Uncovering the Contrasts and Connections in PASC: Viral Load and Cytokine Signatures in Acute COVID-19 versus Post-Acute Sequelae of SARS-CoV-2 (PASC)
Abstract
:1. Introduction
2. Materials and Methods
3. Viral Loads
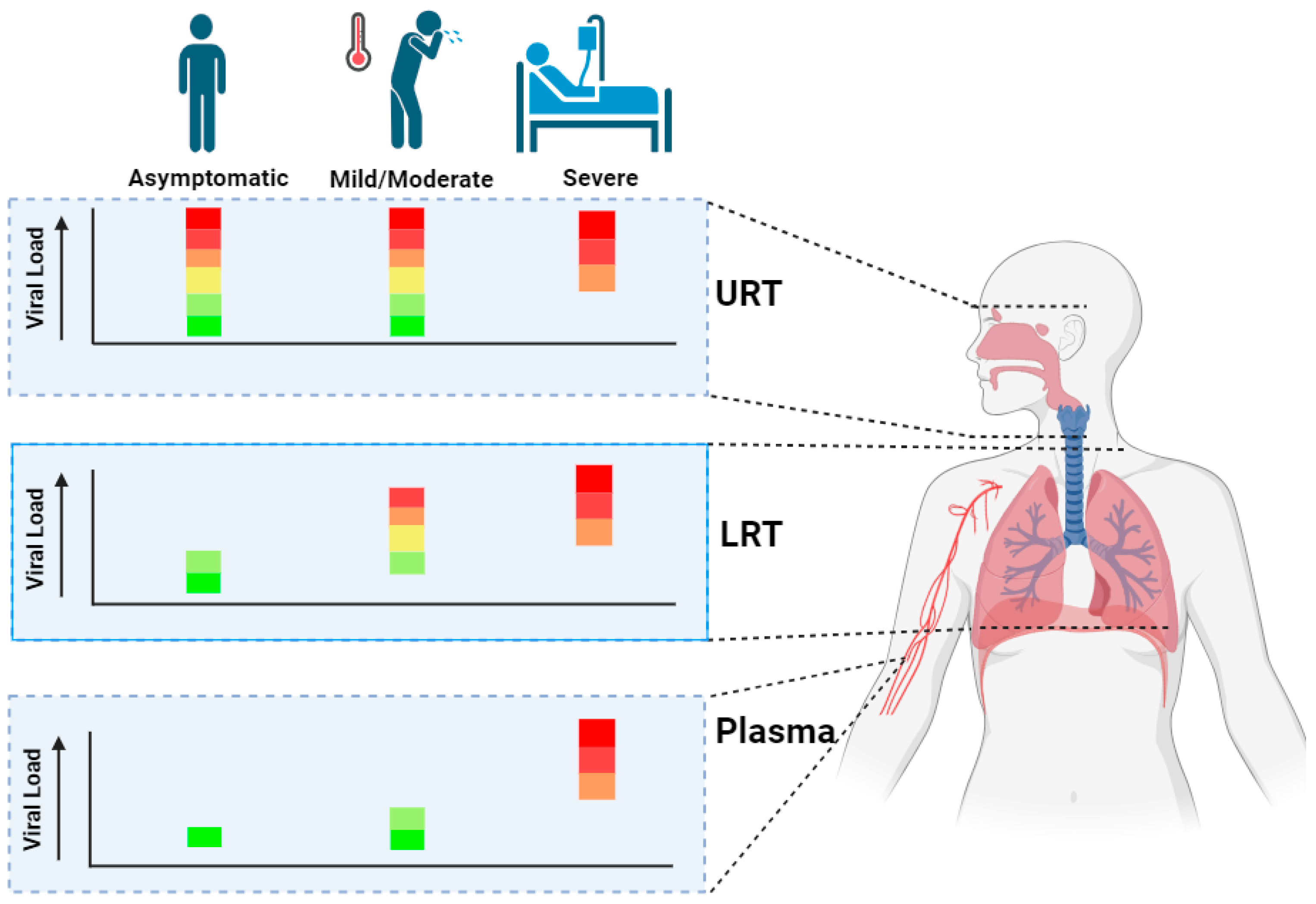
3.1. Viral Loads during Acute COVID-19
3.2. Relation between Acute-Phase Viral Dynamics and PASC Phase
4. Cytokines and Chemokines
4.1. Cytokines and Chemokines during Acute COVID-19
 ) indicates a significant increase in cytokine or chemokine levels as compared with healthy controls. Orange middle line (
) indicates a significant increase in cytokine or chemokine levels as compared with healthy controls. Orange middle line ( ) indicates no significant difference in cytokine or chemokine levels as compared with healthy controls. Green arrow (
) indicates no significant difference in cytokine or chemokine levels as compared with healthy controls. Green arrow ( ) indicates a significant decrease in cytokine or chemokine levels as compared with healthy controls. Empty tables implicate that no literature regarding this cytokine or chemokine in the context of the clinical classification and sample location could be found. References used include [57,58,59,60,61,62,63,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91], and specific references are found in the corresponding part of the figure.
) indicates a significant decrease in cytokine or chemokine levels as compared with healthy controls. Empty tables implicate that no literature regarding this cytokine or chemokine in the context of the clinical classification and sample location could be found. References used include [57,58,59,60,61,62,63,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91], and specific references are found in the corresponding part of the figure.
 ) indicates a significant increase in cytokine or chemokine levels as compared with healthy controls. Orange middle line (
) indicates a significant increase in cytokine or chemokine levels as compared with healthy controls. Orange middle line ( ) indicates no significant difference in cytokine or chemokine levels as compared with healthy controls. Green arrow (
) indicates no significant difference in cytokine or chemokine levels as compared with healthy controls. Green arrow ( ) indicates a significant decrease in cytokine or chemokine levels as compared with healthy controls. Empty tables implicate that no literature regarding this cytokine or chemokine in the context of the clinical classification and sample location could be found. References used include [57,58,59,60,61,62,63,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91], and specific references are found in the corresponding part of the figure.
) indicates a significant decrease in cytokine or chemokine levels as compared with healthy controls. Empty tables implicate that no literature regarding this cytokine or chemokine in the context of the clinical classification and sample location could be found. References used include [57,58,59,60,61,62,63,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91], and specific references are found in the corresponding part of the figure.
