Causal Effects of 25-Hydroxyvitamin D on Metabolic Syndrome and Metabolic Risk Traits: A Bidirectional Two-Sample Mendelian Randomization Study
Abstract
1. Introduction
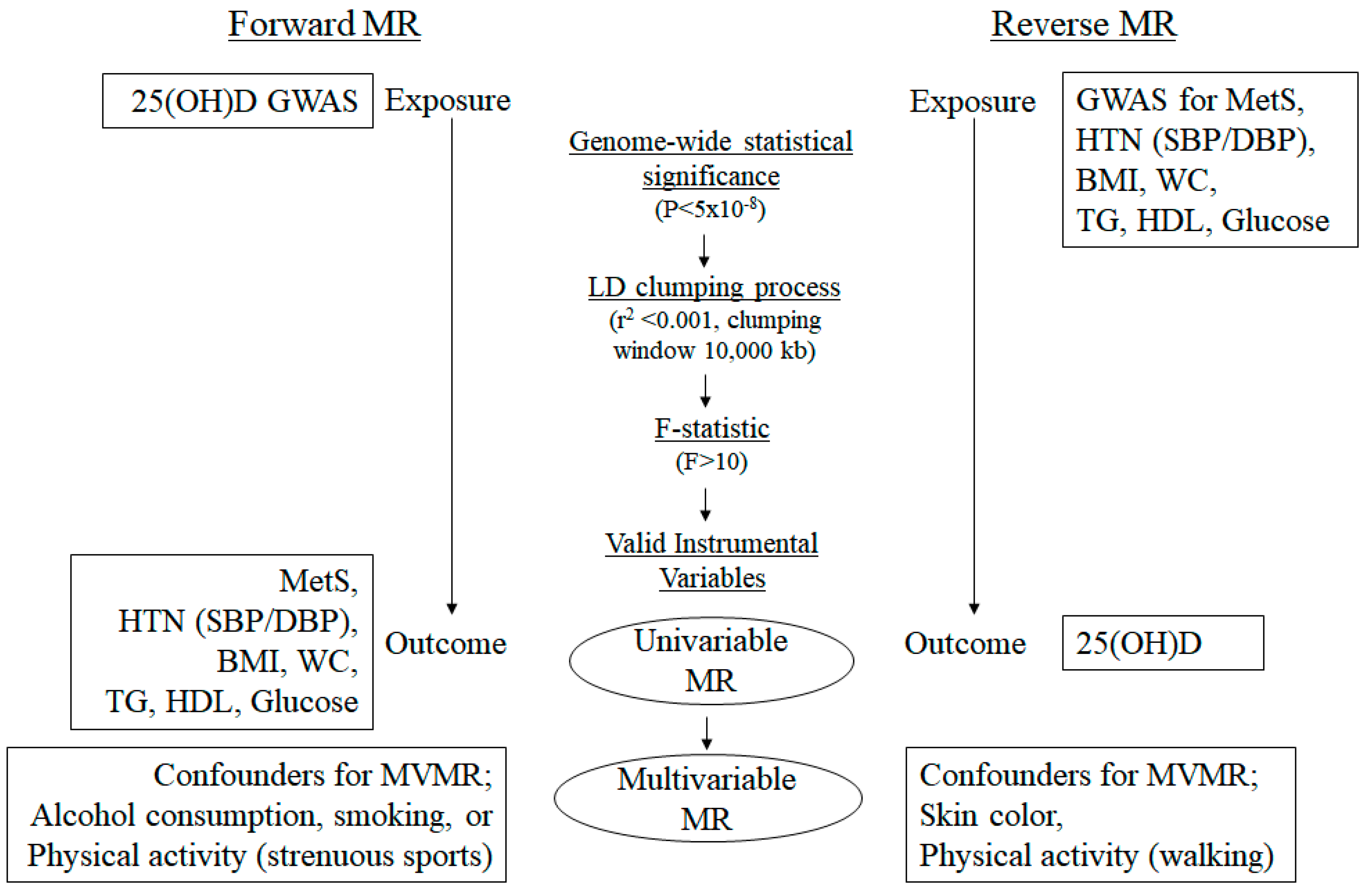
2. Materials and Methods
2.1. Data Resources
2.2. Selection of SNPs as IVs
2.3. MR Study
3. Results
3.1. Genetic IVs in Univariable MR
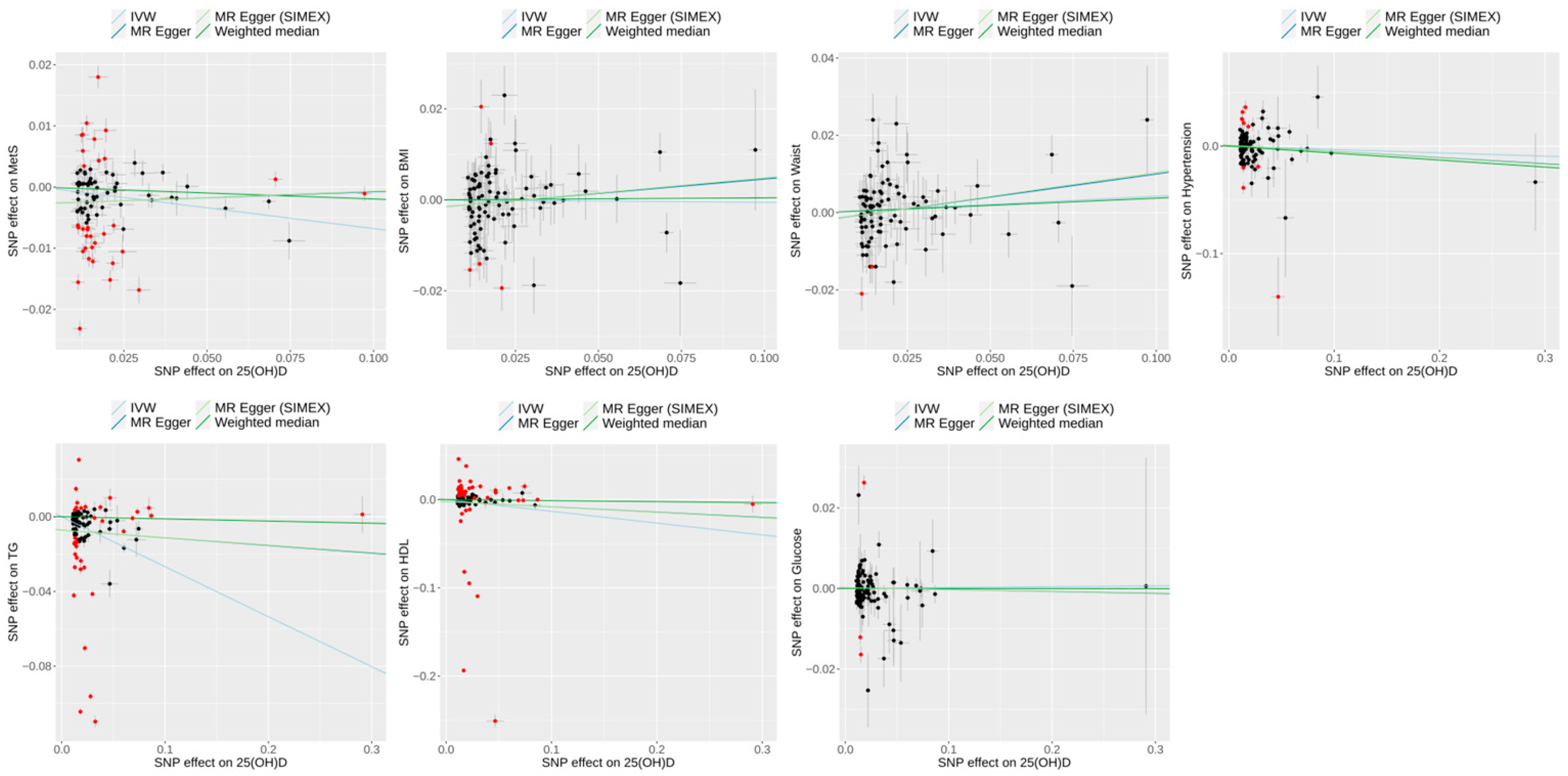
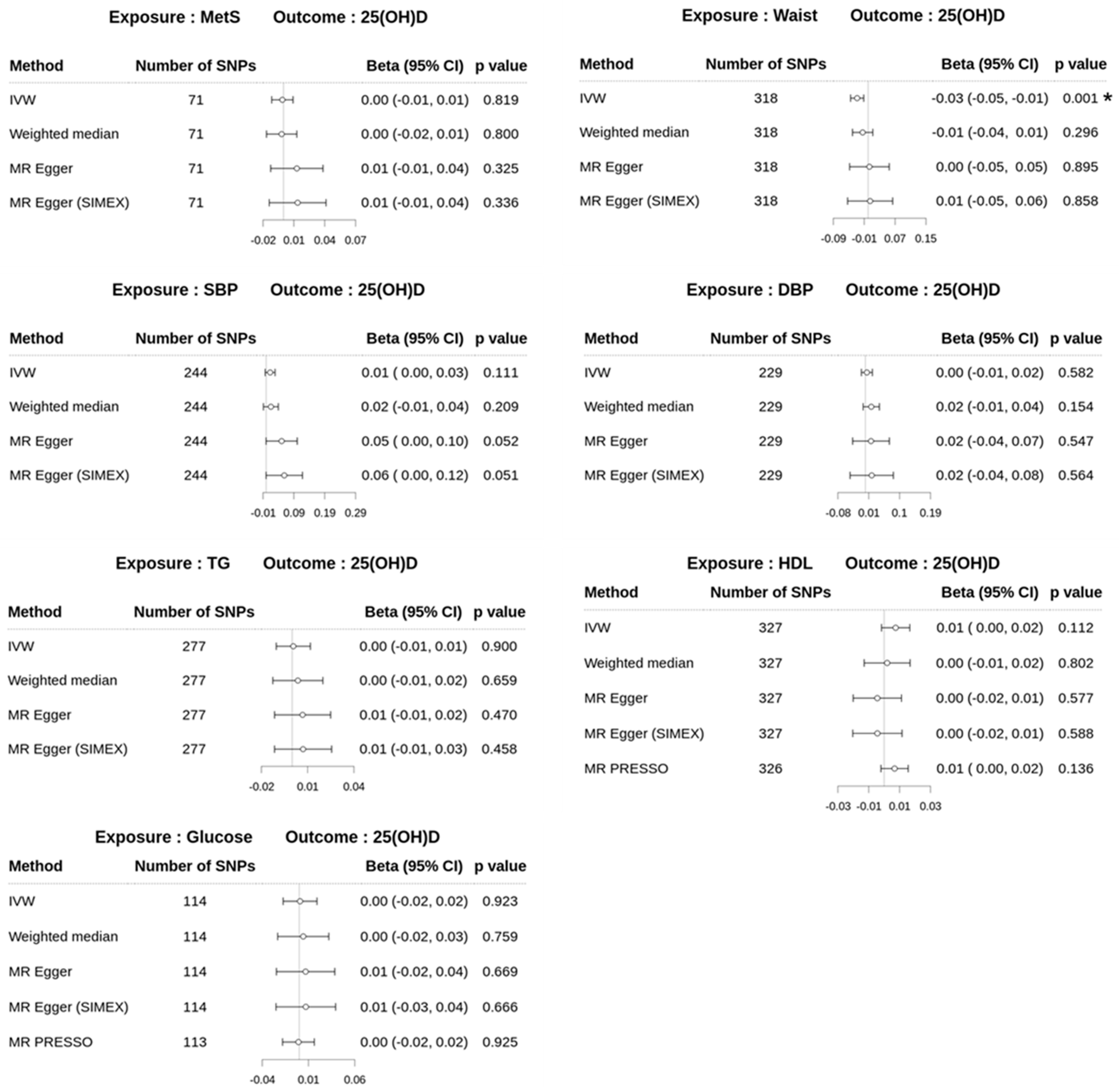
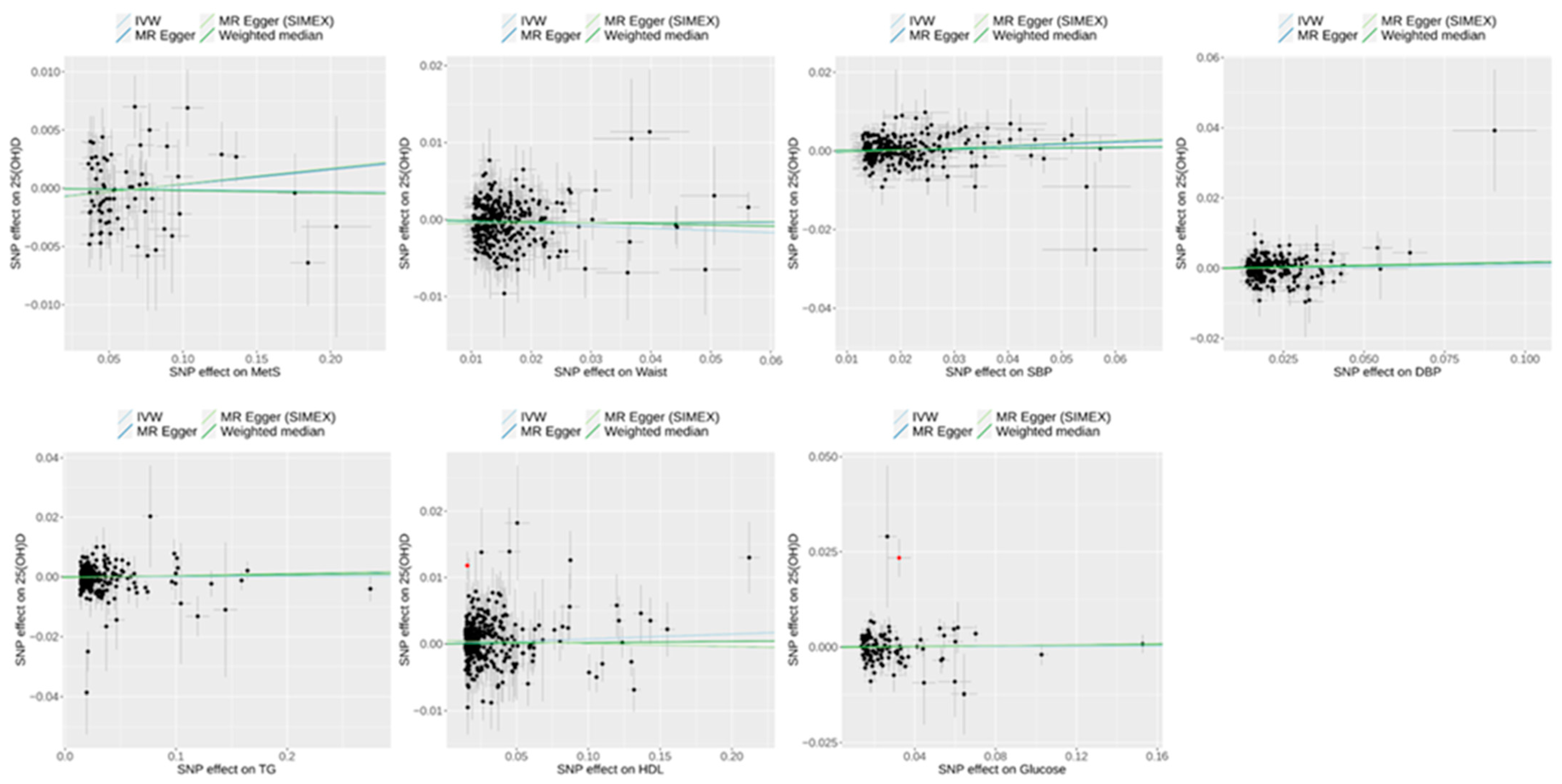
3.2. Heterogeneity and Pleiotropy Tests for IVs in Univariable MR
3.3. Effect of 25(OH)D on Metabolic Syndrome and Its Risk Factors in Univariable MR
3.4. Effect of Metabolic Syndrome on 25(OH)D Levels on Univariable MR
3.5. Multivariable MR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamali, Z.; Ayoobi, F.; Jalali, Z.; Bidaki, R.; Lotfi, M.A.; Esmaeili-Nadimi, A.; Khalili, P. Metabolic syndrome: A population-based study of prevalence and risk factors. Sci. Rep. 2024, 14, 3987. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Pinhas-Hamiel, O.; Levek-Motola, N.; Kaidar, K.; Boyko, V.; Tisch, E.; Mazor-Aronovitch, K.; Graf-Barel, C.; Landau, Z.; Lerner-Geva, L.; Frumkin Ben-David, R. Prevalence of overweight, obesity and metabolic syndrome components in children, adolescents and young adults with type 1 diabetes mellitus. Diabetes Metab. Res. Rev. 2015, 31, 76–84. [Google Scholar] [CrossRef]
- Onyenekwu, C.P.; Dada, A.O.; Babatunde, O.T. The prevalence of metabolic syndrome and its components among overweight and obese Nigerian adolescents and young adults. Niger. J. Clin. Pract. 2017, 20, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Nolan, P.B.; Carrick-Ranson, G.; Stinear, J.W.; Reading, S.A.; Dalleck, L.C. Prevalence of metabolic syndrome and metabolic syndrome components in young adults: A pooled analysis. Prev. Med. Rep. 2017, 7, 211–215. [Google Scholar] [CrossRef]
- Dobrowolski, P.; Prejbisz, A.; Kurylowicz, A.; Baska, A.; Burchardt, P.; Chlebus, K.; Dzida, G.; Jankowski, P.; Jaroszewicz, J.; Jaworski, P.; et al. Metabolic syndrome—A new definition and management guidelines: A joint position paper by the Polish Society of Hypertension, Polish Society for the Treatment of Obesity, Polish Lipid Association, Polish Association for Study of Liver, Polish Society of Family Medicine, Polish Society of Lifestyle Medicine, Division of Prevention and Epidemiology Polish Cardiac Society, “Club 30” Polish Cardiac Society, and Division of Metabolic and Bariatric Surgery Society of Polish Surgeons. Arch. Med. Sci. 2022, 18, 1133–1156. [Google Scholar] [CrossRef] [PubMed]
