DCTPP1 Expression as a Predictor of Chemotherapy Response in Luminal A Breast Cancer Patients
Abstract
:1. Introduction
2. Methods
2.1. Cell Culture
2.2. Real-Time Quantitative Reverse Transcription PCR (RT-qPCR)
2.3. Database Analysis
2.4. Mutation Character Analysis
2.5. Survival Data Analysis
2.6. Statistical Analysis
3. Results
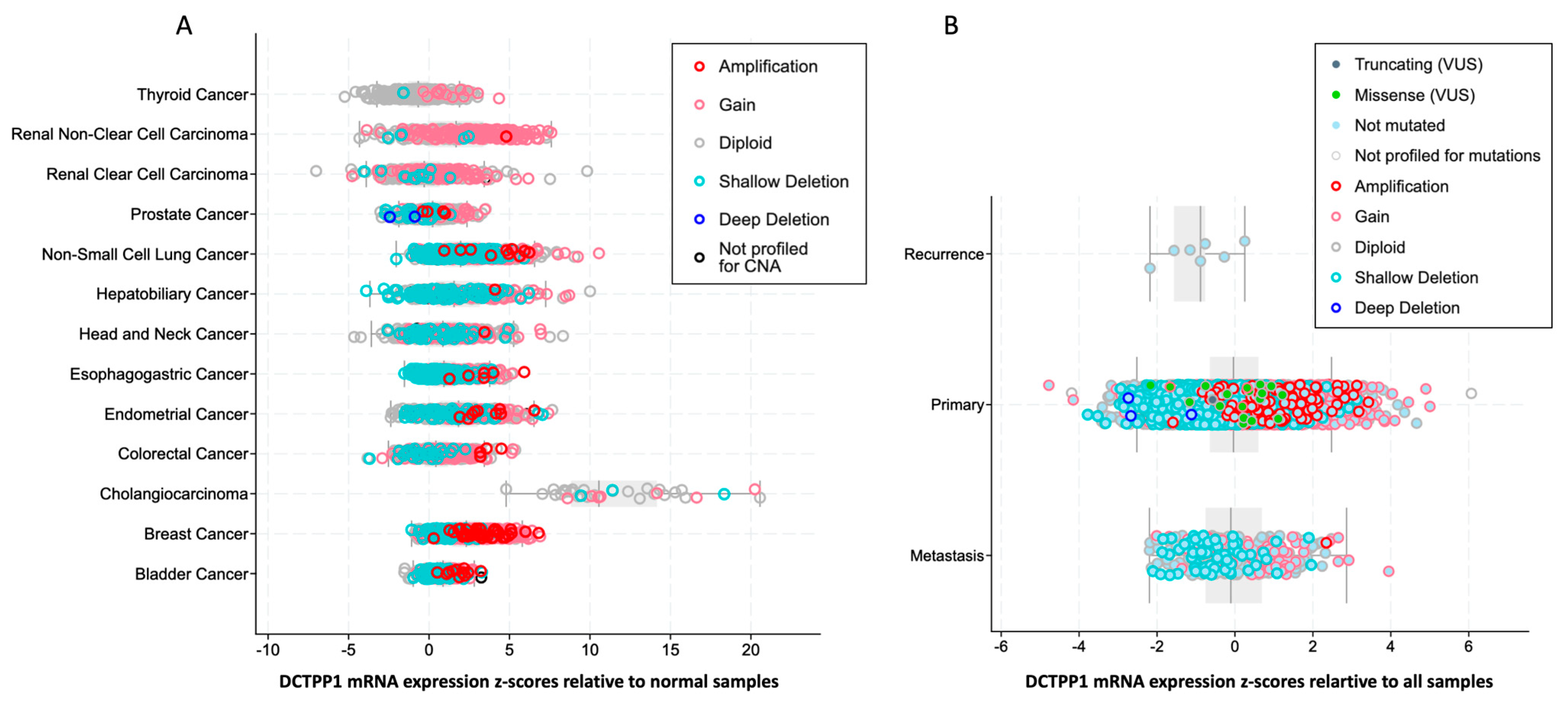
3.1. DCTPP1 Expression Levels and Alteration Frequency in Pan-Cancers
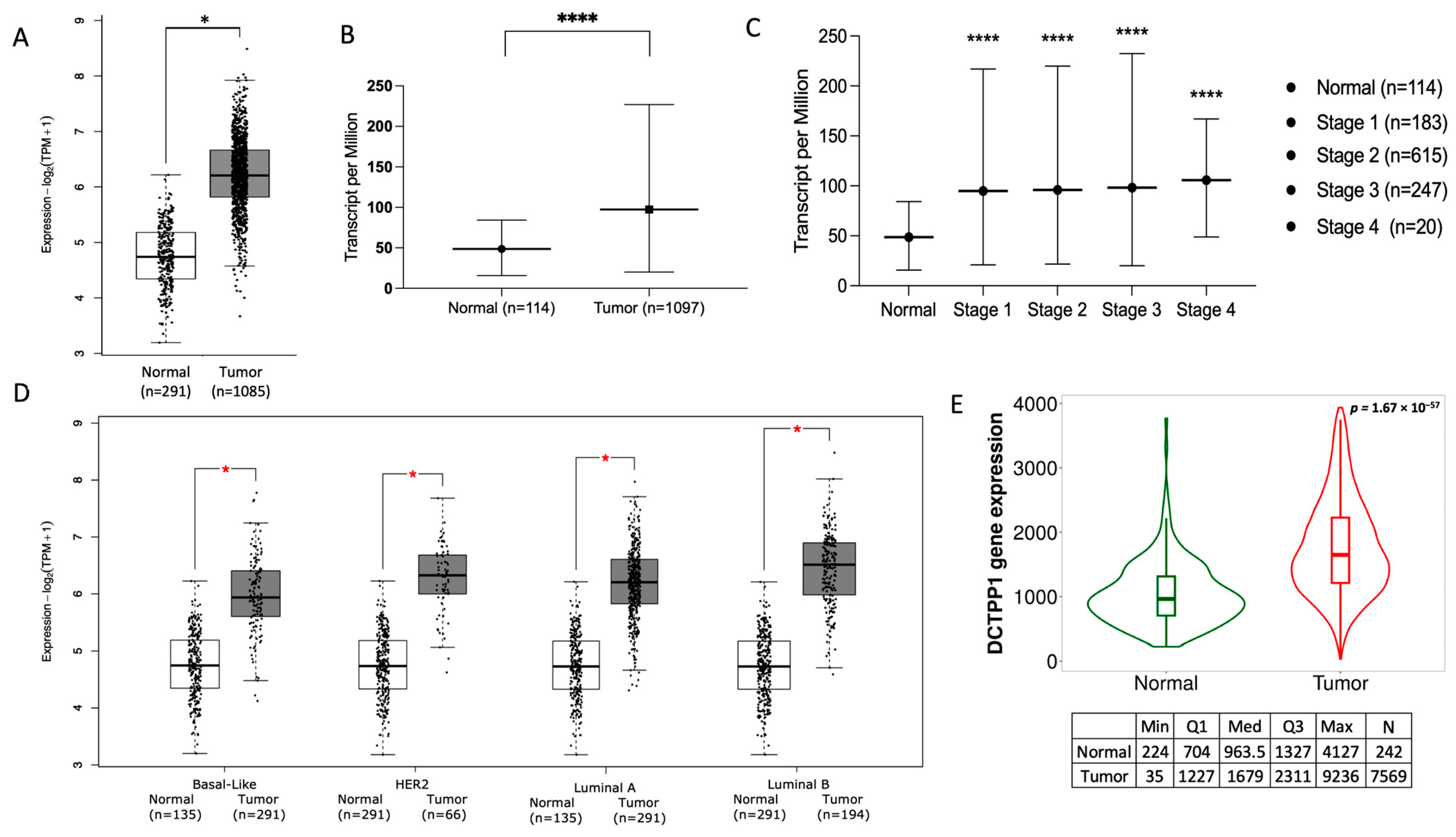
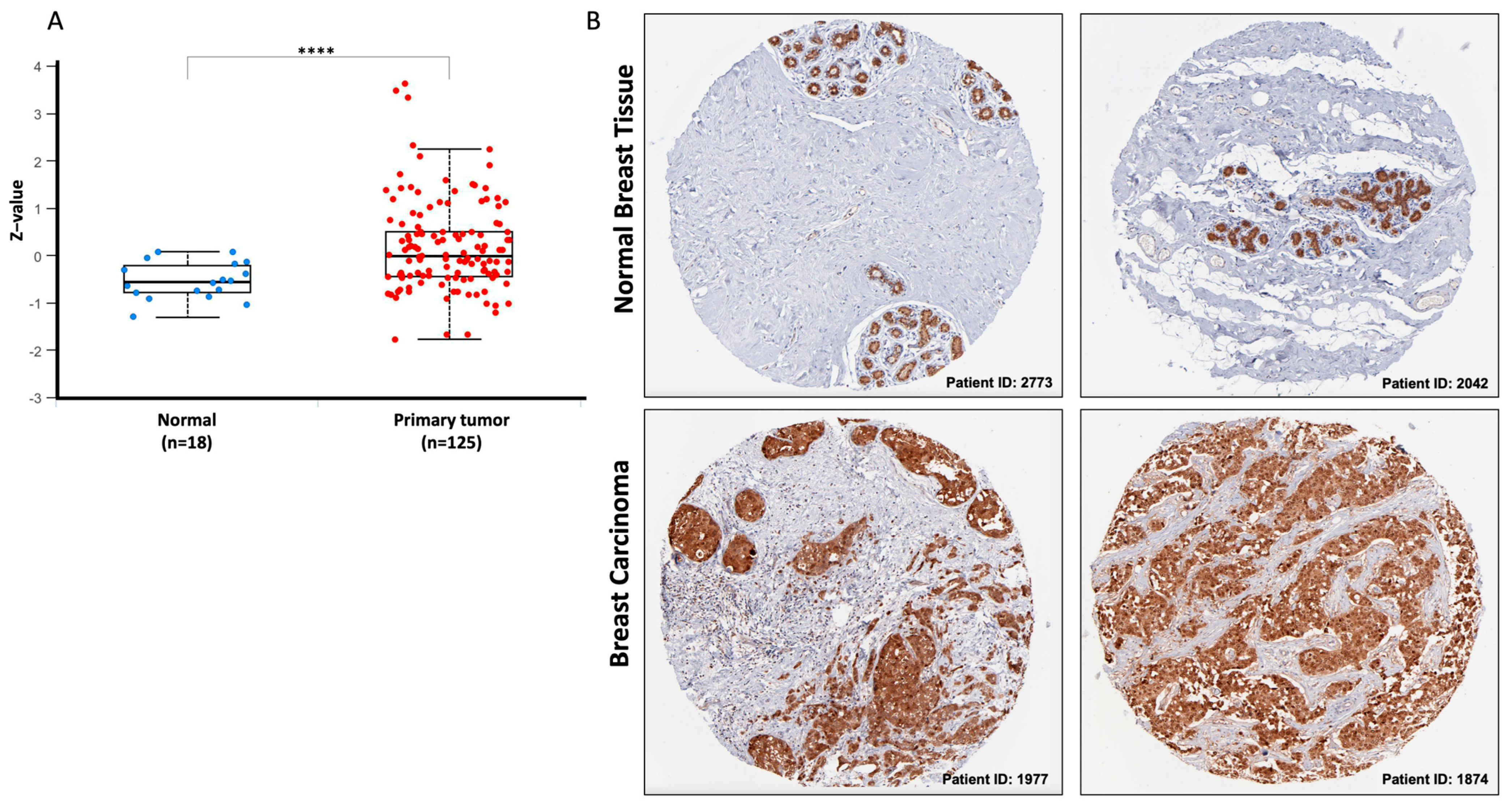
3.2. DCTPP1 Is Up-Regulated across Tumor Grades and Cancer Stages in BRCA
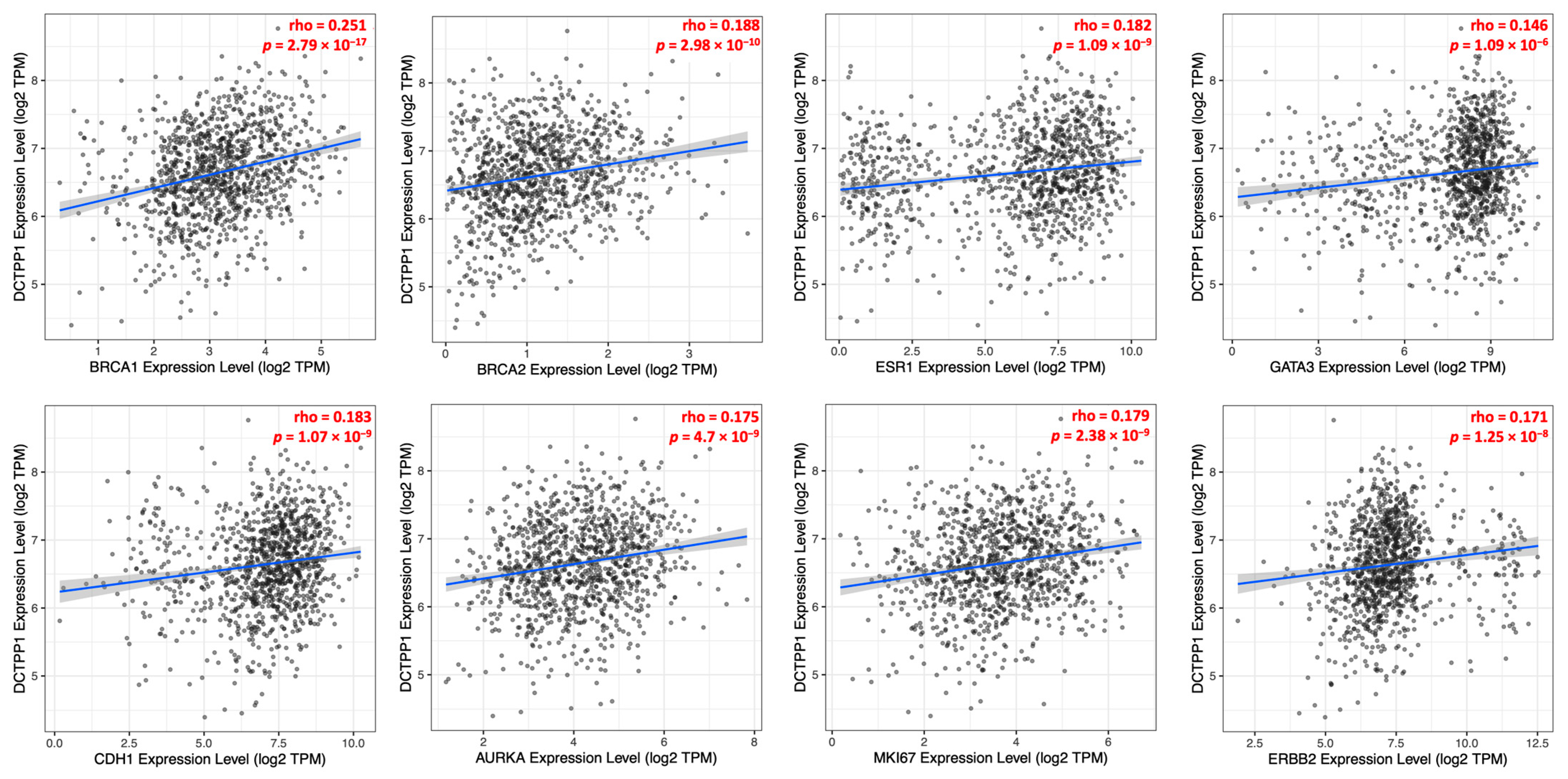
3.3. Correlation between DCTPP1 Gene Expression and Established Markers in ER-Positive, Luminal A BRCA Patients
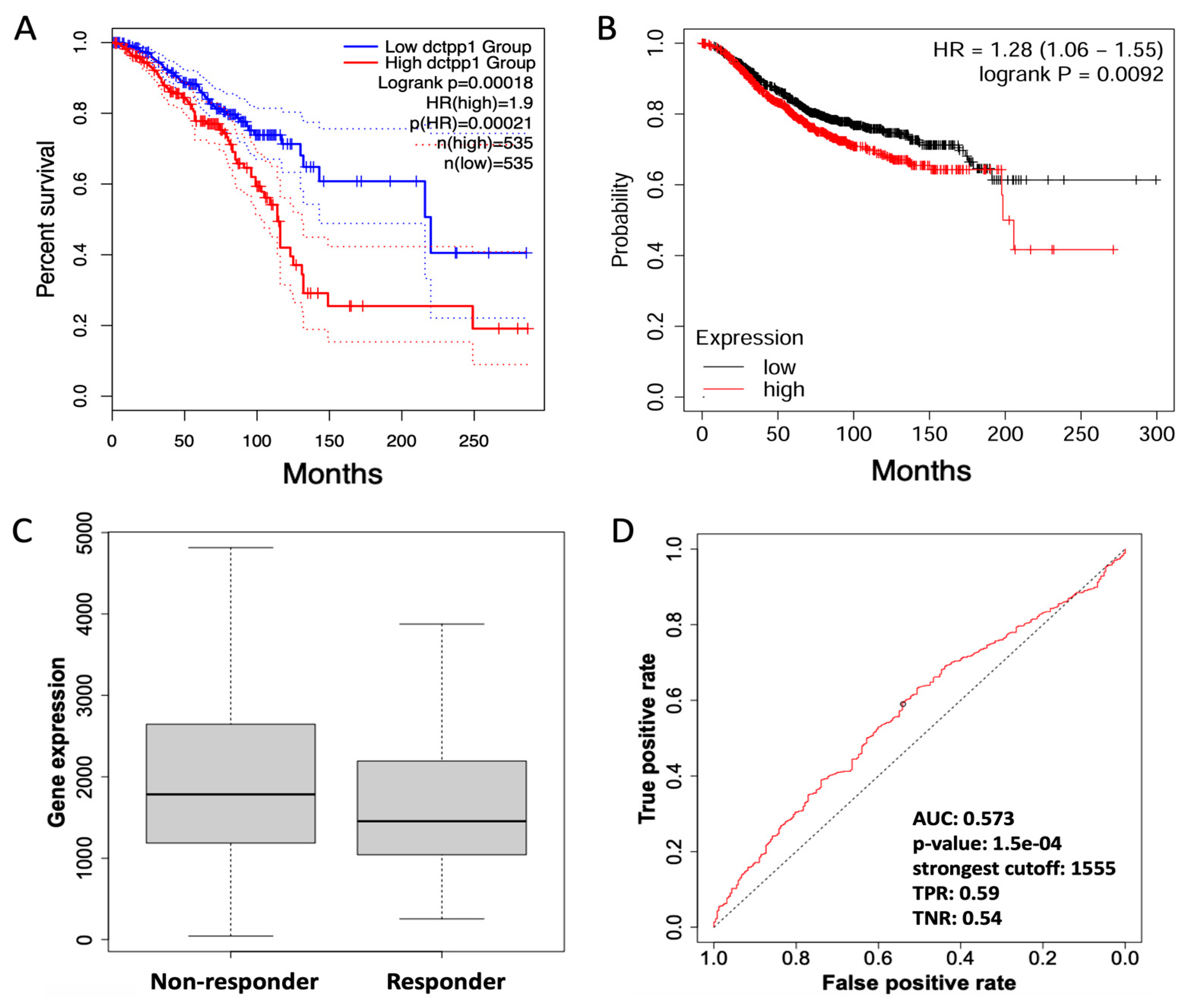
3.4. DCTPP1 as a Potential Marker of Poor Prognosis in Chemotherapy-Treated BRCA Patients