4.2. Relationship between Viral Load and Cyto- and Chemokine Markers across COVID-19 Disease Subsets
4.3. Differences and Similarities in Cytokine Profiles in Acute and PASC

5. In Vitro Models and Preclinical Animal Models to Study Acute COVID-19 and PASC
| Animal | Susceptibility | Severity | Acute Phase | PASC | Ref. |
|---|---|---|---|---|---|
| Transgenic Mice |
|
| K18-hACE2 mice:
|
| [113,114] |
| Syrian Hamsters |
|
|
|
| [112,115,116,117,118] |
| Ferrets |
|
|
|
| [107,108,118,119] |
6. Concluding Remarks and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/deaths?n=c (accessed on 5 March 2024).
- Bulut, C.; Kato, Y. Epidemiology of COVID-19. Turk. J. Med. Sci. 2020, 50, 563–570. [Google Scholar] [CrossRef] [PubMed]
- FAIR Health. Patients Diagnosed with Post-COVID Conditions an Analysis of Private Healthcare Claims Using the Official ICD-10 Diagnostic Code; FAIR Health: New York, NY, USA, 2022. [Google Scholar]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Liew, F.; Efstathiou, C.; Fontanella, S.; Richardson, M.; Saunders, R.; Swieboda, D.; Sidhu, J.K.; Ascough, S.; Moore, S.C.; Mohamed, N.; et al. Large-scale phenotyping of patients with long COVID post-hospitalization reveals mechanistic subtypes of disease. Nat. Immunol. 2024, 25, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Ozonoff, A.; Jayavelu, N.D.; Liu, S.; Melamed, E.; Milliren, C.E.; Qi, J.; Geng, L.N.; McComsey, G.A.; Cairns, C.B.; Baden, L.R.; et al. Features of acute COVID-19 associated with post-acute sequelae of SARS-CoV-2 phenotypes: Results from the IMPACC study. Nat. Commun. 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Pujadas, E.; Chaudhry, F.; McBride, R.; Richter, F.; Zhao, S.; Wajnberg, A.; Nadkarni, G.; Glicksberg, B.S.; Houldsworth, J.; Cordon-Cardo, C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020, 8, e70. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, L.-M.; Wan, L.; Xiang, T.-X.; Le, A.; Liu, J.-M.; Peiris, M.; Poon, L.L.M.; Zhang, W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020, 20, 656–657. [Google Scholar] [CrossRef]
- Tjendra, Y.; Al Mana, A.F.; Espejo, A.P.; Akgun, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Predicting Disease Severity and Outcome in COVID-19 Patients: A Review of Multiple Biomarkers. Arch. Pathol. Lab. Med. 2020, 144, 1465–1474. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Deer, R.R.; Rock, M.A.; Vasilevsky, N.; Carmody, L.; Rando, H.; Anzalone, A.J.; Basson, M.D.; Bennett, T.D.; Bergquist, T.; Boudreau, E.A.; et al. Characterizing Long COVID: Deep Phenotype of a Complex Condition. EBioMedicine 2021, 74. [Google Scholar] [CrossRef]
- Frontera, J.A.; Thorpe, L.E.; Simon, N.M.; de Havenon, A.; Yaghi, S.; Sabadia, S.B.; Yang, D.; Lewis, A.; Melmed, K.; Balcer, L.J.; et al. Post-acute sequelae of COVID-19 symptom phenotypes and therapeutic strategies: A prospective, observational study. PLoS ONE 2022, 17, e0275274. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Sivan, M.; Perlowski, A.; Nikolich, J. Long COVID: A clinical update. Lancet 2024, 404, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Mitsumura, T.; Okamoto, T.; Tosaka, M.; Yamana, T.; Shimada, S.; Iijima, Y.; Sakakibara, R.; Shibata, S.; Honda, T.; Shirai, T.; et al. Association of SARS-CoV-2 RNA Copy Number with the COVID-19 Mortality Rate and Its Effect on the Predictive Performance of Mortality in Severe Cases. Jpn. J. Infect. Dis. 2022, 75, 504–510. [Google Scholar] [CrossRef]
- Hasanoglu, I.; Korukluoglu, G.; Asilturk, D.; Cosgun, Y.; Kalem, A.K.; Altas, A.B.; Kayaaslan, B.; Eser, F.; Kuzucu, E.A.; Guner, R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection 2021, 49, 117–126. [Google Scholar] [CrossRef]
- Puchinger, K.; Castelletti, N.; Rubio-Acero, R.; Geldmacher, C.; Eser, T.M.; Deák, F.; Paunovic, I.; Bakuli, A.; Saathoff, E.; von Meyer, A.; et al. The interplay of viral loads, clinical presentation, and serological responses in SARS-CoV-2—Results from a prospective cohort of outpatient COVID-19 cases. Virology 2022, 569, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Magleby, R.; Westblade, L.F.; Trzebucki, A.; Simon, M.S.; Rajan, M.; Park, J.; Goyal, P.; Safford, M.M.; Satlin, M.J. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality among Hospitalized Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2021, 73, E4197–E4205. [Google Scholar] [CrossRef] [PubMed]
- Boyapati, A.; Wipperman, M.F.; Ehmann, P.J.; Hamon, S.; Lederer, D.J.; Waldron, A.; Flanagan, J.J.; Karayusuf, E.; Bhore, R.; Nivens, M.C.; et al. Baseline Severe Acute Respiratory Syndrome Viral Load Is Associated with Coronavirus Disease 2019 Severity and Clinical Outcomes: Post Hoc Analyses of a Phase 2/3 Trial. J. Infect. Dis. 2021, 224, 1830–1838. [Google Scholar] [CrossRef]
- Souverein, D.; van Stralen, K.; van Lelyveld, S.; van Gemeren, C.; Haverkort, M.; Snijders, D.; Soetekouw, R.; Kapteijns, E.; de Jong, E.; Hermanides, G.; et al. Initial Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load Is Associated with Disease Severity: A Retrospective Cohort Study. Open Forum Infect. Dis. 2022, 9, ofac223. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, D.; Hou, J.; Li, H.; Cao, D.; Guo, M.; Ling, Y.; Gao, M.; Zhou, Y.; Wan, Y.; et al. An inter-correlated cytokine network identified at the center of cytokine storm predicted COVID-19 prognosis. Cytokine 2021, 138, 155365. [Google Scholar] [CrossRef]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 20, 269–270. [Google Scholar] [CrossRef]