- Nazara Otero, C.A.; Pose Reino, A.; Pena Gonzalez, E. Metabolic syndrome: Diagnosis and management. An update. Clin. Investig. Arterioscler. 2016, 28, 230–231. [Google Scholar] [CrossRef]
- Huang, P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009, 2, 231–237. [Google Scholar] [CrossRef]
- American Heart Association; National Heart, Lung; Blood Institute; Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; et al. Diagnosis and management of the metabolic syndrome. An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Executive summary. Cardiol. Rev. 2005, 13, 322–327. [Google Scholar] [CrossRef]
- Expert Panel on Detection, Evaluation; Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef]
- Brown, W.V. Metabolic syndrome and risk of stroke. Clin. Cornerstone 2004, 6 (Suppl. 1), S30–S34. [Google Scholar] [CrossRef] [PubMed]
- Riaz, A.; Asghar, S.; Shahid, S.; Tanvir, H.; Ejaz, M.H.; Akram, M. Prevalence of Metabolic Syndrome and Its Risk Factors Influence on Microvascular Complications in Patients With Type 1 and Type 2 Diabetes Mellitus. Cureus 2024, 16, e55478. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ding, Y.; Tanaka, Y.; Zhang, W. Risk factors contributing to type 2 diabetes and recent advances in the treatment and prevention. Int. J. Med. Sci. 2014, 11, 1185–1200. [Google Scholar] [CrossRef]
- Collaborators, G.B.D.O.; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef]
- Liang, Z.; Wang, Z.; Liu, X.; He, Y. Confronting the global obesity epidemic: Investigating the role and underlying mechanisms of vitamin D in metabolic syndrome management. Front. Nutr. 2024, 11, 1416344. [Google Scholar] [CrossRef]
- Castillo, J.J.; Hazlett, Z.S.; Orlando, R.A.; Garver, W.S. A global evolutionary and metabolic analysis of human obesity gene risk variants. Gene 2017, 627, 412–419. [Google Scholar] [CrossRef]
- Padwal, R.S. Obesity, diabetes, and the metabolic syndrome: The global scourge. Can. J. Cardiol. 2014, 30, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Wranicz, J.; Szostak-Wegierek, D. Health outcomes of vitamin D. Part I. characteristics and classic role. Rocz. Panstw. Zakl. Hig. 2014, 65, 179–184. [Google Scholar]
- Sartini, M.; Del Puente, F.; Oliva, M.; Carbone, A.; Bobbio, N.; Schinca, E.; Giribone, L.; Cristina, M.L. Preventive Vitamin D Supplementation and Risk for COVID-19 Infection: A Systematic Review and Meta-Analysis. Nutrients 2024, 16, 679. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Meng, X. Vitamin D and neurodegenerative diseases. Heliyon 2023, 9, e12877. [Google Scholar] [CrossRef]
