happel, C.M.; von Laue, C.C.; Snyder, P.; Rothbächer, U.; Holstein, T.W. WNT Signalling Molecules Act in Axis Formation in the Diploblastic Metazoan Hydra. Nature 2000, 407, 186–189. [Google Scholar] [CrossRef]
- Broun, M.; Gee, L.; Reinhardt, B.; Bode, H.R. Formation of the Head Organizer in Hydra Involves the Canonical Wnt Pathway. Development 2005, 132, 2907–2916. [Google Scholar] [CrossRef]
- Chera, S.; Ghila, L.; Dobretz, K.; Wenger, Y.; Bauer, C.; Buzgariu, W.; Martinou, J.-C.; Galliot, B. Apoptotic Cells Provide an Unexpected Source of Wnt3 Signaling to Drive Hydra Head Regeneration. Dev. Cell 2009, 17, 279–289. [Google Scholar] [CrossRef]
- Lengfeld, T.; Watanabe, H.; Simakov, O.; Lindgens, D.; Gee, L.; Law, L.; Schmidt, H.A.; Ozbek, S.; Bode, H.; Holstein, T.W. Multiple Wnts Are Involved in Hydra Organizer Formation and Regeneration. Dev. Biol. 2009, 330, 186–199. [Google Scholar] [CrossRef]
- Gee, L.; Hartig, J.; Law, L.; Wittlieb, J.; Khalturin, K.; Bosch, T.C.; Bode, H.R. Beta-Catenin Plays a Central Role in Setting up the Head Organizer in Hydra. Dev. Biol. 2010, 340, 116–124. [Google Scholar] [CrossRef]
- Nakamura, Y.; Tsiairis, C.D.; Özbek, S.; Holstein, T.W. Autoregulatory and Repressive Inputs Localize Hydra Wnt3 to the Head Organizer. Proc. Natl. Acad. Sci. USA 2011, 108, 9137–9142. [Google Scholar] [CrossRef]
- Weidinger, G.; Thorpe, C.J.; Wuennenberg-Stapleton, K.; Ngai, J.; Moon, R.T. The Sp1-Related Transcription Factors Sp5 and Sp5-like Act Downstream of Wnt/Beta-Catenin Signaling in Mesoderm and Neuroectoderm Patterning. Curr. Biol. 2005, 15, 489–500. [Google Scholar] [CrossRef]
- Thorpe, C.J.; Weidinger, G.; Moon, R.T. Wnt/Beta-Catenin Regulation of the Sp1-Related Transcription Factor Sp5l Promotes Tail Development in Zebrafish. Development 2005, 132, 1763–1772. [Google Scholar] [CrossRef]
- Fujimura, N.; Vacik, T.; Machon, O.; Vlcek, C.; Scalabrin, S.; Speth, M.; Diep, D.; Krauss, S.; Kozmik, Z. Wnt-Mediated down-Regulation of Sp1 Target Genes by a Transcriptional Repressor Sp5. J. Biol. Chem. 2007, 282, 1225–1237. [Google Scholar] [CrossRef]
- Park, D.-S.; Seo, J.-H.; Hong, M.; Bang, W.; Han, J.-K.; Choi, S.-C. Role of Sp5 as an Essential Early Regulator of Neural Crest Specification in Xenopus. Dev. Dyn. 2013, 242, 1382–1394. [Google Scholar] [CrossRef]
- Kennedy, M.W.; Chalamalasetty, R.B.; Thomas, S.; Garriock, R.J.; Jailwala, P.; Yamaguchi, T.P. Sp5 and Sp8 Recruit β-Catenin and Tcf1-Lef1 to Select Enhancers to Activate Wnt Target Gene Transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 3545–3550. [Google Scholar] [CrossRef]
- Vogg, M.C.; Beccari, L.; Iglesias Ollé, L.; Rampon, C.; Vriz, S.; Perruchoud, C.; Wenger, Y.; Galliot, B. An Evolutionarily-Conserved Wnt3/β-Catenin/Sp5 Feedback Loop Restricts Head Organizer Activity in Hydra. Nat. Commun. 2019, 10, 312. [Google Scholar] [CrossRef]
- Vogg, M.C.; Ferenc, J.; Buzgariu, W.C.; Perruchoud, C.; Sanchez, P.G.L.; Beccari, L.; Nuninger, C.; Le Cras, Y.; Delucinge-Vivier, C.; Papasaikas, P.; et al. The Transcription Factor Zic4 Promotes Tentacle Formation and Prevents Epithelial Transdifferentiation in Hydra. Sci. Adv. 2022, 8, eabo0694. [Google Scholar] [CrossRef]