- Ynga-Durand, M.; Maaß, H.; Milošević, M.; Krstanović, F.; Matešić, M.P.; Jonjić, S.; Protić, A.; Brizić, I.; Šustić, A.; Čičin-Šain, L. SARS-CoV-2 Viral Load in the Pulmonary Compartment of Critically Ill COVID-19 Patients Correlates with Viral Serum Load and Fatal Outcomes. Viruses 2022, 14, 1292. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.F.; García-Mateo, N.; Motos, A.; Resino, S.; Tamayo, L.; Murua, P.R.; Bustamante-Munguira, E.; Curto, E.G.; Úbeda-Iglesias, A.; Torre, M.d.C.d.l.; et al. Effect of viral storm in patients admitted to intensive care units with severe COVID-19 in Spain: A multicentre, prospective, cohort study. Lancet Microbe 2023, 4, e431–e441. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, A.; Righini, E.; Micheli, V.; Pinoli, P.; Bernasconi, A.; Rizzo, A.; Oreni, L.; Ridolfo, A.L.; Antinori, S.; Ceri, S.; et al. SARS-CoV-2 viremia and COVID-19 mortality: A prospective observational study. PLoS ONE 2023, 18, e0281052.18. [Google Scholar] [CrossRef] [PubMed]
- Karron, R.A.; Hetrich, M.K.; Bin Na, Y.; Knoll, M.D.; Schappell, E.; Meece, J.; Hanson, E.; Tong, S.; Lee, J.S.; Veguilla, V.; et al. Assessment of Clinical and Virological Characteristics of SARS-CoV-2 Infection Among Children Aged 0 to 4 Years and Their Household Members. JAMA Netw. Open 2022, 5, e2227348. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.L.; Bain, W.; Naqvi, A.; Staines, B.; Castanha, P.M.S.; Yang, H.; Boltz, V.F.; Barratt-Boyes, S.; Marques, E.T.A.; Mitchell, S.L.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Viremia Is Associated with Coronavirus Disease 2019 Severity and Predicts Clinical Outcomes. Clin. Infect. Dis. 2022, 74, 1525–1533. [Google Scholar] [CrossRef]
- National Institutes of Health. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 21 March 2024).
- Yu, X.; Sun, S.; Shi, Y.; Wang, H.; Zhao, R.; Sheng, J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit. Care 2020, 24, 1–4. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kim, J.M.; Hong, S.P.; Choi, S.Y.; Yang, M.J.; Ju, Y.S.; Kim, Y.T.; Kim, H.M.; Rahman, T.; Chung, M.K.; et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- McGonagle, D.; Kearney, M.F.; O’Regan, A.; O’Donnell, J.S.; Quartuccio, L.; Watad, A.; Bridgewood, C. Therapeutic implications of ongoing alveolar viral replication in COVID-19. Lancet Rheumatol. 2022, 4, e135–e144. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.-C.; Ng, K.-C.; Ching, R.H.H.; Lai, K.-L.; Kam, T.T.; Gu, H.; Sit, K.-Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Puhach, O.; Adea, K.; Hulo, N.; Sattonnet, P.; Genecand, C.; Iten, A.; Jacquérioz, F.; Kaiser, L.; Vetter, P.; Eckerle, I.; et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat. Med. 2022, 28, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Bertollini, R. Severity of SARS-CoV-2 Reinfections as Compared with Primary Infections. N. Engl. J. Med. 2021, 385, 2487–2489. [Google Scholar] [CrossRef] [PubMed]
- Omololu, A.; Ojelade, B.; Ajayi, O.; Adesomi, T.; Alade, O.; Adebisi, S.; Nwadike, V. “Long COVID”: A case report of persistent symptoms in a patient with prolonged SARS-CoV-2 shedding for over 110 days. SAGE Open Med. Case Rep. 2021, 9, 2050313X211015494. [Google Scholar] [CrossRef]
- Viszlayová, D.; Sojka, M.; Dobrodenková, S.; Szabó, S.; Bilec, O.; Turzová, M.; Ďurina, J.; Baloghová, B.; Borbély, Z.; Kršák, M. SARS-CoV-2 RNA in the Cerebrospinal Fluid of a Patient with Long COVID. Ther. Adv. Infect. Dis. 2021, 8, 204993612110485. [Google Scholar] [CrossRef]
- Stadtmüller, M.; Laubner, A.; Rost, F.; Winkler, S.; Patrasová, E.; Šimůnková, L.; Reinhardt, S.; Beil, J.; Dalpke, A.H.; Yi, B. Emergence and spread of a sub-lineage of SARS-CoV-2 Alpha variant B.1.1.7 in Europe, and with further evolution of spike mutation accumulations shared with the Beta and Gamma variants. Virus Evol. 2022, 8, veac010. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.M.; Jamoulle, M.; Carletto, M.P.; Jamoulle, M.; Carletto, M.P.; Van Holm, B.; Moens, L.; Meyts, I.; Maes, P.; Van Weyenbergh, J. Blood transcriptomics reveal persistent SARS-CoV-2 RNA and candidate biomarkers in Long COVID patients. medrXiv 2024. [Google Scholar] [CrossRef]
- Craddock, V.; Mahajan, A.; Krishnamachary, B.; Spikes, L.; Chalise, P.; Dhillon, N.K. Persistent Presence of Spike protein and Viral RNA in the Circulation of Individuals with Post-Acute Sequelae of COVID-19. medRxiv 2022. [Google Scholar] [CrossRef]
- Su, Y.; Yuan, D.; Chen, D.G.; Ng, R.H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, 881–895.e20. [Google Scholar] [CrossRef]
- Rombauts, A.; Infante, C.; Lagos, M.d.A.M.d.; Alba, J.; Valiente, A.; Donado-Mazarrón, C.; Carretero-Ledesma, M.; Rodríguez-Álvarez, R.; Omatos, S.; Palacios-Baena, Z.R.; et al. Impact of SARS-CoV-2 RNAemia and other risk factors on long-COVID: A prospective observational multicentre cohort study. J. Infect. 2023, 86, 154–225. [Google Scholar] [CrossRef]
- Ram-Mohan, N.; Kim, D.; Rogers, A.J.; Blish, C.A.; Nadeau, K.C.; Blomkalns, A.L.; Yang, S. Association Between SARS-CoV-2 RNAemia and Postacute Sequelae of COVID-19. Open Forum Infect. Dis. 2022, 9, ofab646. [Google Scholar] [CrossRef]
- Pérez, D.A.G.; Fonseca-Agüero, A.; Toledo-Ibarra, G.A.; Gomez-Valdivia, J.d.J.; Díaz-Resendiz, K.J.G.; Benitez-Trinidad, A.B.; Razura-Carmona, F.F.; Navidad-Murrieta, M.S.; Covantes-Rosales, C.E.; Giron-Pérez, M.I. Post-COVID-19 Syndrome in Outpatients and Its Association with Viral Load. Int. J. Environ. Res. Public. Health 2022, 19, 15145. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. In Prevalence of Post COVID-19 Condition Symptoms: A Systematic Review and Meta-Analysis of Cohort Study Data, Stratified by Recruitment Setting Key Facts; European Centre for Disease Prevention and Control: Solna, Sweeden, 2022.