- Soda, M.; Priante, C.; Pesce, C.; De Maio, G.; Lombardo, M. The Impact of Vitamin D on Immune Function and Its Role in Hashimoto’s Thyroiditis: A Narrative Review. Life 2024, 14, 771. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Jin, M.H.; Yoon, J.H. The contribution of vitamin D insufficiency to the onset of steatotic liver disease among individuals with metabolic dysfunction. Sci. Rep. 2024, 14, 6714. [Google Scholar] [CrossRef]
- Park, J.E.; Pichiah, P.B.T.; Cha, Y.S. Vitamin D and Metabolic Diseases: Growing Roles of Vitamin D. J. Obes. Metab. Syndr. 2018, 27, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Khayyatzadeh, S.S.; Mirmoosavi, S.J.; Fazeli, M.; Abasalti, Z.; Avan, A.; Javandoost, A.; Rahmani, F.; Tayefi, M.; Hanachi, P.; Ferns, G.A.; et al. High-dose vitamin D supplementation is associated with an improvement in several cardio-metabolic risk factors in adolescent girls: A nine-week follow-up study. Ann. Clin. Biochem. 2018, 55, 227–235. [Google Scholar] [CrossRef]
- Migliaccio, S.; Di Nisio, A.; Magno, S.; Romano, F.; Barrea, L.; Colao, A.M.; Muscogiuri, G.; Savastano, S. Vitamin D deficiency: A potential risk factor for cancer in obesity? Int. J. Obes. 2022, 46, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Manoppo, J.I.C.; Pateda, V.; Prayogo, C.; Langi, F.; Nurkolis, F.; Tsopmo, A. Relationships of 25-hydroxyvitamin D levels and non-alcoholic fatty liver disease in obese children: A possible strategy to promote early screening of NAFLD. Front. Nutr. 2022, 9, 1025396. [Google Scholar] [CrossRef]
- Wang, N.; Wang, C.; Chen, X.; Wan, H.; Chen, Y.; Chen, C.; Han, B.; Lu, Y. Vitamin D, prediabetes and type 2 diabetes: Bidirectional Mendelian randomization analysis. Eur. J. Nutr. 2020, 59, 1379–1388. [Google Scholar] [CrossRef]
- Ganji, V.; Sukik, A.; Alaayesh, H.; Rasoulinejad, H.; Shraim, M. Serum vitamin D concentrations are inversely related to prevalence of metabolic syndrome in Qatari women. Biofactors 2020, 46, 180–186. [Google Scholar] [CrossRef]
- Dekkers, O.M.; Vandenbroucke, J.P.; Cevallos, M.; Renehan, A.G.; Altman, D.G.; Egger, M. COSMOS-E: Guidance on conducting systematic reviews and meta-analyses of observational studies of etiology. PLoS Med. 2019, 16, e1002742. [Google Scholar] [CrossRef]
