CCZ1 Accelerates the Progression of Cervical Squamous Cell Carcinoma by Promoting MMP2/MMP17 Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Processing
2.2. Tissue Samples
2.3. Differential Expression and Prognosis Correlation Analysis
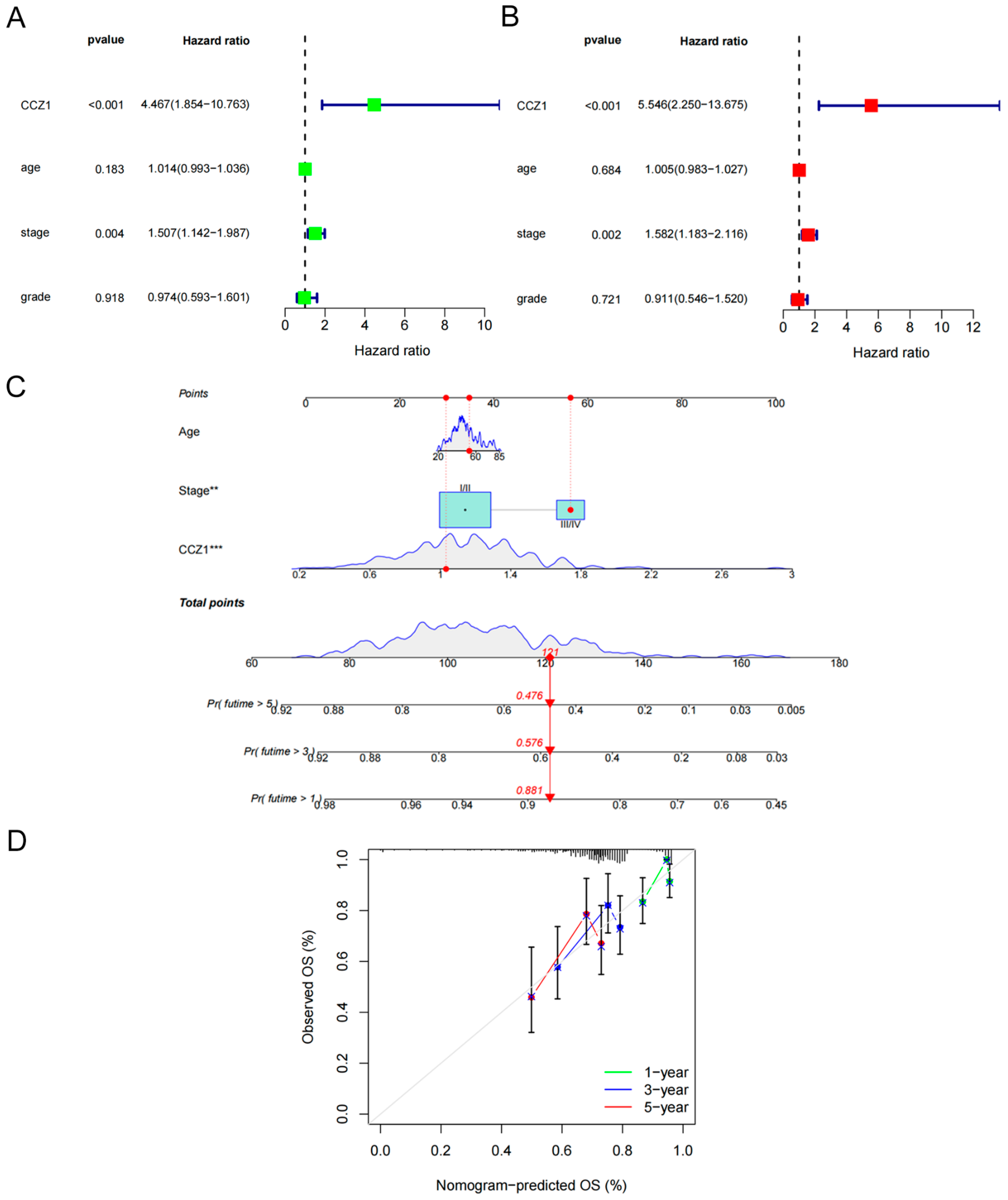
2.4. Construction and Validation of the Nomogram
2.5. Functional Enrichment Analysis
2.6. Cell Culture and Transfection
2.7. Cell Viability Assay
2.8. Colony Formation Assay
2.9. Cell Cycle Analysis
2.10. Transwell Migration/Invasion Assays
2.11. Total RNA Extraction, Quantitative Real-Time PCR (qRT-PCR)
2.12. Western Blot Analysis
2.13. Tumor Xenograft Model
2.14. Immunohistochemistry (IHC) and Quantification
2.15. RNA-Seq
2.16. Statistical Analysis
3. Results
3.1. CCZ1 mRNA Levels Were Elevated in CSCC Tissues, and Increased CCZ1 mRNA Levels Correlated with Poor Prognosis
3.2. Establishment and Validation of a Nomogram Based on CCZ1
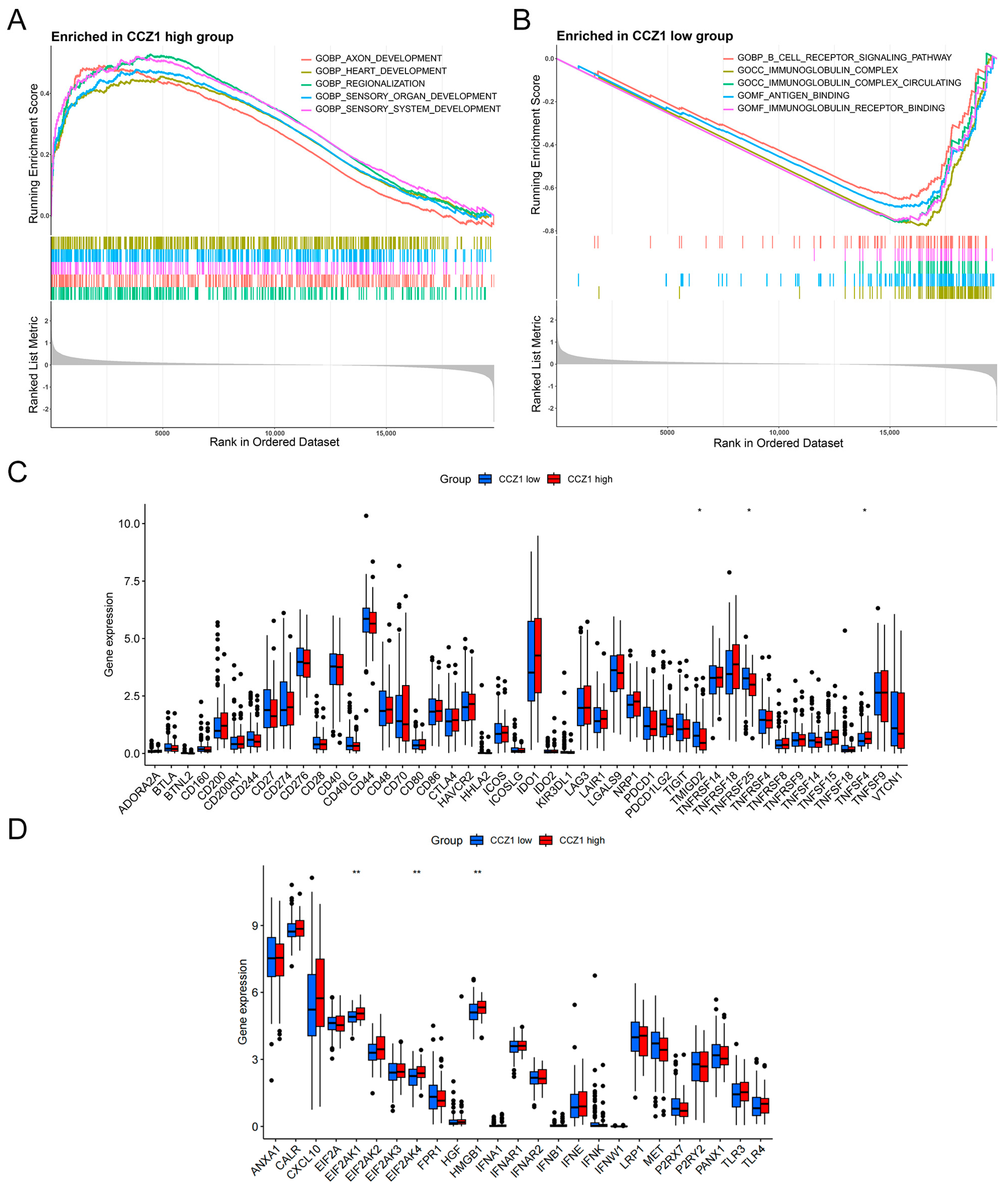
3.3. Association of CCZ1 with Immune Checkpoints and Immunogenic Cell Death Modulators
3.4. CCZ1 Knockdown (KD) Inhibited CSCC Progression In Vitro and In Vivo
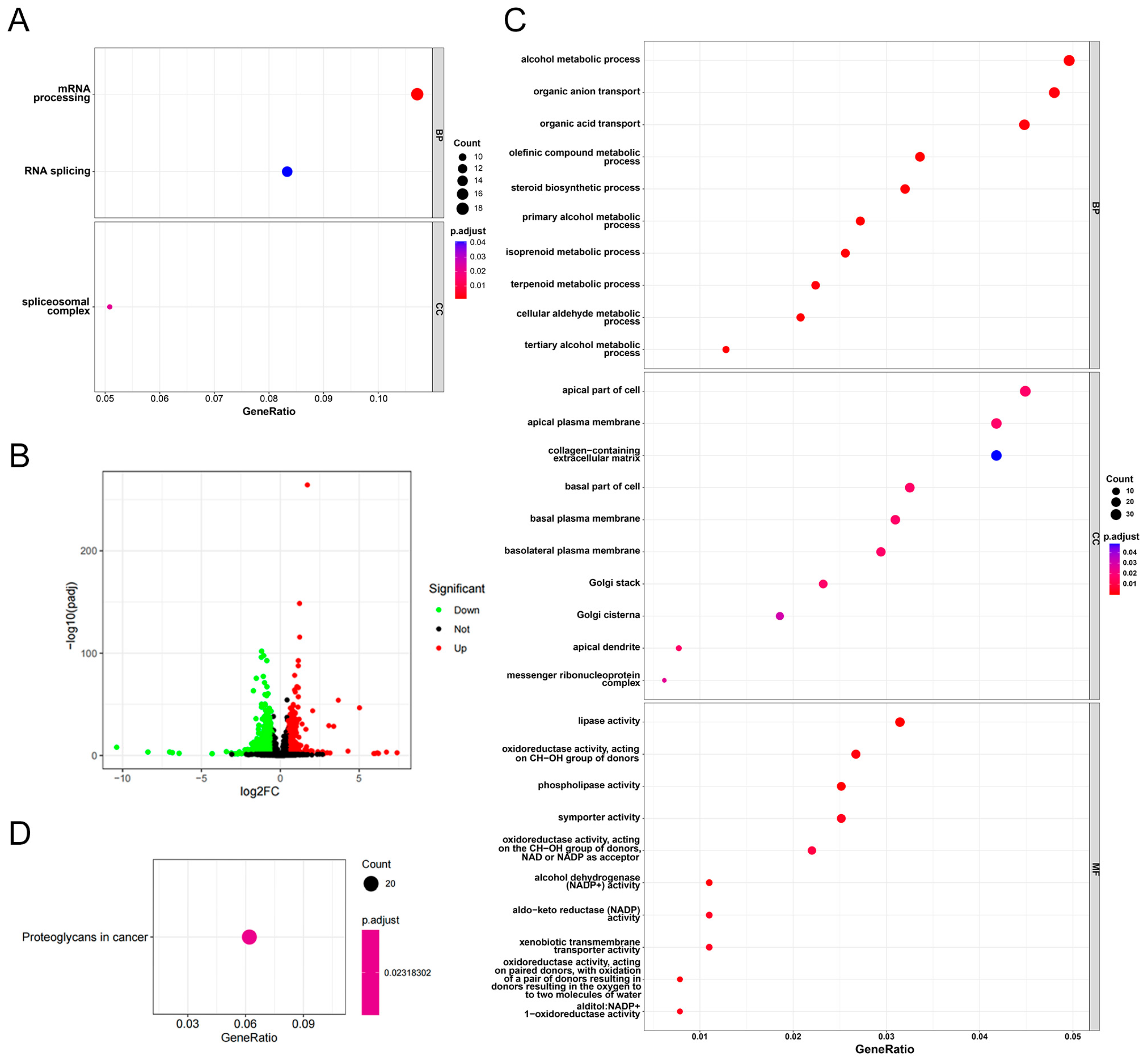
3.5. Functional Enrichment Analysis of CCZ1 in CSCC
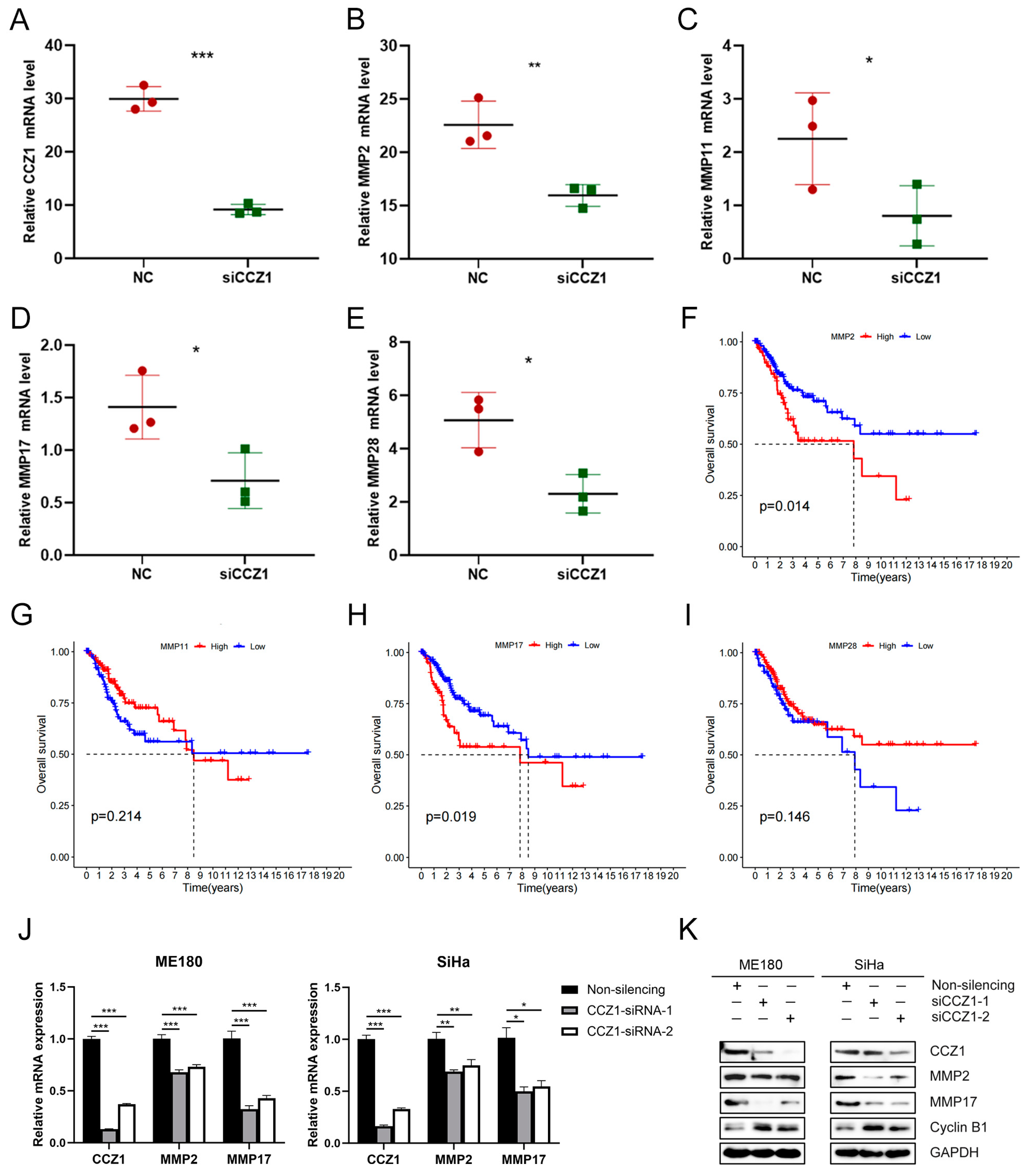
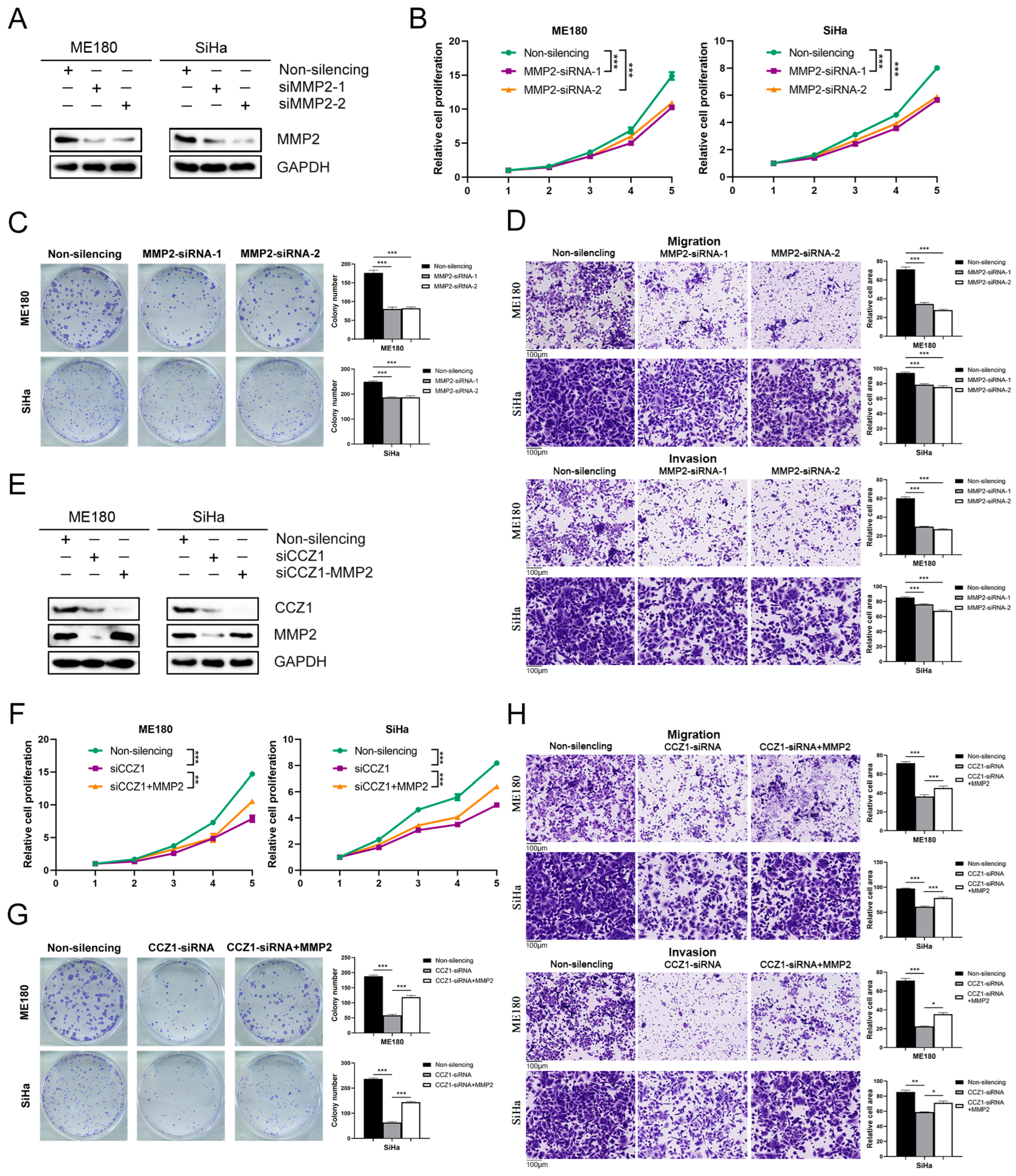
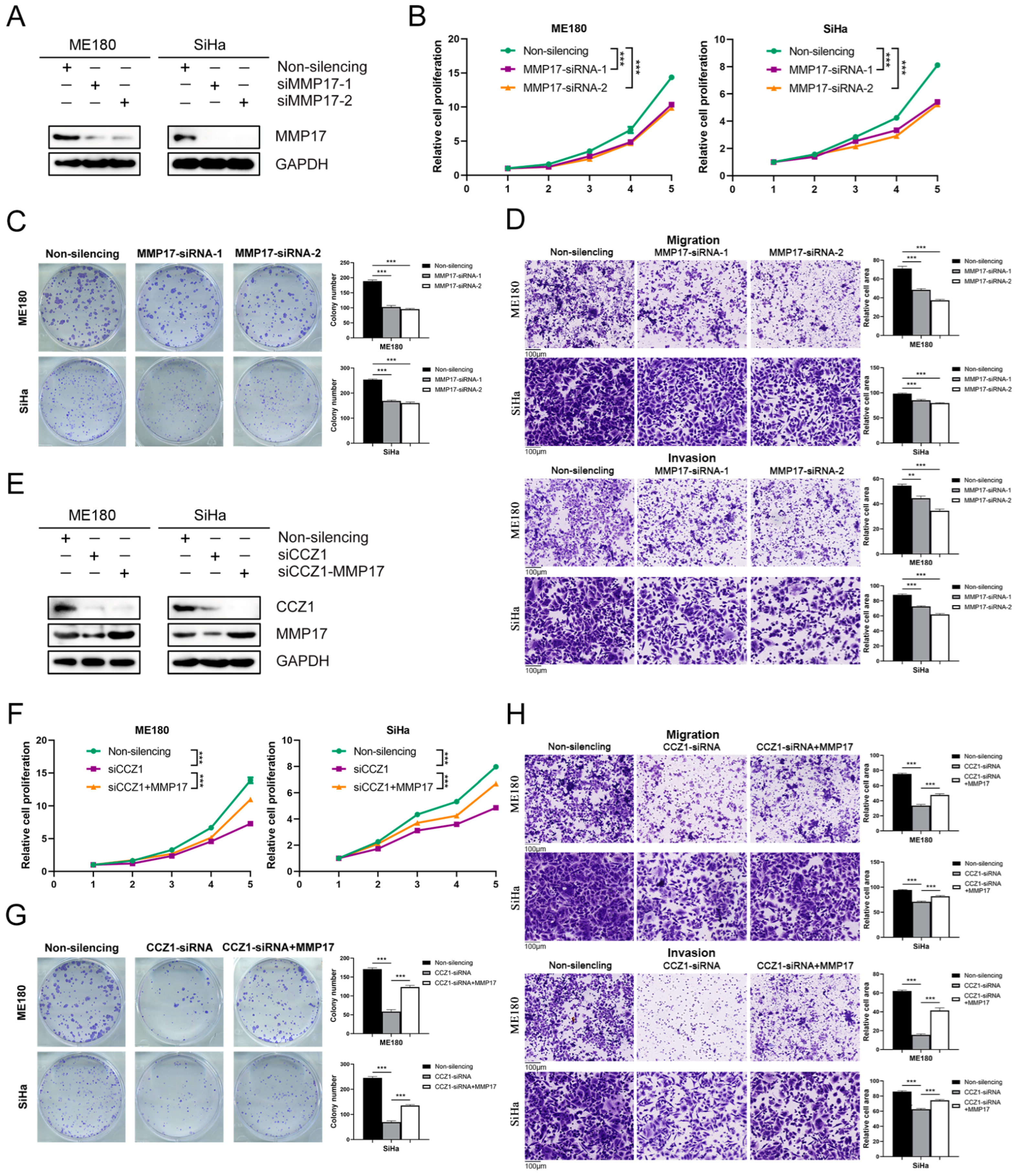
3.6. CCZ1 KD Downregulated MMP2/MMP17 Expression