- Chen, B.; Julg, B.; Mohandas, S.; Bradfute, S.B.; Force, R.M.P.T.; Hospital, M.G.; Mit; Harvard; Angeles, C.H.L.; States, U. Viral persistence, reactivation, and mechanisms of long COVID. eLife 2023, 12, e86015. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 698169. [Google Scholar] [CrossRef]
- Zhang, L.; Richards, A.; Inmaculada Barrasa, M.; Hughes, S.H.; Young, R.A.; Jaenisch, R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Nat. Acad. Sci. USA 2021, 118, e2105968118. [Google Scholar] [CrossRef]
- Randall, R.E.; Griffin, D.E. Within host RNA virus persistence: Mechanisms and consequences. Curr. Opin. Virol. 2017, 23, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-D.; Ding, M.; Dong, X.; Zhang, J.-J.; Azkur, A.K.; Azkur, D.; Gan, H.; Sun, Y.-L.; Fu, W.; Li, W.; et al. Risk factors for severe and critically ill COVID-19 patients: A review. Allergy 2021, 76, 428–455. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Ishikawa, F.; Kondo, M.; Kakiuchi, T. The Role of IL-17 and Related Cytokines in Inflammatory Autoimmune Diseases. Mediat. Inflamm. 2017, 2017, 3908061. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The Role of Interleukin 6 During Viral Infections. Front. Microbiol. 2019, 10, 01057. [Google Scholar] [CrossRef]
- Carlini, V.; Noonan, D.M.; Abdalalem, E.; Goletti, D.; Sansone, C.; Calabrone, L.; Albini, A. The multifaceted nature of IL-10: Regulation, role in immunological homeostasis and its relevance to cancer, COVID-19 and post-COVID conditions. Front. Immunol. 2023, 14, 1161067. [Google Scholar] [CrossRef]
- Zawawi, Z.M.; Kalyanasundram, J.; Zain, R.M.; Thayan, R.; Basri, D.F.; Yap, W.B. Prospective Roles of Tumor Necrosis Factor-Alpha (TNF-α) in COVID-19: Prognosis, Therapeutic and Management. Int. J. Mol. Sci. 2023, 24, 6142. [Google Scholar] [CrossRef]
- Duong-Quy, S.; Vo-Pham-Minh, T.; Tran-Xuan, Q.; Huynh-Anh, T.; Vo-Van, T.; Vu-Tran-Thien, Q.; Nguyen-Nhu, V. Post-COVID-19 Pulmonary Fibrosis: Facts—Challenges and Futures: A Narrative Review. Pulm. Ther. 2023, 20, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Shin, E.-C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021, 53, 750–760. [Google Scholar] [CrossRef]
- Vu, D.-L.; Martinez-Murillo, P.; Pigny, F.; Vono, M.; Meyer, B.; Eberhardt, C.S.; Lemeille, S.; Von Dach, E.; Blanchard-Rohner, G.; Eckerle, I.; et al. Longitudinal Analysis of Inflammatory Response to SARS-CoV-2 in the Upper Respiratory Tract Reveals an Association with Viral Load, Independent of Symptoms. J. Clin. Immunol. 2021, 41, 1723–1732. [Google Scholar] [CrossRef]
- Sidhu, J.K.; Siggins, M.K.; Liew, F.; Russell, C.D.; Uruchurtu, A.S.S.; Davis, C.; Turtle, L.; Moore, S.C.; E Hardwick, H.; Oosthuyzen, W.; et al. Delayed Mucosal Antiviral Responses Despite Robust Peripheral Inflammation in Fatal COVID-19. J. Infect. Dis. 2023, 230, e17–e29. [Google Scholar] [CrossRef]
- Rolland-Debord, C.; Piéroni, L.; Bejar, F.; Milon, A.; Choinier, P.; Blin, E.; Bravais, J.; Halitim, P.; Letellier, A.; Camuset, J.; et al. Cell and cytokine analyses from bronchoalveolar lavage in non-critical COVID-19 pneumonia. Intern. Emerg. Med. 2023, 18, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Li, Q.; Li, L.; Peng, X.; Ling, Z.; Xiao, B.; Feng, J.; Chen, Z.; Chang, D.; Xie, L.; et al. Association of Early Inflammation with Age and Asymptomatic Disease in COVID-19. J. Inflamm. Res. 2021, 14, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Zaid, Y.; Doré, É.; Dubuc, I.; Archambault, A.-S.; Flamand, O.; Laviolette, M.; Flamand, N.; Boilard, É.; Flamand, L. Chemokines and eicosanoids fuel the hyperinflammation within the lungs of patients with severe COVID-19. J. Allergy Clin. Immunol. 2021, 148, 368–380.e3. [Google Scholar] [CrossRef]
- Reynolds, D.; Guillamet, C.V.; Day, A.; Borcherding, N.; Guillamet, R.V.; Choreño-Parra, J.A.; House, S.L.; O’halloran, J.A.; Zúñiga, J.; Ellebedy, A.H.; et al. Comprehensive Immunologic Evaluation of Bronchoalveolar Lavage Samples from Human Patients with Moderate and Severe Seasonal Influenza and Severe COVID-19. J. Immunol. 2021, 207, 1229–1238. [Google Scholar] [CrossRef]
- Grant, R.A.; Poor, T.A.; Sichizya, L.; Diaz, E.; Bailey, J.I.; Soni, S.; Senkow, K.J.; Perez-Leonor, X.G.; Abdala-Valencia, H.; Lu, Z.; et al. Prolonged exposure to lung-derived cytokines is associated with inflammatory activation of microglia in patients with COVID-19. bioRxiv 2023. [Google Scholar] [CrossRef]
- Min, C.-K.; Cheon, S.; Ha, N.-Y.; Sohn, K.M.; Kim, Y.; Aigerim, A.; Shin, H.M.; Choi, J.-Y.; Inn, K.-S.; Kim, J.-H.; et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016, 6, 25359. [Google Scholar] [CrossRef]