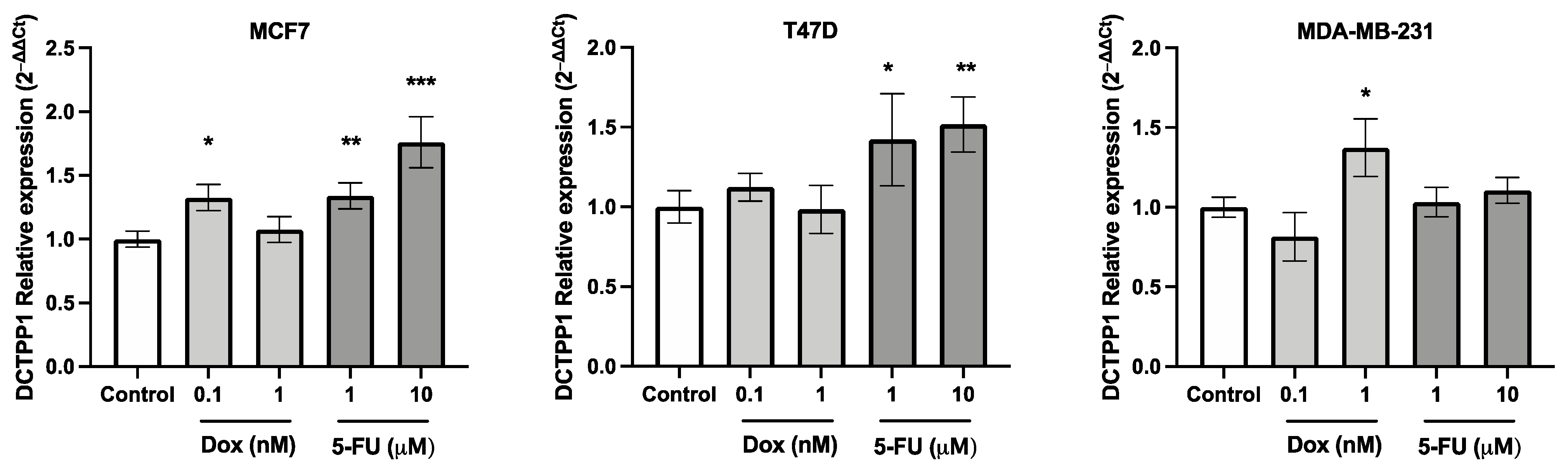
3.5. DCTPP1 Expression Levels in Luminal A BRCA Cell Lines Treated with 5-FU
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Fowler, A.M.; Ulaner, G.A.; Mahajan, A. Molecular Classification of Breast Cancer. PET Clin. 2023, 18, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.J.; Swain, S.M. Luminal A Breast Cancer and Molecular Assays: A Review. Oncologist 2018, 23, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.J.; Stearns, V. Understanding Resistance to Tamoxifen in Hormone Receptor–Positive Breast Cancer. Clin. Chem. 2009, 55, 1453–1455. [Google Scholar] [CrossRef]
- Song, F.F.; Xia, L.L.; Ji, P.; Tang, Y.B.; Huang, Z.M.; Zhu, L.; Zhang, J.; Wang, J.Q.; Zhao, G.P.; Ge, H.L.; et al. Human dCTP pyrophosphatase 1 promotes breast cancer cell growth and stemness through the modulation on 5-methyl-dCTP metabolism and global hypomethylation. Oncogenesis 2015, 4, e159. [Google Scholar] [CrossRef]