3.7. CCZ1 KD Inhibited CSCC Progression by Downregulating MMP2 and MMP17 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjose, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Cohen, C.M.; Wentzensen, N.; Castle, P.E.; Schiffman, M.; Zuna, R.; Arend, R.C.; Clarke, M.A. Racial and Ethnic Disparities in Cervical Cancer Incidence, Survival, and Mortality by Histologic Subtype. J. Clin. Oncol. 2023, 41, 1059–1068. [Google Scholar] [CrossRef]
- Kalliala, I.; Athanasiou, A.; Veroniki, A.A.; Salanti, G.; Efthimiou, O.; Raftis, N.; Bowden, S.; Paraskevaidi, M.; Aro, K.; Arbyn, M.; et al. Incidence and mortality from cervical cancer and other malignancies after treatment of cervical intraepithelial neoplasia: A systematic review and meta-analysis of the literature. Ann. Oncol. 2020, 31, 213–227. [Google Scholar] [CrossRef]
- Guo, C.; Qu, X.; Tang, X.; Song, Y.; Wang, J.; Hua, K.; Qiu, J. Spatiotemporally deciphering the mysterious mechanism of persistent HPV-induced malignant transition and immune remodelling from HPV-infected normal cervix, precancer to cervical cancer: Integrating single-cell RNA-sequencing and spatial transcriptome. Clin. Transl. Med. 2023, 13, e1219. [Google Scholar] [CrossRef]
- Fan, J.; Fu, Y.; Peng, W.; Li, X.; Shen, Y.; Guo, E.; Lu, F.; Zhou, S.; Liu, S.; Yang, B.; et al. Multi-omics characterization of silent and productive HPV integration in cervical cancer. Cell Genom. 2023, 3, 100211. [Google Scholar] [CrossRef]
- Mayadev, J.S.; Ke, G.; Mahantshetty, U.; Pereira, M.D.; Tarnawski, R.; Toita, T. Global challenges of radiotherapy for the treatment of locally advanced cervical cancer. Int. J. Gynecol. Cancer 2022, 32, 436–445. [Google Scholar] [CrossRef]
