RNA Sequencing Reveals Candidate Genes and Pathways Associated with Resistance to MDM2 Antagonist Idasanutlin in TP53 Wild-Type Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Patient Sample Information
2.3. Cell Viability Assay
2.4. RNA Extraction
2.5. RNA Quality Assessment
2.6. RNA-Seq
2.7. Real-Time Reverse Transcriptase Polymerase Chain Reaction Gene Expression Analysis
2.8. Immunoblotting
2.9. Statistical Analysis
3. Results
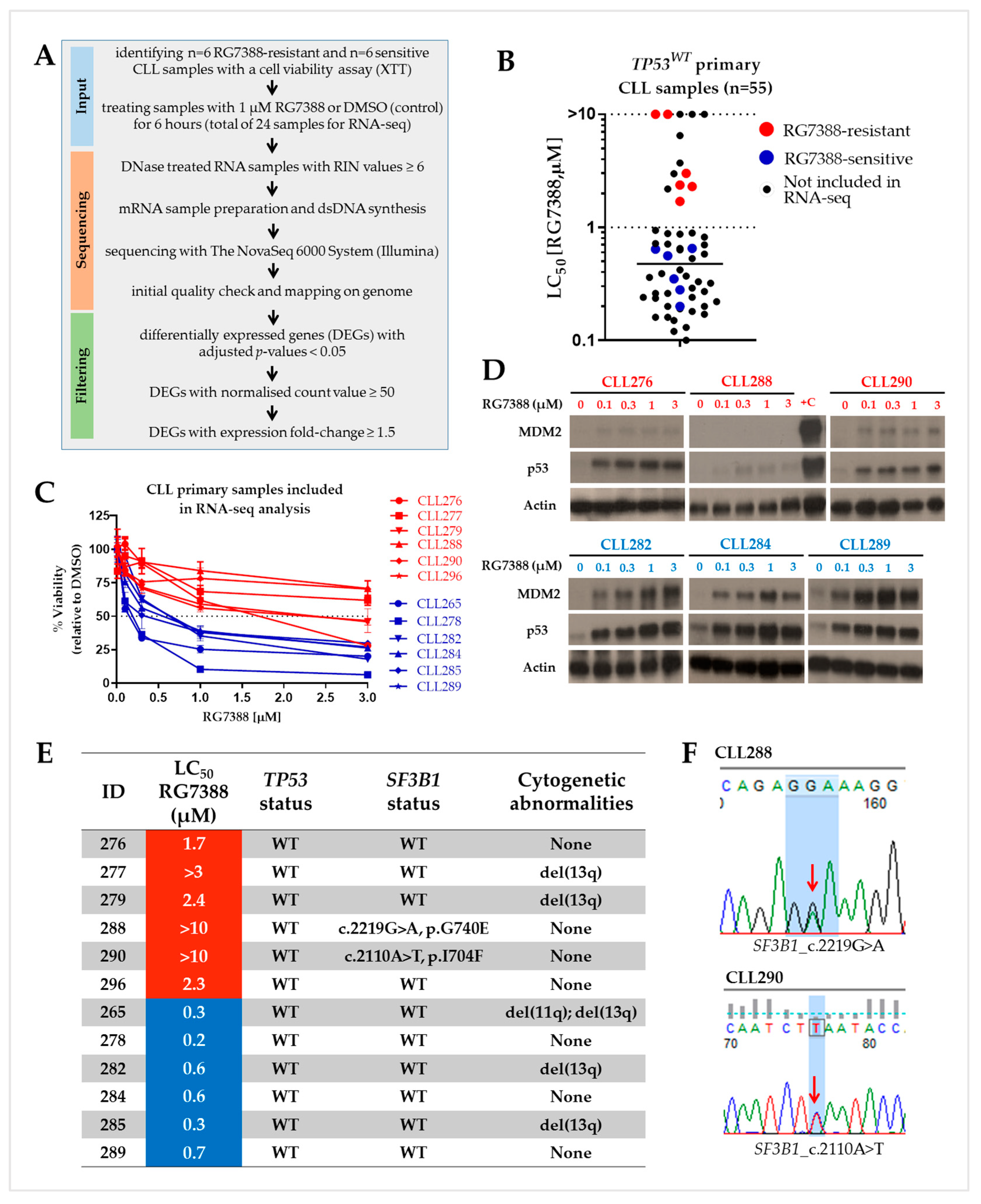
3.1. Selecting TP53 Wild-Type Differentially Responsive Patient Samples for Transcriptome Analysis
3.2. Principal Component Analysis (PCA) and Initial Assessment of Differentially Regulated Genes
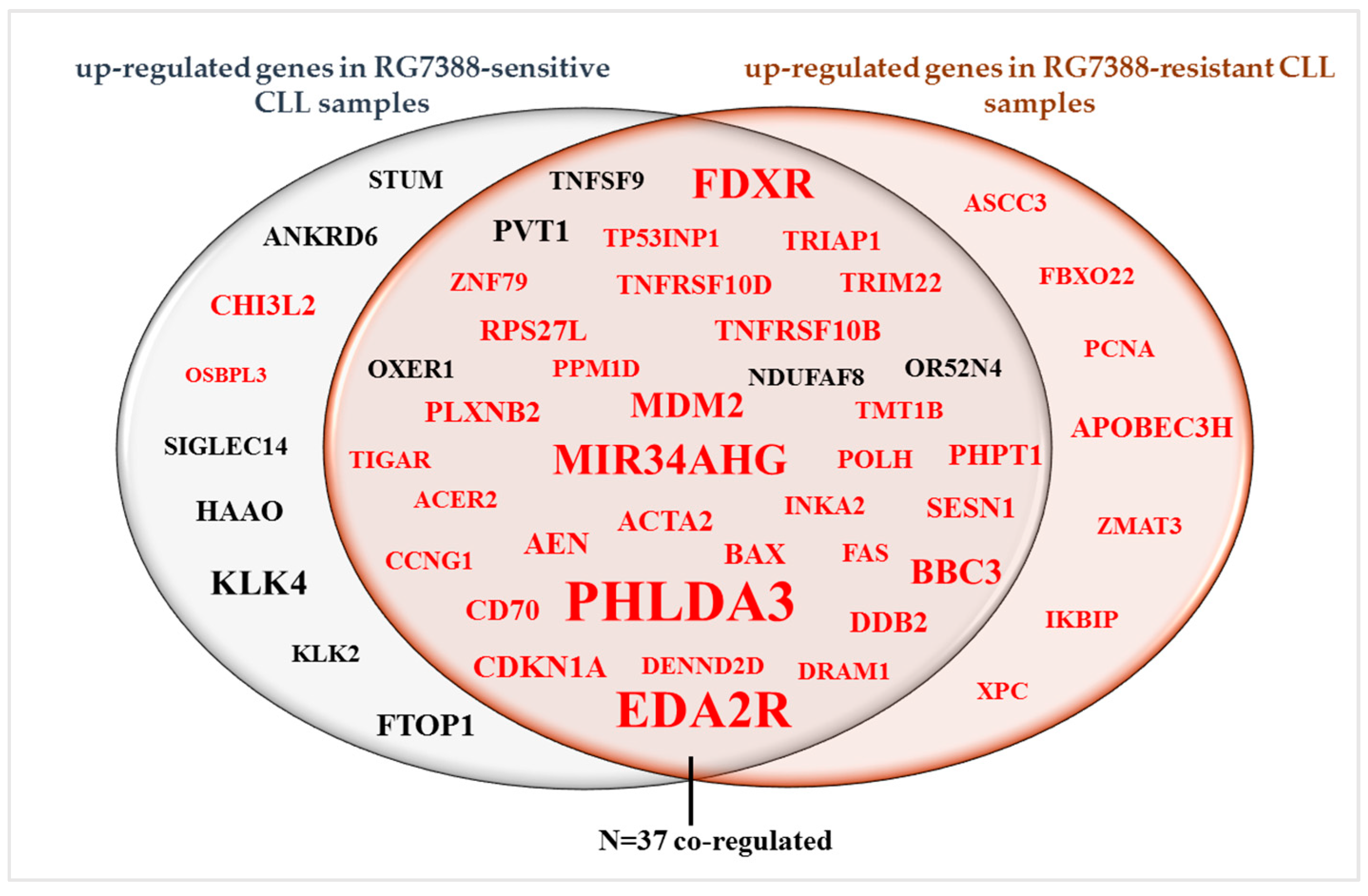
3.3. Differentially Expressed Genes between Control and Treated Samples
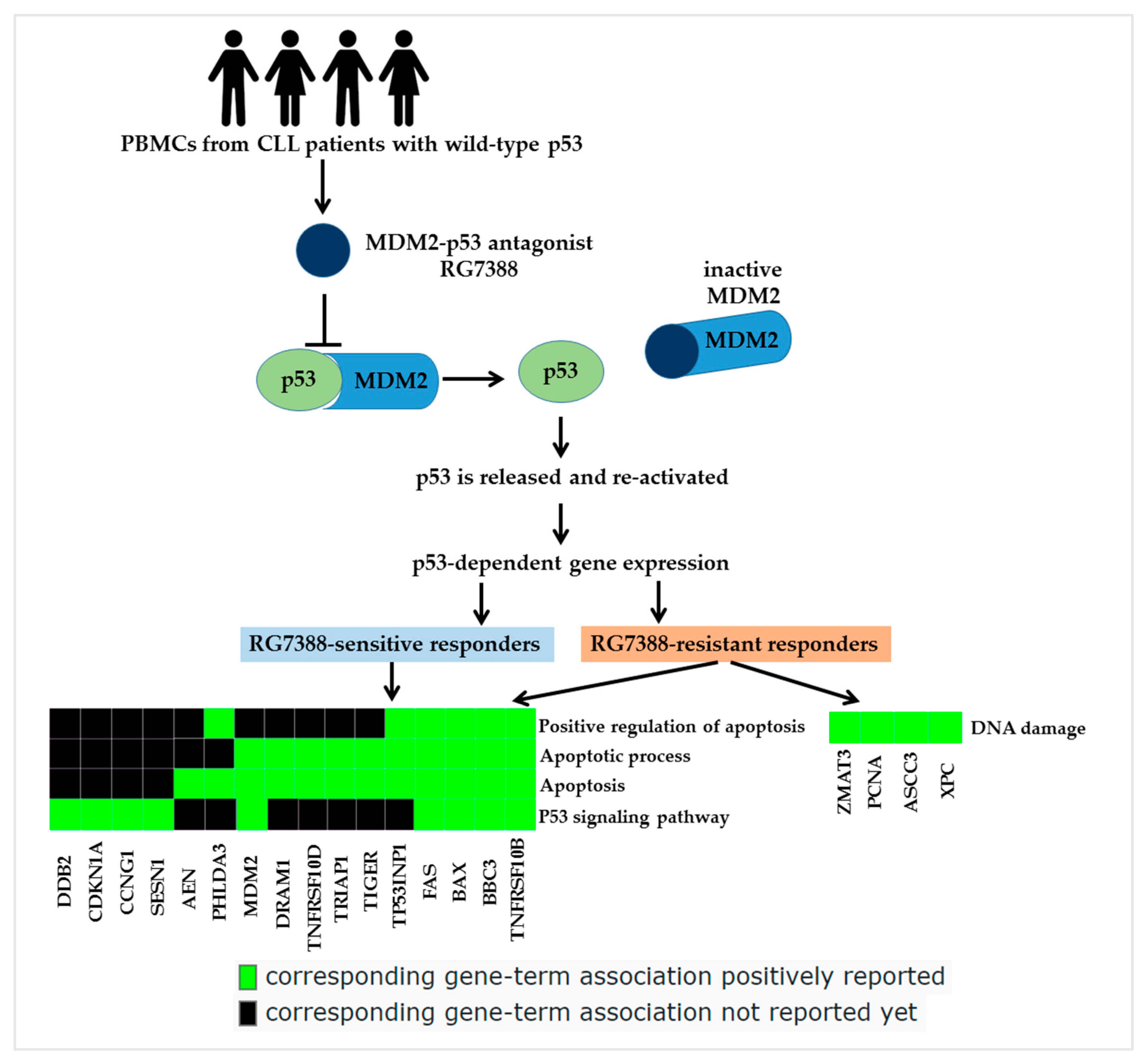
3.4. RNA-Seq Analysis Reveals Candidate Genes and Pathways Associated with Resistance to RG7388
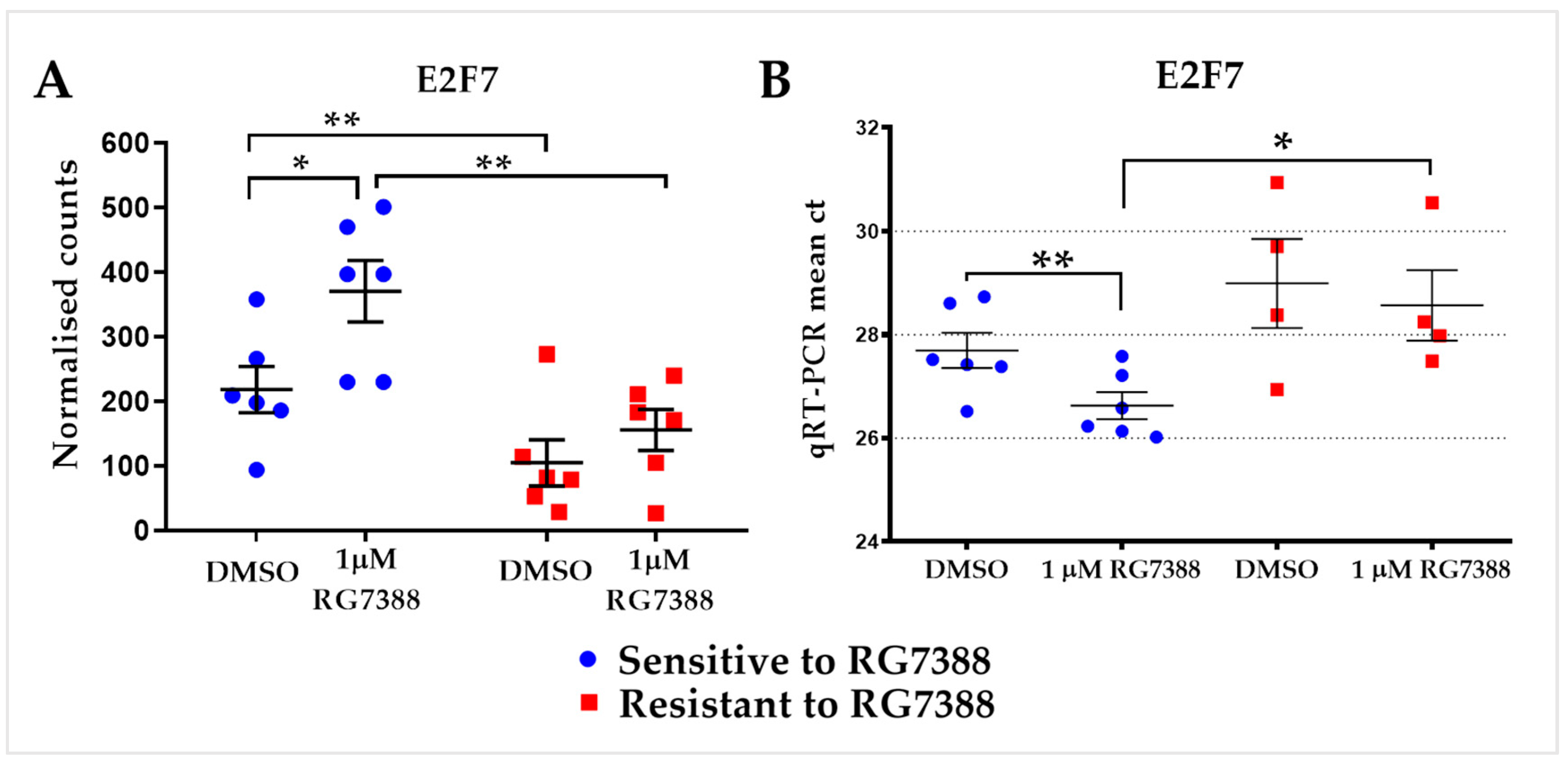
3.5. The RG7388-Resistant Subgroup of CLL Samples Had Lower Basal and Induced Levels of E2F7 Expression Compared to the RG7388-Sensitive Subgroup
3.6. Validation of RNA-Seq Results Using Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ciardullo, C.; Aptullahoglu, E.; Woodhouse, L.; Lin, W.Y.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. Non-genotoxic MDM2 inhibition selectively induces a pro-apoptotic p53 gene signature in chronic lymphocytic leukemia cells. Haematologica 2019, 104, 2429–2442. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.G.; Dail, M.; Garcia, J.S.; Jonas, B.A.; Yee, K.W.L.; Kelly, K.R.; Vey, N.; Assouline, S.; Roboz, G.J.; Paolini, S.; et al. Venetoclax and idasanutlin in relapsed/refractory AML: A nonrandomized, open-label phase 1b trial. Blood 2023, 141, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Aptullahoglu, E.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2023, 24, 11335. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef] [PubMed]
- Aptullahoglu, E.; Ciardullo, C.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. Splicing Modulation Results in Aberrant Isoforms and Protein Products of p53 Pathway Genes and the Sensitization of B Cells to Non-Genotoxic MDM2 Inhibition. Int. J. Mol. Sci. 2023, 24, 2410. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- Marani, M.; Tenev, T.; Hancock, D.; Downward, J.; Lemoine, N.R. Identification of Novel Isoforms of the BH3 Domain Protein Bim Which Directly Activate Bax to Trigger Apoptosis. Mol. Cell. Biol. 2002, 22, 3577–3589. [Google Scholar] [CrossRef]
- Yeung, K.Y.; Ruzzo, W.L. Principal component analysis for clustering gene expression data. Bioinformatics 2001, 17, 763–774. [Google Scholar] [CrossRef]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef]
- Chang, T.-C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Coudert, E.; Gehant, S.; de Castro, E.; Pozzato, M.; Baratin, D.; Neto, T.; A Sigrist, C.J.; Redaschi, N.; Bridge, A. Annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics 2022, 39, btac793. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Pepper, C.; Lin, T.T.; Pratt, G.; Hewamana, S.; Brennan, P.; Hiller, L.; Hills, R.; Ward, R.; Starczynski, J.; Austen, B.; et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood 2008, 112, 3807–3817. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Yang, C.-Y.; Bai, L. MCL-1 inhibition in cancer treatment. OncoTargets Ther. 2018, 11, 7301–7314. [Google Scholar] [CrossRef]
- Lundwall, A.; Band, V.; Blaber, M.; Clements, J.A.; Courty, Y.; Diamandis, E.P.; Fritz, H.; Lilja, H.; Malm, J.; Maltais, L.J.; et al. A comprehensive nomenclature for serine proteases with homology to tissue kallikreins. Biol. Chem. 2006, 387, 637–641. [Google Scholar] [CrossRef][Green Version]
- Yang, F.; Aubele, M.; Walch, A.; Gross, E.; Napieralski, R.; Zhao, S.; Ahmed, N.; Kiechle, M.; Reuning, U.; Dorn, J.; et al. Tissue kallikrein-related peptidase 4 (KLK4), a novel biomarker in triple-negative breast cancer. Biol. Chem. 2017, 398, 1151–1164. [Google Scholar] [CrossRef]
- Obiezu, C.V.; Scorilas, A.; Katsaros, D.; Massobrio, M.; Yousef, G.M.; Fracchioli, S.; Longrais, I.A.R.D.L.; Arisio, R.; Diamandis, E.P. Higher human kallikrein gene 4 (KLK4) expression indicates poor prognosis of ovarian cancer patients. Clin. Cancer Res. 2001, 7, 2380–2386. [Google Scholar]
- Malnic, B.; Godfrey, P.A.; Buck, L.B. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2584–2589. [Google Scholar] [CrossRef]
- Barsotti, A.M.; Beckerman, R.; Laptenko, O.; Huppi, K.; Caplen, N.J.; Prives, C. p53-Dependent induction of PVT1 and miR-1204*. J. Biol. Chem. 2012, 287, 2509–2519. [Google Scholar] [CrossRef] [PubMed]