- Siebert, S.; Farrell, J.A.; Cazet, J.F.; Abeykoon, Y.; Primack, A.S.; Schnitzler, C.E.; Juliano, C.E. Stem Cell Differentiation Trajectories in Hydra Resolved at Single-Cell Resolution. Science 2019, 365, eaav9314. [Google Scholar] [CrossRef]
- Schenkelaars, Q.; Perez-Cortes, D.; Perruchoud, C.; Galliot, B. The Polymorphism of Hydra Microsatellite Sequences Provides Strain-Specific Signatures. PLoS ONE 2020, 15, e0230547. [Google Scholar] [CrossRef]
- Wenger, Y.; Buzgariu, W.; Perruchoud, C.; Loichot, G.; Galliot, B. Generic and Context-Dependent Gene Modulations during Hydra Whole Body Regeneration. BioRxiv 2019, 587147. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Wittlieb, J.; Khalturin, K.; Lohmann, J.U.; Anton-Erxleben, F.; Bosch, T.C. Transgenic Hydra Allow in Vivo Tracking of Individual Stem Cells during Morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 6208–6211. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Galliot, B.; Welschof, M.; Schuckert, O.; Hoffmeister, S.; Schaller, H.C. The cAMP Response Element Binding Protein Is Involved in Hydra Regeneration. Development 1995, 121, 1205–1216. [Google Scholar] [CrossRef]
- Suknovic, N.; Tomczyk, S.; Colevret, D.; Perruchoud, C.; Galliot, B. The ULK1 Kinase, a Necessary Component of the pro-Regenerative and Anti-Aging Machinery in Hydra. Mech. Ageing Dev. 2021, 194, 111414. [Google Scholar] [CrossRef]
- Corish, P.; Tyler-Smith, C. Attenuation of Green Fluorescent Protein Half-Life in Mammalian Cells. Protein Eng. 1999, 12, 1035–1040. [Google Scholar] [CrossRef]
- Harrison, S.M.; Houzelstein, D.; Dunwoodie, S.L.; Beddington, R.S. Sp5, a New Member of the Sp1 Family, Is Dynamically Expressed during Development and Genetically Interacts with Brachyury. Dev. Biol. 2000, 227, 358–372. [Google Scholar] [CrossRef]
- Hatayama, M.; Aruga, J. Role of Zic Family Proteins in Transcriptional Regulation and Chromatin Remodeling. In Zic Family; Aruga, J., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; Volume 1046, pp. 353–380. ISBN 978-981-10-7310-6. [Google Scholar]
- Aruga, J.; Yokota, N.; Hashimoto, M.; Furuichi, T.; Fukuda, M.; Mikoshiba, K. A Novel Zinc Finger Protein, Zic, Is Involved in Neurogenesis, Especially in the Cell Lineage of Cerebellar Granule Cells. J. Neurochem. 1994, 63, 1880–1890. [Google Scholar] [CrossRef]
- Ishiguro, A.; Inoue, T.; Mikoshiba, K.; Aruga, J. Molecular Properties of Zic4 and Zic5 Proteins: Functional Diversity within Zic Family. Biochem. Biophys. Res. Commun. 2004, 324, 302–307. [Google Scholar] [CrossRef]
- Okumura, K.; Hosoe, Y.; Nakajima, N. Zic1 Is a Transcriptional Repressor Through the Lamin A/C Promoter and Has an Intrinsic Repressive Domain. J. Health Sci. 2004, 50, 423–427. [Google Scholar] [CrossRef]
- Ebert, P.J.; Timmer, J.R.; Nakada, Y.; Helms, A.W.; Parab, P.B.; Liu, Y.; Hunsaker, T.L.; Johnson, J.E. Zic1 Represses Math1 Expression via Interactions with the Math1 Enhancer and Modulation of Math1 Autoregulation. Development 2003, 130, 1949–1959. [Google Scholar] [CrossRef]
- Salero, E.; Pérez-Sen, R.; Aruga, J.; Giménez, C.; Zafra, F. Transcription Factors Zic1 and Zic2 Bind and Transactivate the Apolipoprotein E Gene Promoter. J. Biol. Chem. 2001, 276, 1881–1888. [Google Scholar] [CrossRef]
- Yang, Y.; Hwang, C.K.; Junn, E.; Lee, G.; Mouradian, M.M. ZIC2 and Sp3 Repress Sp1-Induced Activation of the HumanD Dopamine Receptor Gene. J. Biol. Chem. 2000, 275, 38863–38869. [Google Scholar] [CrossRef]