- Requena, C.E.; Pérez-Moreno, G.; Ruiz-Pérez, L.M.; Vidal, A.E.; González-Pacanowska, D. The NTP pyrophosphatase DCTPP1 contributes to the homoeostasis and cleansing of the dNTP pool in human cells. Biochem. J. 2014, 459, 171–180. [Google Scholar] [CrossRef]
- Lu, L.D.; Sun, Q.; Fan, X.Y.; Zhong, Y.; Yao, Y.F.; Zhao, G.P. Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J. Biol. Chem. 2010, 285, 28076–28085. [Google Scholar] [CrossRef]
- Niu, M.; Shan, M.; Liu, Y.; Song, Y.; Han, J.G.; Sun, S.; Liang, X.S.; Zhang, G.Q. DCTPP1, an Oncogene Regulated by miR-378a-3p, Promotes Proliferation of Breast Cancer via DNA Repair Signaling Pathway. Front. Oncol. 2021, 11, 641931. [Google Scholar] [CrossRef] [PubMed]
- Morisaki, T.; Yashiro, M.; Kakehashi, A.; Inagaki, A.; Kinoshita, H.; Fukuoka, T.; Kasashima, H.; Masuda, G.; Sakurai, K.; Kubo, N.; et al. Comparative proteomics analysis of gastric cancer stem cells. PLoS ONE 2014, 9, e110736. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Dong, W.; He, H.; Han, Z.; Zhuo, Y.; Mo, R.; Liang, Y.; Zhu, J.; Li, R.; Qu, H.; et al. Autophagy induced by overexpression of DCTPP1 promotes tumor progression and predicts poor clinical outcome in prostate cancer. Int. J. Biol. Macromol. 2018, 118, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, P.; Chen, X.; Gong, D.; Wu, Y.; Huang, L.; Chen, Y. ROS-Induced DCTPP1 Upregulation Contributes to Cisplatin Resistance in Ovarian Cancer. Front. Mol. Biosci. 2022, 9, 838006. [Google Scholar] [CrossRef] [PubMed]