- Derderian, C.; Orunmuyi, A.T.; Olapade-Olaopa, E.O.; Ogunwobi, O.O. PVT1 Signaling Is a Mediator of Cancer Progression. Front. Oncol. 2019, 9, 502. [Google Scholar] [CrossRef]
- Zenz, T.; Mohr, J.; Eldering, E.; Kater, A.P.; Bühler, A.; Kienle, D.; Winkler, D.; Dürig, J.; van Oers, M.H.J.; Mertens, D.; et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 2009, 113, 3801–3808. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Gao, H.; Ji, Y.; Zhou, Q.; Du, Z.; Tian, L.; Jiang, Y.; Yao, K.; Zhou, Z. Targeting p53–MDM2 interaction by small-molecule inhibitors: Learning from MDM2 inhibitors in clinical trials. J. Hematol. Oncol. 2022, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Vilgelm, A.E.; Cobb, P.; Malikayil, K.; Flaherty, D.; Johnson, C.A.; Raman, D.; Saleh, N.; Higgins, B.; Vara, B.A.; Johnston, J.N.; et al. MDM2 Antagonists Counteract Drug-Induced DNA Damage. EBioMedicine 2017, 24, 43–55. [Google Scholar] [CrossRef][Green Version]
- Carvajal, L.A.; Hamard, P.J.; Tonnessen, C.; Manfredi, J.J. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012, 26, 1533–1545. [Google Scholar] [CrossRef]
- Di Stefano, L.; Jensen, M.R.; Helin, K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003, 22, 6289–6298. [Google Scholar] [CrossRef]
- Westendorp, B.; Mokry, M.; Koerkamp, M.J.G.; Holstege, F.C.; Cuppen, E.; de Bruin, A. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 2012, 40, 3511–3523. [Google Scholar] [CrossRef] [PubMed]