- Greenland, S. An introduction To instrumental variables for epidemiologists. Int. J. Epidemiol. 2000, 29, 1102. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Davey Smith, G.; Kundu, D.; Bruckdorfer, K.R.; Ebrahim, S. Those confounded vitamins: What can we learn from the differences between observational versus randomised trial evidence? Lancet 2004, 363, 1724–1727. [Google Scholar] [CrossRef] [PubMed]
- Davey Smith, G. Capitalizing on Mendelian randomization to assess the effects of treatments. J. R. Soc. Med. 2007, 100, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Howe, L.J.; Brumpton, B.; Havdahl, A.; Evans, D.M.; Davey Smith, G. Within family Mendelian randomization studies. Hum. Mol. Genet. 2019, 28, R170–R179. [Google Scholar] [CrossRef]
- Matsuyama, Y.; Tabuchi, T. Does Tobacco Smoking Increase Social Isolation? A Mendelian Randomization Study. Am. J. Epidemiol. 2024, 193, 626–635. [Google Scholar] [CrossRef]
- Harrison, S.; Davies, A.R.; Dickson, M.; Tyrrell, J.; Green, M.J.; Katikireddi, S.V.; Campbell, D.; Munafo, M.; Dixon, P.; Jones, H.E.; et al. The causal effects of health conditions and risk factors on social and socioeconomic outcomes: Mendelian randomization in UK Biobank. Int. J. Epidemiol. 2020, 49, 1661–1681. [Google Scholar] [CrossRef]
- Howe, L.D.; Kanayalal, R.; Harrison, S.; Beaumont, R.N.; Davies, A.R.; Frayling, T.M.; Davies, N.M.; Hughes, A.; Jones, S.E.; Sassi, F.; et al. Effects of body mass index on relationship status, social contact and socio-economic position: Mendelian randomization and within-sibling study in UK Biobank. Int. J. Epidemiol. 2020, 49, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Davey Smith, G.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Kutalik, Z.; Holmes, M.V.; Minelli, C.; et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 2019, 4, 186. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ 2021, 375, n2233. [Google Scholar] [CrossRef]
- Lind, L. Genome-Wide Association Study of the Metabolic Syndrome in UK Biobank. Metab. Syndr. Relat. Disord. 2019, 17, 505–511. [Google Scholar] [CrossRef]
- Jiang, X.; O’Reilly, P.F.; Aschard, H.; Hsu, Y.H.; Richards, J.B.; Dupuis, J.; Ingelsson, E.; Karasik, D.; Pilz, S.; Berry, D.; et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat. Commun. 2018, 9, 260. [Google Scholar] [CrossRef]