- Ou, Z.; Lin, S.; Qiu, J.; Ding, W.; Ren, P.; Chen, D.; Wang, J.; Tong, Y.; Wu, D.; Chen, A.; et al. Single-Nucleus RNA Sequencing and Spatial Transcriptomics Reveal the Immunological Microenvironment of Cervical Squamous Cell Carcinoma. Adv. Sci. 2022, 9, e2203040. [Google Scholar] [CrossRef]
- Gerondopoulos, A.; Langemeyer, L.; Liang, J.R.; Linford, A.; Barr, F.A. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 2012, 22, 2135–2139. [Google Scholar] [CrossRef]
- Kiontke, S.; Langemeyer, L.; Kuhlee, A.; Schuback, S.; Raunser, S.; Ungermann, C.; Kummel, D. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 2017, 8, 14034. [Google Scholar] [CrossRef]
- Yong, X.; Jia, G.; Liu, Z.; Zhou, C.; Yi, J.; Tang, Y.; Chen, L.; Chen, L.; Wang, Y.; Sun, Q.; et al. Cryo-EM structure of the Mon1-Ccz1-RMC1 complex reveals molecular basis of metazoan RAB7A activation. Proc. Natl. Acad. Sci. USA 2023, 120, e2301725120. [Google Scholar] [CrossRef]
- Cai, C.Z.; Zhuang, X.X.; Zhu, Q.; Wu, M.Y.; Su, H.; Wang, X.J.; Iyaswamy, A.; Yue, Z.; Wang, Q.; Zhang, B.; et al. Enhancing autophagy maturation with CCZ1-MON1A complex alleviates neuropathology and memory defects in Alzheimer disease models. Theranostics 2022, 12, 1738–1755. [Google Scholar] [CrossRef]
- Klink, B.U.; Herrmann, E.; Antoni, C.; Langemeyer, L.; Kiontke, S.; Gatsogiannis, C.; Ungermann, C.; Raunser, S.; Kummel, D. Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2121494119. [Google Scholar] [CrossRef]