- Zalmas, L.P.; Zhao, X.; Graham, A.L.; Fisher, R.; Reilly, C.; Coutts, A.S.; La Thangue, N.B. DNA-damage response control of E2F7 and E2F8. Embo Rep. 2008, 9, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, O.; Chicas, A.; Zeng, T.; Zhao, Z.; McCurrach, M.; Wang, X.; Lowe, S.W. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 2012, 26, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, A.; Maiti, B.; Jakoi, L.; Timmers, C.; Buerki, R.; Leone, G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2003, 278, 42041–42049. [Google Scholar] [CrossRef] [PubMed]
- Lain, S.; Lane, D. Improving cancer therapy by non-genotoxic activation of p53. Eur. J. Cancer 2003, 39, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, P.; Song, L.; Jeffrey, P.D.; Chernova, T.A.; Wilkinson, K.D.; Cohen, R.E.; Shi, Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005, 24, 3747–3756. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, C.; Gu, C.; Li, Q.; Wu, N. Function of Deubiquitinating Enzyme USP14 as Oncogene in Different Types of Cancer. Cell Physiol. Biochem. 2016, 38, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Liu, C.; Bai, C.; Han, Y.P.; Cho, W.C.; Li, Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 2013, 14, 10749–10760. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Xu, H.-Z.; Shan, H.-Z.; Liu, M.; Lu, Y.; Fang, Z.-X.; Jin, J.; Jing, B.; Xiao, X.-H.; Gao, S.-M.; et al. Targeting USP47 overcomes tyrosine kinase inhibitor resistance and eradicates leukemia stem/progenitor cells in chronic myelogenous leukemia. Nat. Commun. 2021, 12, 51. [Google Scholar] [CrossRef]
- E Robertson, L.; Plunkett, W.; McConnell, K.; Keating, M.J.; McDonnell, T.J. Bcl-2 expression in chronic lymphocytic leukemia and its correlation with the induction of apoptosis and clinical outcome. Leukemia 1996, 10, 456–459. [Google Scholar]