- Manousaki, D.; Mitchell, R.; Dudding, T.; Haworth, S.; Harroud, A.; Forgetta, V.; Shah, R.L.; Luan, J.; Langenberg, C.; Timpson, N.J.; et al. Genome-wide Association Study for Vitamin D Levels Reveals 69 Independent Loci. Am. J. Hum. Genet. 2020, 106, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, S.; Kim, B.; Kim, D.S.; Kim, J.; Ahn, Y.; Kim, H.; Song, M.; Shim, I.; Jung, S.H.; et al. Multivariate genomic analysis of 5 million people elucidates the genetic architecture of shared components of the metabolic syndrome. Nat. Genet. 2024, 56, 2380–2391. [Google Scholar] [CrossRef] [PubMed]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Magi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.E.; Clarke, S.L.; Wu, K.H.; Kanoni, S.; Zajac, G.J.M.; Ramdas, S.; Surakka, I.; Ntalla, I.; Vedantam, S.; Winkler, T.W.; et al. Author Correction: The power of genetic diversity in genome-wide association studies of lipids. Nature 2023, 618, E19–E20. [Google Scholar] [CrossRef]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef]
- Pierce, B.L.; Ahsan, H.; Vanderweele, T.J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 2011, 40, 740–752. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G.; Collaboration, C.C.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Improving bias and coverage in instrumental variable analysis with weak instruments for continuous and binary outcomes. Stat. Med. 2012, 31, 1582–1600. [Google Scholar] [CrossRef]
- Sanderson, E.; Spiller, W.; Bowden, J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat. Med. 2021, 40, 5434–5452. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Lee, S.; Won, S. Causal Evaluation of Laboratory Markers in Type 2 Diabetes on Cancer and Vascular Diseases Using Various Mendelian Randomization Tools. Front. Genet. 2020, 11, 597420. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Slob, E.A.W.; Groenen, P.J.F.; Thurik, A.R.; Rietveld, C.A. A note on the use of Egger regression in Mendelian randomization studies. Int. J. Epidemiol. 2017, 46, 2094–2097. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.R.; ReproGen, C.; Psychiatric Genomics, C.; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium; Duncan, L.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef]
- Zheng, J.; Richardson, T.G.; Millard, L.A.C.; Hemani, G.; Elsworth, B.L.; Raistrick, C.A.; Vilhjalmsson, B.; Neale, B.M.; Haycock, P.C.; Smith, G.D.; et al. PhenoSpD: An integrated toolkit for phenotypic correlation estimation and multiple testing correction using GWAS summary statistics. Gigascience 2018, 7, giy090. [Google Scholar] [CrossRef]