- Vuger, A.T.; Tiscoski, K.; Apolinario, T.; Cardoso, F. Anthracyclines in the treatment of early breast cancer friend or foe? Breast 2022, 65, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Y.; Lin, H.J.; Hsiao, L.C.; Lin, Y.F.; Chang, C.S.; Liu, D.Z. Reduction of breast tumor drug resistance by 2,3,5,4’-tetrahydroxystilbene for exhibition synergic chemotherapeutic effect. PLoS ONE 2021, 16, e0260533. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.; Briasoulis, E.; Linardou, H.; Bafaloukos, D.; Papadimitriou, C. Taxane resistance in breast cancer: Mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 2012, 38, 890–903. [Google Scholar] [CrossRef]
- Xia, L.L.; Tang, Y.B.; Song, F.F.; Xu, L.; Ji, P.; Wang, S.J.; Zhu, J.M.; Zhang, Y.; Zhao, G.P.; Wang, Y.; et al. DCTPP1 attenuates the sensitivity of human gastric cancer cells to 5-fluorouracil by up-regulating MDR1 expression epigenetically. Oncotarget 2016, 7, 68623–68637. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Fekete, J.T.; Győrffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef]
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Eroles, P.; Bosch, A.; Alejandro Pérez-Fidalgo, J.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Shi, Y.; Ye, P.; Long, X. Differential Expression Profiles of the Transcriptome in Breast Cancer Cell Lines Revealed by Next Generation Sequencing. Cell Physiol. Biochem. 2017, 44, 804–816. [Google Scholar] [CrossRef]
- Hunakova, L.; Horvathova, E.; Majerova, K.; Bobal, P.; Otevrel, J.; Brtko, J. Genotoxic Effects of Tributyltin and Triphenyltin Isothiocyanates, Cognate RXR Ligands: Comparison in Human Breast Carcinoma MCF 7 and MDA-MB-231 Cells. Int. J. Mol. Sci. 2019, 20, 1198. [Google Scholar] [CrossRef]
- Yang, S.; Zhou, Q.; Yang, X. Caspase-3 status is a determinant of the differential responses to genistein between MDA-MB-231 and MCF-7 breast cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 903–911. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz, J.P.; Soto-Jiménez, D.; Calaf, G.M. DCTPP1 Expression as a Predictor of Chemotherapy Response in Luminal A Breast Cancer Patients. Biomedicines 2024, 12, 1732. https://doi.org/10.3390/biomedicines12081732
Muñoz JP, Soto-Jiménez D, Calaf GM. DCTPP1 Expression as a Predictor of Chemotherapy Response in Luminal A Breast Cancer Patients. Biomedicines. 2024; 12(8):1732. https://doi.org/10.3390/biomedicines12081732
Chicago/Turabian StyleMuñoz, Juan P., Diego Soto-Jiménez, and Gloria M. Calaf. 2024. "DCTPP1 Expression as a Predictor of Chemotherapy Response in Luminal A Breast Cancer Patients" Biomedicines 12, no. 8: 1732. https://doi.org/10.3390/biomedicines12081732