- Pepper, C.; Hoy, T.; Bentley, D.P. Bcl-2/Bax ratios in chronic lymphocytic leukaemia and their correlation with in vitro apoptosis and clinical resistance. Br. J. Cancer 1997, 76, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Molica, S.; Dattilo, A.; Giulino, C.; Levato, D.; Levato, L. Increased bcl-2/bax ratio in B-cell chronic lymphocytic leukemia is associated with a progressive pattern of disease. Haematologica 1998, 83, 1122–1124. [Google Scholar] [PubMed]


| Genes | Primer Forward (5′-3′) | Primer Reverse (5′-3′) |
|---|---|---|
| GAPDH | CGACCACTTTGTCAAGCTCA | GGGTCTTACTCCTTGGAGGC |
| PUMA (BBC3) | ACCTCAACGCACAGTACGA | CTGGGTAAGGGCAGGAGTC |
| BCL2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| MCL1 | GTGCCTTTGTGGCTAAACACT | AGTCCCGTTTTGTCCTTACGA |
| BCL2L1 (BCL-XL) | QIAGEN QUANTITECT PRIMER ASSAY QT00236712 | |
| PMAIP1 (NOXA) | TGCTACACAATGTGGCGTC | ACTTGGACATGGCCTCCCTTA |
| 1 BCL2L11 (BIM) | TAAGTTCTGAGTGTGACCGAGA | GCTCTGTCTGTAGGGAGGTAGG |
| E2F7 | ATATCTTTGTGTGCAGTCTCCTG | AAGACGGCAGCTGACCTGA |
| DEGs | Subgroup | Annotation Cluster | Enriched Pathway | Count | p-Value | p-adj (Benjamini–Hochberg) |
|---|---|---|---|---|---|---|
| Up | Upregulated genes only in resistant CLLs (n = 7) Enrichment score: 2.04 | 1 UniProt | DNA damage | 4 | 5.0 × 10−4 | 7.5 × 10−3 |
| Co-upregulated genes in both sensitive and resistant CLLs (n = 37) Enrichment score: 6.14 | UniProt | Apoptosis | 12 | 2.5 × 10−9 | 6.9 × 10−8 | |
| 2 QuickGO | Apoptotic process | 10 | 6.2 × 10−7 | 1.3 × 10−4 | ||