- Wong, C.K.; Lam, C.W.K.; Wu, A.K.L.; Ip, W.K.; Lee, N.L.S.; Chan, I.H.S.; Lit, L.C.W.; Hui, D.S.C.; Chan, M.H.M.; Chung, S.S.C.; et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004, 136, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, A.; Wan, Y.; Liu, X.; Qiu, C.; Xi, X.; Ren, Y.; Wang, J.; Dong, Y.; Bao, M.; et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. USA 2014, 111, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Cremoni, M.; Allouche, J.; Graça, D.; Zorzi, K.; Fernandez, C.; Teisseyre, M.; Benzaken, S.; Ruetsch-Chelli, C.; Esnault, V.L.M.; Dellamonica, J.; et al. Low baseline IFN-γ response could predict hospitalization in COVID-19 patients. Front. Immunol. 2022, 13, 953502. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gebo, K.A.; Abraham, A.G.; Habtehyimer, F.; Patel, E.U.; Laeyendecker, O.; Gniadek, T.J.; Fernandez, R.E.; Baker, O.R.; Ram, M.; et al. Dynamics of inflammatory responses after SARS-CoV-2 infection by vaccination status in the USA: A prospective cohort study. Lancet Microbe 2023, 4, e692–e703. [Google Scholar] [CrossRef]
- Martins, M.L.; da Silva-Malta, M.C.F.; Araújo, A.L.; Gonçalves, F.A.; Botelho, M.d.L.; de Oliveira, I.R.; Boy, L.d.S.M.F.; Moreira, H.M.; Barbosa-Stancioli, E.F.; Ribeiro, M.A.; et al. A potent inflammatory response is triggered in asymptomatic blood donors with recent SARS-CoV-2 infection. Rev. Soc. Bras. Med. Trop. 2022, 55, e0239–2022. [Google Scholar] [CrossRef]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Tjan, L.H.; Furukawa, K.; Nagano, T.; Kiriu, T.; Nishimura, M.; Arii, J.; Hino, Y.; Iwata, S.; Nishimura, Y.; Mori, Y. Early Differences in Cytokine Production by Severity of Coronavirus Disease 2019. J. Infect. Dis. 2021, 223, 1145–1149. [Google Scholar] [CrossRef]
- Masood, K.I.; Yameen, M.; Ashraf, J.; Shahid, S.; Mahmood, S.F.; Nasir, A.; Nasir, N.; Jamil, B.; Ghanchi, N.K.; Khanum, I.; et al. Upregulated type I interferon responses in asymptomatic COVID-19 infection are associated with improved clinical outcome. Sci. Rep. 2021, 11, 22958. [Google Scholar] [CrossRef]
- Tripathy, A.S.; Vishwakarma, S.; Trimbake, D.; Gurav, Y.K.; Potdar, V.A.; Mokashi, N.D.; Patsute, S.D.; Kaushal, H.; Choudhary, M.L.; Tilekar, B.N.; et al. Pro-inflammatory CXCL-10, TNF-α, IL-1β, and IL-6: Biomarkers of SARS-CoV-2 infection. Arch. Virol. 2021, 166, 3301–3310. [Google Scholar] [CrossRef]
- Smith, N.; Goncalves, P.; Charbit, B.; Grzelak, L.; Beretta, M.; Planchais, C.; Bruel, T.; Rouilly, V.; Bondet, V.; Hadjadj, J.; et al. Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat. Immunol. 2021, 22, 1428–1439. [Google Scholar] [CrossRef]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.-W.; Zhou, Z.; Wang, J.-L.; Deng, Y.-F.; Jing, H.; Qiu, Y. Viral loads, lymphocyte subsets and cytokines in asymptomatic, mildly and critical symptomatic patients with SARS-CoV-2 infection: A retrospective study. Virol. J. 2021, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.-S.; Kim, J.Y.; Kim, M.-C.; Park, S.Y.; Kim, B.-N.; Bae, S.; Cha, H.H.; Jung, J.; Lee, M.J.; Choi, S.-H.; et al. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses. Am. J. Trop. Med. Hyg. 2020, 103, 2412–2418. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Chen, Z.; Lui, G.; Wong, C.K.; Wong, W.T.; Ng, R.W.Y.; Tso, E.Y.K.; Fung, K.S.C.; Chan, V.; Yeung, A.C.M.; et al. Longitudinal Cytokine Profile in Patients with Mild to Critical COVID-19. Front. Immunol. 2021, 12, 763292. [Google Scholar] [CrossRef]
- Huang, W.; Li, M.; Luo, G.; Wu, X.; Su, B.; Zhao, L.; Zhang, S.; Chen, X.; Jia, M.; Zhu, J.; et al. The Inflammatory Factors Associated with Disease Severity to Predict COVID-19 Progression. J. Immunol. 2021, 206, 1597–1608. [Google Scholar] [CrossRef]
- Calabrese, F.; Lunardi, F.; Baldasso, E.; Pezzuto, F.; Kilitci, A.; Olteanu, G.-E.; Del Vecchio, C.; Fortarezza, F.; Boscolo, A.; Schiavon, M.; et al. Comprehensive bronchoalveolar lavage characterization in COVID-19 associated acute respiratory distress syndrome patients: A prospective cohort study. Respir. Res. 2023, 24, 152. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]
- Bost, P.; De Sanctis, F.; Canè, S.; Ugel, S.; Donadello, K.; Castellucci, M.; Eyal, D.; Fiore, A.; Anselmi, C.; Barouni, R.M.; et al. Deciphering the state of immune silence in fatal COVID-19 patients. Nat. Commun. 2021, 12, 1428. [Google Scholar] [CrossRef]
- Pandolfi, L.; Fossali, T.; Frangipane, V.; Bozzini, S.; Morosini, M.; D’amato, M.; Lettieri, S.; Urtis, M.; Di Toro, A.; Saracino, L.; et al. Broncho-alveolar inflammation in COVID-19 patients: A correlation with clinical outcome. BMC Pulm. Med. 2020, 20, 301. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Liu, J.; Hu, J.; Yang, Y.; Zhou, Y. Analysis of Risk Factors for 24 Patients With COVID-19 Developing from Moderate to Severe Condition. Front. Cell. Infect. Microbiol. 2020, 10, 548582. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, G.; Long, X.; Hou, H.; Wei, J.; Cao, Y.; Tan, J.; Liu, W.; Meng, F.; Huang, L.; et al. Dynamics of Blood Viral Load Is Strongly Associated with Clinical Outcomes in Coronavirus Disease 2019 (COVID-19) Patients—A Prospective Cohort Study. J. Mol. Diagn. 2021, 23, 10–18. [Google Scholar] [CrossRef]