- Schmitt, E.B.; Nahas-Neto, J.; Bueloni-Dias, F.; Poloni, P.F.; Orsatti, C.L.; Petri Nahas, E.A. Vitamin D deficiency is associated with metabolic syndrome in postmenopausal women. Maturitas 2018, 107, 97–102. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, E.Y.; Lee, J.H.; Kim, J.E.; Kim, K.J.; Rhee, Y.; Kim, H.C.; Youm, Y.; Kim, C.O. Associations of serum 25-hydroxyvitamin D with metabolic syndrome and its components in elderly men and women: The Korean Urban Rural Elderly cohort study. BMC Geriatr. 2019, 19, 102. [Google Scholar] [CrossRef]
- Huang, C.Y.; Chang, H.H.; Lu, C.W.; Tseng, F.Y.; Lee, L.T.; Huang, K.C. Vitamin D status and risk of metabolic syndrome among non-diabetic young adults. Clin. Nutr. 2015, 34, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Heil, D.P. Associations of vitamin D status with markers of metabolic health: A community-based study in Shanghai, China. Diabetes Metab. Syndr. 2018, 12, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, A.; Wierzbicka, J.; Zmijewski, M.A. Vitamin D in the skin physiology and pathology. Acta Biochim. Pol. 2016, 63, 17–29. [Google Scholar] [CrossRef]
- Carlberg, C. The physiology of vitamin D-far more than calcium and bone. Front. Physiol. 2014, 5, 335. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, R. Vitamin D: Review of physiology and clinical uses. Minervamedic Endocrinol. 2023, 48, 88–105. [Google Scholar] [CrossRef]
- Khaledi, K.; Hoseini, R.; Gharzi, A. The impact of vitamin D on type 2 diabetes management: Boosting PTP1B gene expression and physical activity benefits in rats. Genes Nutr. 2024, 19, 4. [Google Scholar] [CrossRef]
- Mehri, Z.; Salehi-Abargouei, A.; Shahvazi, S.; Samadi, M.; Zare, F.; Nadjarzadeh, A. The association between vitamin D status and metabolic syndrome and its components among female teachers residing in Yazd city. Endocrinol. Diabetes Nutr. (Engl. Ed.) 2019, 66, 628–638. [Google Scholar] [CrossRef]
- Skaaby, T.; Husemoen, L.L.; Martinussen, T.; Thyssen, J.P.; Melgaard, M.; Thuesen, B.H.; Pisinger, C.; Jorgensen, T.; Johansen, J.D.; Menne, T.; et al. Vitamin D status, filaggrin genotype, and cardiovascular risk factors: A Mendelian randomization approach. PLoS ONE 2013, 8, e57647. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Y.; Weng, P.; Xia, F.; Li, Q.; Zhai, H.; Wang, N.; Lu, Y. Association of 25-hydroxyvitamin D with cardiometabolic risk factors and metabolic syndrome: A mendelian randomization study. Nutr. J. 2019, 18, 61. [Google Scholar] [CrossRef]