- Sakurada, T.; Mima, K.; Kurisaki, A.; Sugino, H.; Yamauchi, T. Neuronal Cell Type-Specific Promoter of the α CaM Kinase II Gene Is Activated by Zic2, a Zic Family Zinc Finger Protein. Neurosci. Res. 2005, 53, 323–330. [Google Scholar] [CrossRef]
- Sanchez-Ferras, O.; Bernas, G.; Laberge-Perrault, E.; Pilon, N. Induction and Dorsal Restriction of Paired-Box 3 (Pax3) Gene Expression in the Caudal Neuroectoderm Is Mediated by Integration of Multiple Pathways on a Short Neural Crest Enhancer. BBA Gene Reg. Mec. 2014, 1839, 546–558. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; He, L.; Huang, G.; Du, Y.; Zhang, G.; Yan, X.; Xia, P.; Ye, B.; Wang, S.; et al. ZIC2-Dependent OCT4 Activation Drives Self-Renewal of Human Liver Cancer Stem Cells. J. Clin. Investig. 2015, 125, 3795–3808. [Google Scholar] [CrossRef]
- Zhu, L.; Harutyunyan, K.G.; Peng, J.L.; Wang, J.; Schwartz, R.J.; Belmont, J.W. Identification of a Novel Role of ZIC3 in Regulating Cardiac Development. Hum. Mol. Genet. 2007, 16, 1649–1660. [Google Scholar] [CrossRef]
- Lim, L.S.; Hong, F.H.; Kunarso, G.; Stanton, L.W. The Pluripotency Regulator Zic3 Is a Direct Activator of the Nanog Promoter in ESCs. Stem Cells 2010, 28, 1961–1969. [Google Scholar] [CrossRef]
- Satow, R.; Nakamura, T.; Kato, C.; Endo, M.; Tamura, M.; Batori, R.; Tomura, S.; Murayama, Y.; Fukami, K. ZIC5 Drives Melanoma Aggressiveness by PDGFD-Mediated Activation of FAK and STAT3. Cancer Res. 2017, 77, 366–377. [Google Scholar] [CrossRef]
- Yagi, K.; Satou, Y.; Satoh, N. A Zinc Finger Transcription Factor, ZicL, Is a Direct Activator of Brachyury in the Notochord Specification of Ciona intestinalis. Development 2004, 131, 1279–1288. [Google Scholar] [CrossRef]
- Yagi, K.; Satoh, N.; Satou, Y. Identification of Downstream Genes of the Ascidian Muscle Determinant Gene Ci-Macho1. Dev. Biol. 2004, 274, 478–489. [Google Scholar] [CrossRef]
- Sawada, K.; Fukushima, Y.; Nishida, H. Macho-1 Functions as Transcriptional Activator for Muscle Formation in Embryos of the Ascidian Halocynthia Roretzi. Gene Expr. Patterns 2005, 5, 429–437. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Morin, P.J.; Van Wichen, D.; De Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC−/− Colon Carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed]
- Huggins, I.J.; Bos, T.; Gaylord, O.; Jessen, C.; Lonquich, B.; Puranen, A.; Richter, J.; Rossdam, C.; Brafman, D.; Gaasterland, T.; et al. The WNT Target SP5 Negatively Regulates WNT Transcriptional Programs in Human Pluripotent Stem Cells. Nat. Commun. 2017, 8, 1034. [Google Scholar] [CrossRef]
- Takano, J.; Sugiyama, T. Genetic Analysis of Developmental Mechanisms in Hydra. VIII. Head-Activation and Head-Inhibition Potentials of a Slow-Budding Strain (L4). J. Embryol. Exp. Morphol. 1983, 78, 141–168. [Google Scholar] [CrossRef]
- Achermann, J.; Sugiyama, T. Genetic Analysis of Developmental Mechanisms in Hydra. X. Morphogenetic Potentials of a Regeneration-Deficient Strain (Reg-16). Dev. Biol. 1985, 107, 13–27. [Google Scholar] [CrossRef]
- Takano, J.; Sugiyama, T. Genetic Analysis of Developmental Mechanisms in Hydra. XII. Analysis of Chimaeric Hydra Produced from a Normal and a Slow-Budding Strain (L4). J. Embryol. Exp. Morphol. 1984, 80, 155–173. [Google Scholar]
