Calcium-Enhanced Medium-Based Delivery of Splice Modulating Antisense Oligonucleotides in 2D and 3D hiPSC-Derived Neuronal Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Neural Progenitor Cell Cultures
2.2. Neural Differentiation and Maturation
2.3. Organoid Preparation
2.4. Calcium Chloride-Enhanced Medium (CEM), Gymnotic Uptake, and Lipofectamine-Mediated ASO Uptake in 2D Neuronal Cell Culture
2.5. Antisense Oligonucleotides
2.6. NEPA 3D Electroporation
2.7. Calcium Chloride-Enhanced Media (CEM)-Mediated ASO Uptake in 3D Neuronal Cell Culture
2.8. RNA Isolation, cDNA Synthesis, and RT-PCR
2.9. Immunocytochemistry
2.10. Organoid Embedding
2.11. Immunohistochemistry on Organoid Sections
2.12. Transfection Efficiency of Antisense Oligonucleotide
3. Results
3.1. CEM Improves Survival in hiPSC-Derived Neuronal Cultures
3.2. CEM Delivery Is More Efficient in Neurons Compared to Astrocytes
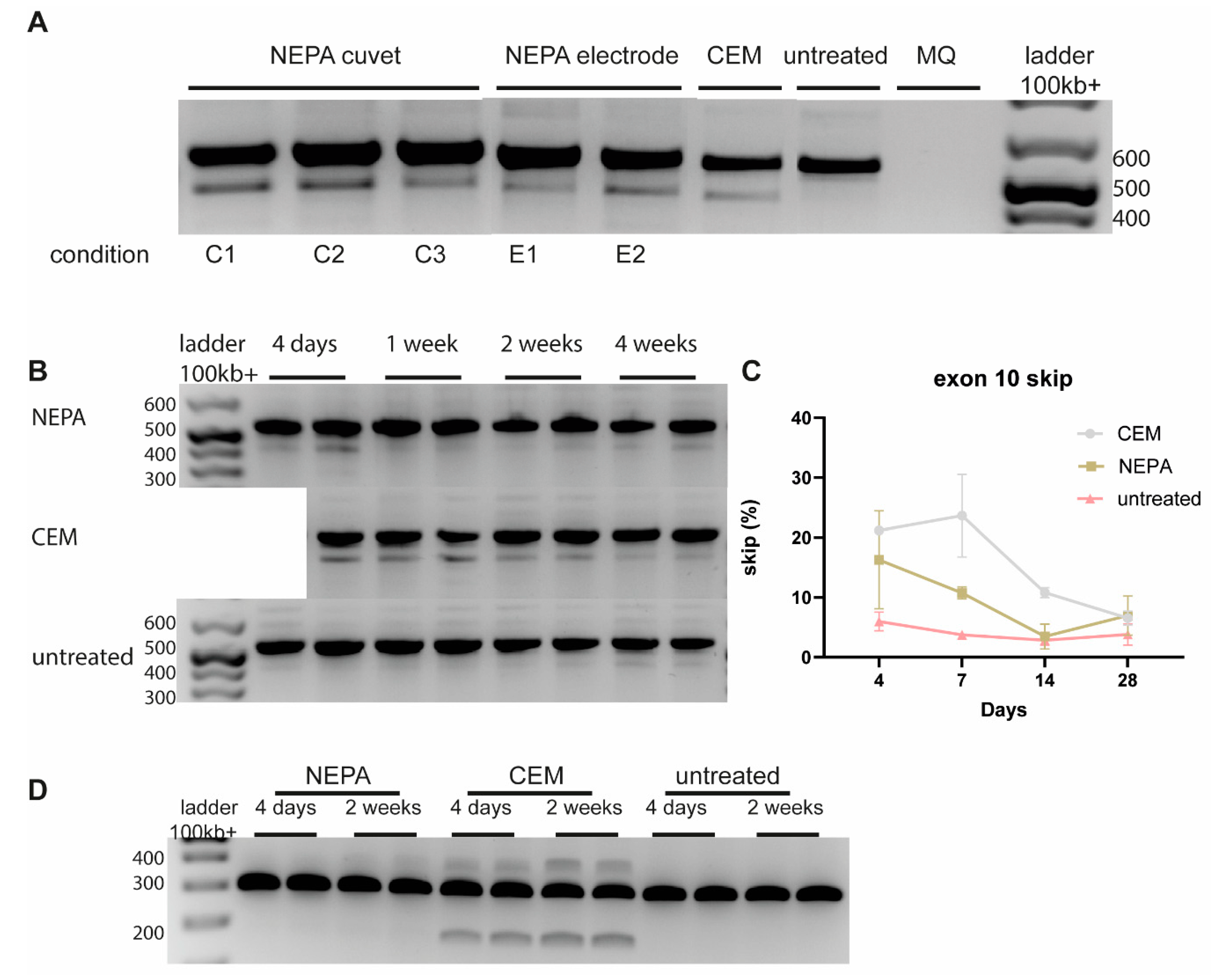
3.3. Dose-Dependent CEM-Mediated ASO Uptake in D15 Brain Organoids
3.4. Long-Term On-Target Effect of CEM-Mediated ASO Uptake in D35 Brain Organoids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A. FDA Approval of Nusinersen for Spinal Muscular Atrophy Makes 2016 the Year of Splice Modulating Oligonucleotides. Nucleic Acid Ther. 2017, 27, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Kordasiewicz, H.B.; Cleveland, D.W. Antisense Drugs Make Sense for Neurological Diseases. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 831–852. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef]
- Li, L.; Chao, J.; Shi, Y. Modeling neurological diseases using iPSC-derived neural cells: iPSC modeling of neurological diseases. Cell Tissue Res. 2018, 371, 143–151. [Google Scholar] [CrossRef]
- Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Induced Pluripotent Stem Cells (iPSCs)-Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells 2021, 10, 2319. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef]
- Wang, H. Modeling Neurological Diseases with Human Brain Organoids. Front. Synaptic Neurosci. 2018, 10, 15. [Google Scholar] [CrossRef]
- Eichmuller, O.L.; Knoblich, J.A. Human cerebral organoids—A new tool for clinical neurology research. Nat. Rev. Neurol. 2022, 18, 661–680. [Google Scholar] [CrossRef]
- Hori, S.; Yamamoto, T.; Waki, R.; Wada, S.; Wada, F.; Noda, M.; Obika, S. Ca2+ enrichment in culture medium potentiates effect of oligonucleotides. Nucleic Acids Res. 2015, 43, e128. [Google Scholar] [CrossRef] [PubMed]
- Bowles, K.R.; Silva, M.C.; Whitney, K.; Bertucci, T.; Berlind, J.E.; Lai, J.D.; Garza, J.C.; Boles, N.C.; Mahali, S.; Strang, K.H.; et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 2021, 184, 4547–4563. [Google Scholar] [CrossRef] [PubMed]