- MacCann, R.; Leon, A.A.G.; Gonzalez, G.; Carr, M.J.; Feeney, E.R.; Yousif, O.; Cotter, A.G.; de Barra, E.; Sadlier, C.; Doran, P.; et al. Dysregulated early transcriptional signatures linked to mast cell and interferon responses are implicated in COVID-19 severity. Front. Immunol. 2023, 14, 1166574. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.G.; Lorenc, A.; del Barrio, I.D.M.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.d.M.B.d.; Soares, C.P.; Monteiro, F.R.; Mello, R.; Amaral, J.B.D.; Aguiar, A.S.; Soledade, M.P.; Sucupira, C.; De Paulis, M.; Andrade, J.B.; et al. In Nasal Mucosal Secretions, Distinct IFN and IgA Responses Are Found in Severe and Mild SARS-CoV-2 Infection. Front. Immunol. 2021, 12, 595343. [Google Scholar] [CrossRef] [PubMed]
- Voiriot, G.; Dorgham, K.; Bachelot, G.; Fajac, A.; Morand-Joubert, L.; Parizot, C.; Gerotziafas, G.; Farabos, D.; Trugnan, G.; Eguether, T.; et al. Identification of bronchoalveolar and blood immune-inflammatory biomarker signature associated with poor 28-day outcome in critically ill COVID-19 patients. Sci. Rep. 2022, 12, 9502. [Google Scholar] [CrossRef]
- Joly, C.; Desjardins, D.; Porcher, R.; Péré, H.; Bruneau, T.; Zhang, Q.; Bastard, P.; Cobat, A.; Resmini, L.; Lenoir, O.; et al. More rapid blood interferon α2 decline in fatal versus surviving COVID-19 patients. Front. Immunol. 2023, 14, 1250214. [Google Scholar] [CrossRef]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.-S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef]
- Durstenfeld, M.S.; Hsue, P.Y.; Peluso, M.J.; Deeks, S.G. Findings from Mayo Clinic’s Post-COVID Clinic: PASC Phenotypes Vary by Sex and Degree of IL-6 Elevation. Mayo Clin. Proc. 2022, 97, 430–432. [Google Scholar] [CrossRef]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef]
- Talla, A.; Vasaikar, S.V.; Lemos, M.P.; Moodie, Z.; Lee, P.M.-P.; Henderson, K.E.; Cohen, K.W.; Czartoski, J.L.; Lai, L.; Suthar, M.S.; et al. Longitudinal immune dynamics of mild COVID-19 define signatures of recovery and persistence. bioRxiv 2021. [Google Scholar] [CrossRef]
- Torres-Ruiz, J.; Lomelín-Gascón, J.; Luna, J.L.; Castro, A.S.V.; Pérez-Fragoso, A.; Nuñez-Aguirre, M.; Alcalá-Carmona, B.; Absalón-Aguilar, A.; Balderas-Miranda, J.T.; Maravillas-Montero, J.L.; et al. Novel clinical and immunological features associated with persistent post-acute sequelae of COVID-19 after six months of follow-up: A pilot study. Infect. Dis. 2023, 55, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Kovarik, J.J.; Bileck, A.; Hagn, G.; Meier-Menches, S.M.; Frey, T.; Kaempf, A.; Hollenstein, M.; Shoumariyeh, T.; Skos, L.; Reiter, B.; et al. A multi-omics based anti-inflammatory immune signature characterizes long COVID-19 syndrome. iScience 2023, 26, 105717. [Google Scholar] [CrossRef]
- Peluso, M.J.; Lu, S.; Tang, A.F.; Durstenfeld, M.S.; Ho, H.-E.; Goldberg, S.A.; Forman, C.A.; E Munter, S.; Hoh, R.; Tai, V.; et al. Markers of Immune Activation and Inflammation in Individuals with Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2021, 224, 1839–1848. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; das Neves, P.F.M.; Lima, S.S.; Lopes, J.d.C.; Torres, M.K.d.S.; Vallinoto, I.M.V.C.; de Brito, M.T.F.M.; da Silva, A.L.S.; Leite, M.d.M.; da Costa, F.P.; et al. Cytokine Profiles Associated with Acute COVID-19 and Long COVID-19 Syndrome. Front. Cell. Infect. Microbiol. 2022, 12, 922422. [Google Scholar] [CrossRef]
- Pretorius, E.; Vlok, M.; Venter, C.; Bezuidenhout, J.A.; Laubscher, G.J.; Steenkamp, J.; Kell, D.B. Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 2021, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Pontelli, M.C.; Castro, Í.A.; Martins, R.B.; La Serra, L.; Veras, F.P.; Nascimento, D.C.; Silva, C.M.; Cardoso, R.S.; Rosales, R.; Gomes, R.; et al. SARS-CoV-2 productively infects primary human immune system cells in vitro and in COVID-19 patients. J. Mol. Cell Biol. 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Leon, J.; Michelson, D.A.; Olejnik, J.; Chowdhary, K.; Oh, H.S.; Hume, A.J.; Galvan-Pena, S.; Zhu, Y.; Chen, F.; Vijaykumar, B.; et al. A virus-specific monocyte inflammatory phenotype is induced by SARS-CoV-2 at the immune-epithelial interface. Proc. Natl. Acad. Sci. USA 2022, 119, e2116853118. [Google Scholar] [CrossRef]
- de Dios-Figueroa, G.T.; Aguilera-Marquez, J.d.R.; Camacho-Villegas, T.A.; Lugo-Fabres, P.H. 3D Cell Culture Models in COVID-19 Times: A Review of 3D Technologies to Understand and Accelerate Therapeutic Drug Discovery. Biomedicines 2021, 9, 602. [Google Scholar] [CrossRef]
- Clevers, H. COVID-19: Organoids go viral. Nat. Rev. Mol. Cell Biol. 2020, 21, 355–356. [Google Scholar] [CrossRef]
- Marshall, L.J.; Bailey, J.; Cassotta, M.; Herrmann, K.; Pistollato, F. Poor Translatability of Biomedical Research Using Animals—A Narrative Review. Altern. Lab. Anim. 2023, 51, 102–135. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Yuen, K.-Y. Animal models in SARS-CoV-2 research. Nat. Methods 2022, 19, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wu, Y.; Rui, X.; Yang, Y.; Ling, C.; Liu, S.; Liu, S.; Wang, Y. Animal models for COVID-19: Advances, gaps and perspectives. Signal Transduct. Target. Ther. 2022, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Jansen, E.B.; Orvold, S.N.; Swan, C.L.; Yourkowski, A.; Thivierge, B.M.; Francis, M.E.; Ge, A.; Rioux, M.; Darbellay, J.; Howland, J.G.; et al. After the virus has cleared—Can preclinical models be employed for Long COVID research? PLoS Pathog. 2022, 18. [Google Scholar] [CrossRef] [PubMed]