- Technau, U.; Bode, H.R. HyBra1, a Brachyury Homologue, Acts during Head Formation in Hydra. Development 1999, 126, 999–1010. [Google Scholar] [CrossRef]
- Schwaiger, M.; Andrikou, C.; Dnyansagar, R.; Murguia, P.F.; Paganos, P.; Voronov, D.; Zimmermann, B.; Lebedeva, T.; Schmidt, H.A.; Genikhovich, G.; et al. An Ancestral Wnt–Brachyury Feedback Loop in Axial Patterning and Recruitment of Mesoderm-Determining Target Genes. Nat. Ecol. Evol. 2022, 6, 1921–1939. [Google Scholar] [CrossRef]
- Kaloulis, K.; Chera, S.; Hassel, M.; Gauchat, D.; Galliot, B. Reactivation of Developmental Programs: The cAMP-Response Element-Binding Protein Pathway Is Involved in Hydra Head Regeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 2363–2368. [Google Scholar] [CrossRef]
- Chera, S.; Ghila, L.; Wenger, Y.; Galliot, B. Injury-Induced Activation of the MAPK/CREB Pathway Triggers Apoptosis-Induced Compensatory Proliferation in Hydra Head Regeneration: MAPK-Dependent Injury-Induced Apoptosis in Hydra. Dev. Growth Differ. 2011, 53, 186–201. [Google Scholar] [CrossRef]
- Tursch, A.; Bartsch, N.; Mercker, M.; Schlüter, J.; Lommel, M.; Marciniak-Czochra, A.; Özbek, S.; Holstein, T.W. Injury-Induced MAPK Activation Triggers Body Axis Formation in Hydra by Default Wnt Signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2204122119. [Google Scholar] [CrossRef] [PubMed]
- Guder, C.; Pinho, S.; Nacak, T.G.; Schmidt, H.A.; Hobmayer, B.; Niehrs, C.; Holstein, T.W. An Ancient Wnt-Dickkopf Antagonism in Hydra. Development 2006, 133, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Augustin, R.; Franke, A.; Khalturin, K.; Kiko, R.; Siebert, S.; Hemmrich, G.; Bosch, T.C. Dickkopf Related Genes Are Components of the Positional Value Gradient in Hydra. Dev. Biol. 2006, 296, 62–70. [Google Scholar] [CrossRef]
- Lommel, M.; Strompen, J.; Hellewell, A.L.; Balasubramanian, G.P.; Christofidou, E.D.; Thomson, A.R.; Boyle, A.L.; Woolfson, D.N.; Puglisi, K.; Hartl, M.; et al. Hydra Mesoglea Proteome Identifies Thrombospondin as a Conserved Component Active in Head Organizer Restriction. Sci. Rep. 2018, 8, 11753. [Google Scholar] [CrossRef]
- Ziegler, B.; Yiallouros, I.; Trageser, B.; Kumar, S.; Mercker, M.; Kling, S.; Fath, M.; Warnken, U.; Schnölzer, M.; Holstein, T.W.; et al. The Wnt-Specific Astacin Proteinase HAS-7 Restricts Head Organizer Formation in Hydra. BMC Biol. 2021, 19, 120. [Google Scholar] [CrossRef]
- Smith, K.M.; Gee, L.; Bode, H.R. HyAlx, an Aristaless-Related Gene, Is Involved in Tentacle Formation in Hydra. Development 2000, 127, 4743–4752. [Google Scholar] [CrossRef]
- Munder, S.; Tischer, S.; Grundhuber, M.; Buchels, N.; Bruckmeier, N.; Eckert, S.; Seefeldt, C.A.; Prexl, A.; Kasbauer, T.; Bottger, A. Notch-Signalling Is Required for Head Regeneration and Tentacle Patterning in Hydra. Dev. Biol. 2013, 383, 146–157. [Google Scholar] [CrossRef]
- Pan, Q.; Mercker, M.; Klimovich, A.; Wittlieb, J.; Marciniak-Czochra, A.; Böttger, A. Genetic Interference with HvNotch Provides New Insights into the Role of the Notch-Signalling Pathway for Developmental Pattern Formation in Hydra. Sci. Rep. 2024, 14, 8553. [Google Scholar] [CrossRef]
- Melotti, A.; Mas, C.; Kuciak, M.; Lorente-Trigos, A.; Borges, I.; Ruiz I Altaba, A. The River Blindness Drug Ivermectin and Related Macrocyclic Lactones Inhibit WNT-TCF Pathway Responses in Human Cancer. EMBO Mol. Med. 2014, 6, 1263–1278. [Google Scholar] [CrossRef]