- Borchers, A.C.; Janz, M.; Schafer, J.H.; Moeller, A.; Kummel, D.; Paululat, A.; Ungermann, C.; Langemeyer, L. Regulatory sites in the Mon1-Ccz1 complex control Rab5 to Rab7 transition and endosome maturation. Proc. Natl. Acad. Sci. USA 2023, 120, e2303750120. [Google Scholar] [CrossRef]
- Visse, R.; Nagase, H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: Structure, function, and biochemistry. Circ. Res. 2003, 92, 827–839. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- de Almeida, L.G.N.; Thode, H.; Eslambolchi, Y.; Chopra, S.; Young, D.; Gill, S.; Devel, L.; Dufour, A. Matrix Metalloproteinases: From Molecular Mechanisms to Physiology, Pathophysiology, and Pharmacology. Pharmacol. Rev. 2022, 74, 712–768. [Google Scholar] [CrossRef]
- Chow, C.F.W.; Guo, X.; Asthana, P.; Zhang, S.; Wong, S.K.K.; Fallah, S.; Che, S.; Gurung, S.; Wang, Z.; Lee, K.B.; et al. Body weight regulation via MT1-MMP-mediated cleavage of GFRAL. Nat. Metab. 2022, 4, 203–212. [Google Scholar] [CrossRef]
- Hsu, P.L.; Chien, C.W.; Tang, Y.A.; Lin, B.W.; Lin, S.C.; Lin, Y.S.; Chen, S.Y.; Sun, H.S.; Tsai, S.J. Targeting BRD3 eradicates nuclear TYRO3-induced colorectal cancer metastasis. Sci. Adv. 2023, 9, eade3422. [Google Scholar] [CrossRef]
- Kayagaki, N.; Kawasaki, A.; Ebata, T.; Ohmoto, H.; Ikeda, S.; Inoue, S.; Yoshino, K.; Okumura, K.; Yagita, H. Metalloproteinase-mediated release of human Fas ligand. J. Exp. Med. 1995, 182, 1777–1783. [Google Scholar] [CrossRef]