- Teixeira, J.S.; Bull Ferreira Campos, A.; Cordeiro, A.; Pereira, S.E.; Saboya, C.J.; Ramalho, A. Vitamin D nutritional status and its relationship with metabolic changes in adolescents and adults with severe obesity. Nutr. Hosp. 2018, 35, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.; Xiao, L.; Imayama, I.; Duggan, C.R.; Bain, C.; Foster-Schubert, K.E.; Kong, A.; Campbell, K.L.; Wang, C.Y.; Neuhouser, M.L.; et al. Effects of weight loss on serum vitamin D in postmenopausal women. Am. J. Clin. Nutr. 2011, 94, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Bertolini, L.; Scala, L.; Cigolini, M.; Zenari, L.; Falezza, G.; Arcaro, G. Associations between serum 25-hydroxyvitamin D3 concentrations and liver histology in patients with non-alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 517–524. [Google Scholar] [CrossRef]
- Xiao, J.; Lv, J.; Wang, S.; Zhou, Y.; Chen, L.; Lu, J.; Zhang, X.; Wang, X.; Gu, Y.; Lu, Q. Association of serum 25-hydroxyvitamin D with metabolic syndrome and type 2 diabetes: A one sample Mendelian randomization study. BMC Geriatr. 2021, 21, 391. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S.; Berry, D.J.; Lu, C.; Tikkanen, E.; Pilz, S.; Hiraki, L.T.; Cooper, J.D.; Dastani, Z.; Li, R.; Houston, D.K.; et al. Causal relationship between obesity and vitamin D status: Bi-directional Mendelian randomization analysis of multiple cohorts. PLoS Med. 2013, 10, e1001383. [Google Scholar] [CrossRef]
- Peterson, C.A.; Belenchia, A.M. Vitamin D deficiency & childhood obesity: A tale of two epidemics. Mo. Med. 2014, 111, 49–53. [Google Scholar]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Revez, J.A.; Lin, T.; Qiao, Z.; Xue, A.; Holtz, Y.; Zhu, Z.; Zeng, J.; Wang, H.; Sidorenko, J.; Kemper, K.E.; et al. Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat. Commun. 2020, 11, 1647. [Google Scholar] [CrossRef]
- Hiraki, L.T.; Major, J.M.; Chen, C.; Cornelis, M.C.; Hunter, D.J.; Rimm, E.B.; Simon, K.C.; Weinstein, S.J.; Purdue, M.P.; Yu, K.; et al. Exploring the genetic architecture of circulating 25-hydroxyvitamin D. Genet. Epidemiol. 2013, 37, 92–98. [Google Scholar] [CrossRef]
- Bouillon, R. Genetic and environmental determinants of vitamin D status. Lancet 2010, 376, 148–149. [Google Scholar] [CrossRef]