- Molenaar, M.; van de Wetering, M.; Oosterwegel, M.; Peterson-Maduro, J.; Godsave, S.; Korinek, V.; Roose, J.; Destrée, O.; Clevers, H. XTcf-3 Transcription Factor Mediates β-catenin-Induced Axis Formation in Xenopus Embryos. Cell 1996, 86, 391–399. [Google Scholar] [CrossRef]
- Ma, G.; Yasunaga, J.; Fan, J.; Yanagawa, S.; Matsuoka, M. HTLV-1 bZIP Factor Dysregulates the Wnt Pathways to Support Proliferation and Migration of Adult T-cell Leukemia Cells. Oncogene 2013, 32, 4222–4230. [Google Scholar] [CrossRef]



| Species | Target Genes | Zic TF | ZIC-Binding Site Sequence | References | |
|---|---|---|---|---|---|
| MM | Gli—consensus | Zic1–5 | TGGGTGGTC | [46,47] | |
| MM | Lamin A/C | Zic1 | CCACCCCCT | [48] | |
| MM | Math1 | Zic1 | GCTCCCCGGGGAGCT | [49] | |
| MM | ApoE | _A | Zic1/2 | GGACTGTGGGGGGTGGTCAA | [50] |
| _B | AAACTGTGGGGGGTGGTCAA | ||||
| _C | GGACTGTGGGGGGTGAAAAA | ||||
| _D | CTATCCCTGGGGGAGGGGGC | ||||
| HS | D1A | ZIC2 | CCCCCAGGGCA | [51] | |
| MM | CamK II | Zic2 | GTGTGGGC | [52] | |
| MM | Pax3 | Zic2 | CTGCTGGGG | [53] | |
| MM | Oct4 | Zic2 | 2550~−2430 | [54] | |
| HS | a-ACTIN | ZIC3 | GGAGGG | [55] | |
| MM | Nanog-C, Nanog-T | Zic3 | CC(C/T)GCTGGG CCTGCTGGG | [56] | |
| HS | E-CADHERIN (CDH1) | ZIC5 | −283~−71 | [57] | |
| CI | Brachyury | ZicL | CCAGCTGTG | [58] | |
| CI | E(SPL)/HAIRY-B | Macho1 | 5′-GCCCCCCGCTG-3′ | [59] | |
| HR | synthetic | Macho1 | GACCCCCCA | [60] | |
| HV | Sp5 | −672 | Zic | AACCTGGCCTGC | [33,34] |
| −383 | GGCAGGTGCCGGC | this work (Figure 7 and Figure S2) | |||
| HV | Wnt3 | −1781 | Zic | GCCCGCGCTCTCC | this work (Figure 7 and Figure S20A) |
| −1177 | GACAGCGGGTG | ||||
| HV | Zic4 | −1863 | Zic4 | ATCGCCCCCTCTCGCT | [34] |
| −1825 | ACGGGCATTGGCGTGA | this work (Figure 7 and Figure S20B) | |||
| −1736 | GAGGTGACCCATGCTG | ||||
| −1291 | AGGAAAGGGGTGCTACA | ||||
| −107 +77 | TGCTCCCGT TACCCCGCTA | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iglesias Ollé, L.; Perruchoud, C.; Sanchez, P.G.L.; Vogg, M.C.; Galliot, B. The Wnt/β-catenin/TCF/Sp5/Zic4 Gene Network That Regulates Head Organizer Activity in Hydra Is Differentially Regulated in Epidermis and Gastrodermis. Biomedicines 2024, 12, 1274. https://doi.org/10.3390/biomedicines12061274
Iglesias Ollé L, Perruchoud C, Sanchez PGL, Vogg MC, Galliot B. The Wnt/β-catenin/TCF/Sp5/Zic4 Gene Network That Regulates Head Organizer Activity in Hydra Is Differentially Regulated in Epidermis and Gastrodermis. Biomedicines. 2024; 12(6):1274. https://doi.org/10.3390/biomedicines12061274
Chicago/Turabian StyleIglesias Ollé, Laura, Chrystelle Perruchoud, Paul Gerald Layague Sanchez, Matthias Christian Vogg, and Brigitte Galliot. 2024. "The Wnt/β-catenin/TCF/Sp5/Zic4 Gene Network That Regulates Head Organizer Activity in Hydra Is Differentially Regulated in Epidermis and Gastrodermis" Biomedicines 12, no. 6: 1274. https://doi.org/10.3390/biomedicines12061274
APA StyleIglesias Ollé, L., Perruchoud, C., Sanchez, P. G. L., Vogg, M. C., & Galliot, B. (2024). The Wnt/β-catenin/TCF/Sp5/Zic4 Gene Network That Regulates Head Organizer Activity in Hydra Is Differentially Regulated in Epidermis and Gastrodermis. Biomedicines, 12(6), 1274. https://doi.org/10.3390/biomedicines12061274