| QuickGO | Positive regulation of apoptotic process | 6 | 2.4 × 10−4 | 2.5 × 10−2 | ||
| 3 KEGG | P53 signaling pathway | 9 | 3.8 × 10−12 | 3.8 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aptullahoglu, E.; Nakjang, S.; Wallis, J.P.; Marr, H.; Marshall, S.; Willmore, E.; Lunec, J. RNA Sequencing Reveals Candidate Genes and Pathways Associated with Resistance to MDM2 Antagonist Idasanutlin in TP53 Wild-Type Chronic Lymphocytic Leukemia. Biomedicines 2024, 12, 1388. https://doi.org/10.3390/biomedicines12071388
Aptullahoglu E, Nakjang S, Wallis JP, Marr H, Marshall S, Willmore E, Lunec J. RNA Sequencing Reveals Candidate Genes and Pathways Associated with Resistance to MDM2 Antagonist Idasanutlin in TP53 Wild-Type Chronic Lymphocytic Leukemia. Biomedicines. 2024; 12(7):1388. https://doi.org/10.3390/biomedicines12071388
Chicago/Turabian StyleAptullahoglu, Erhan, Sirintra Nakjang, Jonathan P. Wallis, Helen Marr, Scott Marshall, Elaine Willmore, and John Lunec. 2024. "RNA Sequencing Reveals Candidate Genes and Pathways Associated with Resistance to MDM2 Antagonist Idasanutlin in TP53 Wild-Type Chronic Lymphocytic Leukemia" Biomedicines 12, no. 7: 1388. https://doi.org/10.3390/biomedicines12071388
APA StyleAptullahoglu, E., Nakjang, S., Wallis, J. P., Marr, H., Marshall, S., Willmore, E., & Lunec, J. (2024). RNA Sequencing Reveals Candidate Genes and Pathways Associated with Resistance to MDM2 Antagonist Idasanutlin in TP53 Wild-Type Chronic Lymphocytic Leukemia. Biomedicines, 12(7), 1388. https://doi.org/10.3390/biomedicines12071388