- Buijsen, R.A.M.; Hu, M.; Saez-Gonzalez, M.; Notopoulou, S.; Mina, E.; Koning, W.; Gardiner, S.L.; van der Graaf, L.M.; Daoutsali, E.; Pepers, B.A.; et al. Spinocerebellar Ataxia Type 1 Characteristics in Patient-Derived Fibroblast and iPSC-Derived Neuronal Cultures. Mov. Disord. 2023, 38, 1428–1442. [Google Scholar] [CrossRef]
- Capaldi, D.C.; Scozzari, A.N. Manufacturing and Analytical Processes for 2-O-(2-Methoxyethyl)-Modified Oligonucleotides. In Antisense Drug Technology, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Evers, M.M.; Tran, H.D.; Zalachoras, I.; Meijer, O.C.; den Dunnen, J.T.; van Ommen, G.J.; Aartsma-Rus, A.; van Roon-Mom, W.M. Preventing formation of toxic N-terminal huntingtin fragments through antisense oligonucleotide-mediated protein modification. Nucleic Acid Ther. 2014, 24, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Daoutsali, E.; Hailu, T.T.; Buijsen, R.A.M.; Pepers, B.A.; van der Graaf, L.M.; Verbeek, M.M.; Curtis, D.; de Vlaam, T.; van Roon-Mom, W.M.C. Antisense Oligonucleotide-Induced Amyloid Precursor Protein Splicing Modulation as a Therapeutic Approach for Dutch-Type Cerebral Amyloid Angiopathy. Nucleic Acid Ther. 2021, 31, 351–363. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- Stein, C.A.; Hansen, J.B.; Lai, J.; Wu, S.; Voskresenskiy, A.; Hog, A.; Worm, J.; Hedtjarn, M.; Souleimanian, N.; Miller, P.; et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010, 38, e3. [Google Scholar] [CrossRef]
- Mortberg, M.A.; Gentile, J.E.; Nadaf, N.M.; Vanderburg, C.; Simmons, S.; Dubinsky, D.; Slamin, A.; Maldonado, S.; Petersen, C.L.; Jones, N.; et al. A single-cell map of antisense oligonucleotide activity in the brain. Nucleic Acids Res. 2023, 51, 7109–7124. [Google Scholar] [CrossRef]
- Wan, L.; Kral, A.J.; Voss, D.; Krainer, A.R. Preclinical Screening of Splice-Switching Antisense Oligonucleotides in PDAC Organoids. bioRxiv 2023. Update in Nucleic Acid Ther. 2024, 34, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Tynianskaia, L.; Esiyok, N.; Huttner, W.B.; Heide, M. Targeted Microinjection and Electroporation of Primate Cerebral Organoids for Genetic Modification. J. Vis. Exp. 2023, 193, e65176. [Google Scholar] [CrossRef] [PubMed]



| ASO | Chemistry | Sequence | |
|---|---|---|---|
| 1 | H40 (FAM-labeled) | 2′-O-methoxyethyl-modified phosphorothioate | 5′-UCC UUU CAU CUC UGG GCU C-3′ |
| 2 | HTT-AON12.1 | 2′-O-methoxyethyl-modified phosphorothioate | 5′-GUC CCA UCA UUC AGG UCC AU-3′ |
| 3 | APP-AT040 | 2′-O-methoxyethyl-modified phosphorothioate | 5′-CUU CUG CAA AGA ACA CCU UG-3′ |
| Set Parameters | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Poring Pulse | Transfer Pulse | ||||||||||
| V | Length (ms) | Interval (ms) | No. | D. Rate (%) | Polarity | V | Length (ms) | Interval (ms) | No. | D. Rate (%) | Polarity |