- Rosenke, K.; Okumura, A.; Lewis, M.C.; Feldmann, F.; Meade-White, K.; Bohler, W.F.; Griffin, A.J.; Rosenke, R.; Shaia, C.; Jarvis, M.A.; et al. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. J. Clin. Investig. 2022, 7, e160108. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Jochmans, D.; Vangeel, L.; De Jonghe, S.; Augustijns, P.; Mols, R.; Weynand, B.; Wattanakul, T.; Hoglund, R.M.; et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat. Commun. 2022, 13, 719. [Google Scholar] [CrossRef]
- Jeong, J.H.; Chokkakula, S.; Min, S.C.; Kim, B.K.; Choi, W.-S.; Oh, S.; Yun, Y.S.; Kang, D.H.; Lee, O.-J.; Kim, E.-G.; et al. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antivir. Res. 2022, 208, 105430. [Google Scholar] [CrossRef]
- Frere, J.J.; Serafini, R.A.; Pryce, K.D.; Zazhytska, M.; Oishi, K.; Golynker, I.; Panis, M.; Zimering, J.; Horiuchi, S.; Hoagland, D.A.; et al. SARS-CoV-2 Infection in Hamsters and Humans Results in Lasting and Unique Systemic Perturbations after Recovery. Sci. Translational Med. 2022, 14, eabq3059. [Google Scholar] [CrossRef]
- Yinda, C.K.; Port, J.R.; Bushmaker, T.; Offei Owusu, I.; Purushotham, J.N.; Avanzato, V.A.; Fischer, R.J.; Schulz, J.E.; Holbrook, M.G.; Hebner, M.J.; et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021, 17, e1009195. [Google Scholar] [CrossRef]
- Gressett, T.E.; Leist, S.R.; Ismael, S.; Talkington, G.; Dinnon, K.H.; Baric, R.S.; Bix, G. Mouse Adapted SARS-CoV-2 Model Induces “Long-COVID” Neuropathology in BALB/c Mice. bioRxiv 2023. [Google Scholar] [CrossRef]
- Handley, A.; Ryan, K.A.; Davies, E.R.; Bewley, K.R.; Carnell, O.T.; Challis, A.; Coombes, N.S.; Fotheringham, S.A.; Gooch, K.E.; Charlton, M.; et al. SARS-CoV-2 Disease Severity in the Golden Syrian Hamster Model of Infection Is Related to the Volume of Intranasal Inoculum. Viruses 2023, 15, 748. [Google Scholar] [CrossRef] [PubMed]
- Reyna, R.A.; Kishimoto-Urata, M.; Urata, S.; Makishima, T.; Paessler, S.; Maruyama, J. Recovery of anosmia in hamsters infected with SARS-CoV-2 is correlated with repair of the olfactory epithelium. Sci. Rep. 2022, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Bertzbach, L.D.; Dietert, K.; Abdelgawad, A.; Vladimirova, D.; Kunec, D.; Hoffmann, D.; Beer, M.; Gruber, A.D.; Trimpert, J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses 2020, 12, 779. [Google Scholar] [CrossRef] [PubMed]
- Briand, F.; Sencio, V.; Robil, C.; Heumel, S.; Deruyter, L.; Machelart, A.; Barthelemy, J.; Bogard, G.; Hoffmann, E.; Infanti, F.; et al. Diet-Induced Obesity and NASH Impair Disease Recovery in SARS-CoV-2-Infected Golden Hamsters. Viruses 2022, 14, 2067. [Google Scholar] [CrossRef]
- van de Ven, K.; van Dijken, H.; Wijsman, L.; Gomersbach, A.; Schouten, T.; Kool, J.; Lenz, S.; Roholl, P.; Meijer, A.; van Kasteren, P.B.; et al. Pathology and Immunity After SARS-CoV-2 Infection in Male Ferrets Is Affected by Age and Inoculation Route. Front. Immunol. 2021, 12, 750229. [Google Scholar] [CrossRef]
| Term | Description |
|---|---|
| PASC or Long COVID | WHO definition of PASC is adopted for this review. This defines PASC as the continuation or development of new symptoms 3 months after the initial SARS-CoV-2 infection, with these symptoms lasting for at least 2 months with no other explanation. |
| Acute phases spectrum | The phases, asymptomatic, mild, moderate, severe and critical are most often used. However, in this review, the spectrum includes asymptomatic, mild/moderate and severe phase. Rationale for this deviation is that is the review focuses on the differences and progression from non-severe phases (asymptomatic, mild/moderate) to the severe phases. |
| Asymptomatic | Individuals positive for COVID-19 in absence of infection related symptoms. |
| Mild/Moderate | Individuals who display mild symptoms including fever, muscle pain and anosmia and moderate symptoms including tachypnea and mild pneumonia without the need for hospitalization. |
| Severe | Hospitalized patients with clinical signs of pneumonia, severe tachypnea, severe dyspnea and critical symptoms including respiratory and organ failure and coma. |
| Cytokine storm | State of deregulated immune system, characterized by an excessive production and release of cytokines and chemokines. Release may cause damage to various tissues including lung. Cytokine storm may subsequently trigger acute respiratory stress syndrome leading to organ failure or even death. |
| Viral load | Commonly quantitatively expressed as concentration in viral copies/mL in plasma. Here, viral load magnitude is defined by the copy number or Ct value determined via (RT-q)PCR. This definition is widely used in the papers cited in this review and is therefore adopted here. |
| Viremia | Classically defined as the presence of (non-) infectious whole virus or immune neutralized whole virus in serum. Publications referred to here do not use viral culture or staining to determine presence of virus in serum. In turn, the majority uses (RT-q)PCR and adopt the Ct value or quantified copy number as a benchmark for viremia. Therefore, in case the review refers to viremia or viral load, remember that these are Ct values representing RNA copy numbers. |