- Chopra, S.; Overall, C.M.; Dufour, A. Matrix metalloproteinases in the CNS: Interferons get nervous. Cell Mol. Life Sci. 2019, 76, 3083–3095. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.Z.; Hu, A.X.; Liu, X.S. DUSP19 regulates IL-1beta-induced apoptosis and MMPs expression in rat chondrocytes through JAK2/STAT3 signaling pathway. Biomed. Pharmacother. 2017, 96, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Chilla, A.; Anceschi, C.; Frediani, E.; Scavone, F.; Del Rosso, T.; Pelagio, G.; Tufaro, A.; De Palma, G.; Del Rosso, M.; Fibbi, G.; et al. Inhibition of MMPs supports amoeboid angiogenesis hampering VEGF-targeted therapies via MLC and ERK 1/2 signaling. J. Transl. Med. 2023, 21, 102. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiao, L.; Feng, D.; Yuan, Y.; Yang, X.; Li, J.; Jiang, D.; Chen, H.; Meng, Q.; Chen, R.; et al. Human apical-out nasal organoids reveal an essential role of matrix metalloproteinases in airway epithelial differentiation. Nat. Commun. 2024, 15, 143. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Sleeboom, J.J.F.; van Tienderen, G.S.; Schenke-Layland, K.; van der Laan, L.J.W.; Khalil, A.A.; Verstegen, M.M.A. The extracellular matrix as hallmark of cancer and metastasis: From biomechanics to therapeutic targets. Sci. Transl. Med. 2024, 16, eadg3840. [Google Scholar] [CrossRef] [PubMed]
- Muniz-Bongers, L.R.; McClain, C.B.; Saxena, M.; Bongers, G.; Merad, M.; Bhardwaj, N. MMP2 and TLRs modulate immune responses in the tumor microenvironment. JCI Insight 2021, 6, 144913. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Zhang, Y.; Liu, D.; Chen, J. MMP2 is a immunotherapy related biomarker and correlated with cancer-associated fibroblasts infiltrate in melanoma. Cancer Cell Int. 2023, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Paye, A.; Truong, A.; Yip, C.; Cimino, J.; Blacher, S.; Munaut, C.; Cataldo, D.; Foidart, J.M.; Maquoi, E.; Collignon, J.; et al. EGFR activation and signaling in cancer cells are enhanced by the membrane-bound metalloprotease MT4-MMP. Cancer Res. 2014, 74, 6758–6770. [Google Scholar] [CrossRef]
- Morgan, E.L.; Scarth, J.A.; Patterson, M.R.; Wasson, C.W.; Hemingway, G.C.; Barba-Moreno, D.; Macdonald, A. E6-mediated activation of JNK drives EGFR signalling to promote proliferation and viral oncoprotein expression in cervical cancer. Cell Death Differ. 2021, 28, 1669–1687. [Google Scholar] [CrossRef]
- Kalogera, E.; Nevala, W.K.; Finnes, H.D.; Suman, V.J.; Schimke, J.M.; Strand, C.A.; Kottschade, L.A.; Kudgus, R.A.; Buhrow, S.A.; Becher, L.R.; et al. A Phase I Trial of Nab-Paclitaxel/Bevacizumab (AB160) Nano-Immunoconjugate Therapy for Gynecologic Malignancies. Clin. Cancer Res. 2024, 30, 2623–2635. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Vanoni, G.; Loap, P.; Dubot, C.; Timperi, E.; Minsat, M.; Bazire, L.; Durdux, C.; Fourchotte, V.; Laas, E.; et al. Nivolumab plus chemoradiotherapy in locally-advanced cervical cancer: The NICOL phase 1 trial. Nat. Commun. 2023, 14, 3698. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, Q.; Liao, J.Y.; Song, E.; Xia, Q.; Pan, J.; Li, Y.; Li, J.; Zhou, B.; Ye, Y.; et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell 2020, 180, 1081–1097. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tang, T.; Zhang, G.; Liang, T. Identification of tumor antigens and immune subtypes of cholangiocarcinoma for mRNA vaccine development. Mol. Cancer 2021, 20, 50. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, A.; De Veylder, L. The Dual Face of Cyclin B1. Trends Plant Sci. 2018, 23, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, J.T.; Fabian, K.P.; Padget, M.R.; Lopez, D.C.; Hoke, A.T.; Allen, C.T.; Hermsen, M.; London, N.R., Jr.; Hodge, J.W. Exploiting the immunogenic potential of standard of care radiation or cisplatin therapy in preclinical models of HPV-associated malignancies. J. Immunother. Cancer 2022, 10, e005752. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Cui, Z.; Zheng, J.; Wu, Q.; Yu, H.; You, Y.; Zheng, C.; Sun, Y. Significance of Immunogenic Cell Death-Related Prognostic Gene Signature in Cervical Cancer Prognosis and Anti-Tumor Immunity. J. Inflamm. Res. 2023, 16, 2189–2207. [Google Scholar] [CrossRef]
- Huang, V.; Place, R.F.; Portnoy, V.; Wang, J.; Qi, Z.; Jia, Z.; Yu, A.; Shuman, M.; Yu, J.; Li, L.C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012, 40, 1695–1707. [Google Scholar] [CrossRef]
- Xie, B.; Wang, S.; Jiang, N.; Li, J.J. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019, 443, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.R.; Banjara, B.; Ohemeng, A.; Davidson, A.M.; Boue, S.M.; Burow, M.E.; Tilghman, S.L. Novel Therapeutic Combination Targets the Growth of Letrozole-Resistant Breast Cancer through Decreased Cyclin B1. Nutrients 2023, 15, 1632. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R. The matrix in cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Baidya, S.K.; Adhikari, N.; Jha, T. An updated patent review of matrix metalloproteinase (MMP) inhibitors (2021-present). Expert. Opin. Ther. Pat. 2023, 33, 631–649. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Yuan, Z.; Liu, J.; Deng, L.; Zhao, Y.; Wang, S.; Tang, E.; Yang, X.; Li, N.; An, J.; et al. CCZ1 Accelerates the Progression of Cervical Squamous Cell Carcinoma by Promoting MMP2/MMP17 Expression. Biomedicines 2024, 12, 1468. https://doi.org/10.3390/biomedicines12071468
Yu J, Yuan Z, Liu J, Deng L, Zhao Y, Wang S, Tang E, Yang X, Li N, An J, et al. CCZ1 Accelerates the Progression of Cervical Squamous Cell Carcinoma by Promoting MMP2/MMP17 Expression. Biomedicines. 2024; 12(7):1468. https://doi.org/10.3390/biomedicines12071468
Chicago/Turabian StyleYu, Jing, Zhenlong Yuan, Jing Liu, Lu Deng, Yuting Zhao, Shengnan Wang, Enyu Tang, Xi Yang, Ning Li, Jusheng An, and et al. 2024. "CCZ1 Accelerates the Progression of Cervical Squamous Cell Carcinoma by Promoting MMP2/MMP17 Expression" Biomedicines 12, no. 7: 1468. https://doi.org/10.3390/biomedicines12071468
APA StyleYu, J., Yuan, Z., Liu, J., Deng, L., Zhao, Y., Wang, S., Tang, E., Yang, X., Li, N., An, J., & Wu, L. (2024). CCZ1 Accelerates the Progression of Cervical Squamous Cell Carcinoma by Promoting MMP2/MMP17 Expression. Biomedicines, 12(7), 1468. https://doi.org/10.3390/biomedicines12071468