| 225 | 1.5 | 50 | 4 | 10 | + | 20 | 50 | 50 | 5 | 40 | +/− |
| Target | Forward/Reverse primer (5′-3′) | |
|---|---|---|
| RT-PCR | HTT APP | CATCACACACAGCACCAAGA/ TCTCACATCTCTGTCTGGAACC CAAGACGGAGGAGATCTCTGA/ TTGGATTTTCGTAGCCGTTC |
| Manufacturer | Catalogue Number | Dilution | |
|---|---|---|---|
| Primary antibodies | |||
| Mouse-a-MAP2 | Sigma-Aldrich, Burlington, MA, USA | MAB364 | 1:500 |
| Rabbit-a-GFAP | Sigma-Aldrich Burlington, MA, USA | Z0334 | 1:500 |
| Mouse-a-Nestin | Stemcell Technologies Vancouver, BC, Canada | 60091 | 1:250 |
| Rabbit-a-PAX6 | Biolegend, San Diego, CA, USA | 901301 | 1:250 |
| Secondary antibodies | |||
| DAPI | Sigma Aldrich, Burlington, MA, USA | D9542 | 1:1000 |
| Goat-a-Mouse-Alexa594 | Invitrogen, Waltham, MA, USA | A11005 | 1:500 |
| Goat-a-Rabbit-Alexa594 | Invitrogen, Waltham, MA, USA | A11012 | 1:500 |
| Goat-a-Mouse-Alexa488 | Invitrogen, Waltham, MA, USA | A11001 | 1:500 |
| Goat-a-Rabbit-Alexa750 | Abcam, Cambridge, UK | ab175732 | 1:500 |
| Channel | Label | Color | Wavelength | Target% | Exposure Type | Exposure Time (s) |
|---|---|---|---|---|---|---|
| Ch1 | DAPI | Blue | 386 | 25 | Fixed | 0.00612 |
| Ch2 | ASO | Green | 485 | 25 | Fixed | 0.03745 |
| Ch3 | MAP2 | Red | 560 | 25 | Fixed | 0.02687 |
| Ch4 | GFAP | Yellow | 740 | 25 | Fixed | 0.05654 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buijsen, R.A.M.; van der Graaf, L.M.; Kuijper, E.C.; Pepers, B.A.; Daoutsali, E.; Weel, L.; Raz, V.; Parfitt, D.A.; van Roon-Mom, W.M.C. Calcium-Enhanced Medium-Based Delivery of Splice Modulating Antisense Oligonucleotides in 2D and 3D hiPSC-Derived Neuronal Models. Biomedicines 2024, 12, 1933. https://doi.org/10.3390/biomedicines12091933
Buijsen RAM, van der Graaf LM, Kuijper EC, Pepers BA, Daoutsali E, Weel L, Raz V, Parfitt DA, van Roon-Mom WMC. Calcium-Enhanced Medium-Based Delivery of Splice Modulating Antisense Oligonucleotides in 2D and 3D hiPSC-Derived Neuronal Models. Biomedicines. 2024; 12(9):1933. https://doi.org/10.3390/biomedicines12091933
Chicago/Turabian StyleBuijsen, Ronald A. M., Linda M. van der Graaf, Elsa C. Kuijper, Barry A. Pepers, Elena Daoutsali, Lotte Weel, Vered Raz, David A. Parfitt, and Willeke M. C. van Roon-Mom. 2024. "Calcium-Enhanced Medium-Based Delivery of Splice Modulating Antisense Oligonucleotides in 2D and 3D hiPSC-Derived Neuronal Models" Biomedicines 12, no. 9: 1933. https://doi.org/10.3390/biomedicines12091933
APA StyleBuijsen, R. A. M., van der Graaf, L. M., Kuijper, E. C., Pepers, B. A., Daoutsali, E., Weel, L., Raz, V., Parfitt, D. A., & van Roon-Mom, W. M. C. (2024). Calcium-Enhanced Medium-Based Delivery of Splice Modulating Antisense Oligonucleotides in 2D and 3D hiPSC-Derived Neuronal Models. Biomedicines, 12(9), 1933. https://doi.org/10.3390/biomedicines12091933