| Ct value | Cycle threshold (Ct) value in PCR is the number of cycles needed to indicate that a sample contains the target RNA or DNA. The Ct value negatively correlates with the initial amount of target RNA or DNA, i.e., in this review a low Ct value indicates a high viral load. |
| Persistent infection | Prolonged presence of SARS-CoV-2 (non-)infectious virus or RNA remnants that is detected for an extended period beyond acute infection. Persistence could be associated with display of symptoms or in absence of symptoms. |
| URT & LRT | Upper respiratory tract (URT) that includes nasal cavity, pharynx and larynx. Lower respiratory tract (LRT) that includes trachea and lungs. |
| RNA Load | Copy number of SARS-CoV-2 RNA, detected by (RT-q)PCR on samples from various origin including serum, BAL, nasopharyngeal swabs. |
| Inoculum | The virus quantity or concentration introduced at the time of host infection. |
| Study Population Characteristics | Acute-Phase Viral Load Correlation or Association with PASC Phase | Ref. |
|---|---|---|
| Hospitalized (n = 47) Vaccinated Strain variant unknown | Plasma of PASC patients tested positive more frequently (55% vs. 29%) and had higher average viral loads compared to previously infected persons without PASC. In PASC-positive patients, plasma viral load remained unchanged or increased, while in PASC-negative individuals, it decreased or became undetectable over time. | [40] |
| Mix of mild-to-critical patients. Exact numbers unknown. Unvaccinated * Wuhan-Hu-1 strain | Detectable plasma viral load was associated with memory-related issues, whereas there were no associations with anosmia or fatigue. | [41] |
| Hospitalized (n = 129) Severe/Critical (≥90% pneumonia) | Viremia at time of hospitalization resulted in a higher chance of PASC symptom development as compared to hospital-admitted patients without viremia. | [42] |
| Mild/moderate (n = 119) Severe/Critical (n = 8) Unvaccinated * Wuhan-Hu-1 strain ** | Patients with detectable plasma viral load at enrollment were more likely to report PASC symptoms one month after confirmed infection as compared to individuals without detectable plasma viral loads (83% vs. 41%). | [43] |
| Mild/moderate (n = 70) Severe/Critical (n = 6) Vaccination status *** Strain variant unknown | Viral URT loads correlated with an increased number of experienced PASC symptoms. | [44] |
| Study Population Characteristics * | Viral Load | Cytokine | Ref. | |
|---|---|---|---|---|
| Origin | Origin | Correlation with *** | ||
| Non-stratified ** Mild/Moderate (n = 26) Severe/critical (n = 11) | URT | Plasma | IL-6, IL-10 | [76] |
| TNF-α, INF-γ | ||||
| Non-stratified ** Asymptomatic (n = 6) Mild/Moderate (n = 17) Severe/critical (n = 8) | URT | Plasma | CCL-2 | [77] |
| IFN-α, IFN-γ, TNF-α, IL-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, IL-17A | ||||
| Non-stratified ** Asymptomatic (n = 4) Mild/Moderate (n = 58) Severe/critical (n = 8) | URT | Plasma | CCL-2, VEGF, G-CSF | [82] |
| IL-1β, IL-1ra, IL-2, IL-2Rα, IL-6, IL-7, IL-8, IL-9, IL-10, IL-13, IL-15, IL-17, IL-18, IFN-α2, IFN-γ, TNF-α Whole list; see reference | ||||
| Hospitalized Severe/critical (n = 88) | Plasma | Plasma | IL-6, IL-8, IP10, CCL-2 | [32] |
| IFN-γ, IL-1RA | ||||
| URT | Plasma | IL-6, IL-8, IP10, CCL-2, IFN-γ | ||
| IL-1RA | ||||
| LRT | Plasma | IL-6 | ||
| IL-8, IP10, CCL-2, IFN-γ, IL-1RA | ||||
| Non-stratified ** Mild/moderate (n = 15) Severe/critical (n = 34) | Plasma | Plasma | IL-6, CCL-2, CCL-19 | [74] |
| IFN-α2 | ||||
| URT | URT | IFN-γ, IL-33 | ||
| IL-10 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Compeer, B.; Neijzen, T.R.; van Lelyveld, S.F.L.; Martina, B.E.E.; Russell, C.A.; Goeijenbier, M. Uncovering the Contrasts and Connections in PASC: Viral Load and Cytokine Signatures in Acute COVID-19 versus Post-Acute Sequelae of SARS-CoV-2 (PASC). Biomedicines 2024, 12, 1941. https://doi.org/10.3390/biomedicines12091941
Compeer B, Neijzen TR, van Lelyveld SFL, Martina BEE, Russell CA, Goeijenbier M. Uncovering the Contrasts and Connections in PASC: Viral Load and Cytokine Signatures in Acute COVID-19 versus Post-Acute Sequelae of SARS-CoV-2 (PASC). Biomedicines. 2024; 12(9):1941. https://doi.org/10.3390/biomedicines12091941
Chicago/Turabian StyleCompeer, Brandon, Tobias R. Neijzen, Steven F. L. van Lelyveld, Byron E. E. Martina, Colin A. Russell, and Marco Goeijenbier. 2024. "Uncovering the Contrasts and Connections in PASC: Viral Load and Cytokine Signatures in Acute COVID-19 versus Post-Acute Sequelae of SARS-CoV-2 (PASC)" Biomedicines 12, no. 9: 1941. https://doi.org/10.3390/biomedicines12091941
APA StyleCompeer, B., Neijzen, T. R., van Lelyveld, S. F. L., Martina, B. E. E., Russell, C. A., & Goeijenbier, M. (2024). Uncovering the Contrasts and Connections in PASC: Viral Load and Cytokine Signatures in Acute COVID-19 versus Post-Acute Sequelae of SARS-CoV-2 (PASC). Biomedicines, 12(9), 1941. https://doi.org/10.3390/biomedicines12091941