- Yao, S.; Hong, C.C.; Bandera, E.V.; Zhu, Q.; Liu, S.; Cheng, T.D.; Zirpoli, G.; Haddad, S.A.; Lunetta, K.L.; Ruiz-Narvaez, E.A.; et al. Demographic, lifestyle, and genetic determinants of circulating concentrations of 25-hydroxyvitamin D and vitamin D-binding protein in African American and European American women. Am. J. Clin. Nutr. 2017, 105, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, T.; Kaaks, R.; Teucher, B.; Hirche, F.; Dierkes, J.; Weikert, C.; Katzke, V.; Boeing, H.; Stangl, G.I.; Buijsse, B. Dietary, lifestyle, and genetic determinants of vitamin D status: A cross-sectional analysis from the European Prospective Investigation into Cancer and Nutrition (EPIC)-Germany study. Eur. J. Nutr. 2014, 53, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, D.A.; Hanifa, Y.; Witt, K.D.; Venton, T.R.; Rowe, M.; Timms, P.M.; Hypponen, E.; Walton, R.T.; Griffiths, C.J.; Martineau, A.R. Environmental and genetic determinants of vitamin D status among older adults in London, UK. J. Steroid. Biochem. Mol. Biol. 2016, 164, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Hendi, N.N.; Al-Sarraj, Y.; Ismail Umlai, U.K.; Suhre, K.; Nemer, G.; Albagha, O. Genetic determinants of Vitamin D deficiency in the Middle Eastern Qatari population: A genome-wide association study. Front. Nutr. 2023, 10, 1242257. [Google Scholar] [CrossRef]
- AlAnouti, F.; Abboud, M.; Papandreou, D.; Mahboub, N.; Haidar, S.; Rizk, R. Effects of Vitamin D Supplementation on Lipid Profile in Adults with the Metabolic Syndrome: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2020, 12, 3352. [Google Scholar] [CrossRef]
- Luxwolda, M.F.; Kuipers, R.S.; Kema, I.P.; Dijck-Brouwer, D.A.; Muskiet, F.A. Traditionally living populations in East Africa have a mean serum 25-hydroxyvitamin D concentration of 115 nmol/l. Br. J. Nutr. 2012, 108, 1557–1561. [Google Scholar] [CrossRef]
- Majumdar, V.; Nagaraja, D.; Christopher, R. Vitamin D status and metabolic syndrome in Asian Indians. Int. J. Obes. 2011, 35, 1131–1134. [Google Scholar] [CrossRef]
- Qi, K.J.; Zhao, Z.T.; Zhang, W.; Yang, F. The impacts of vitamin D supplementation in adults with metabolic syndrome: A systematic review and meta-analysis of randomized controlled trials. Front. Pharmacol. 2022, 13, 1033026. [Google Scholar] [CrossRef]



| Traits | Data Sources | No. of Participants | Population | No. of Variants | Reference |
|---|---|---|---|---|---|
| Dataset 1 | |||||
| 25(OH)D level | SUNLIGHT Consortium | 79,366 | European | 2,579,296 | PMID: [40] 29343764 |
| Metabolic syndrome | UK Biobank (UKB) | 291,107 | European | 9,463,307 | PMID: [39] 31589552 |
| Waist circumference | UKB | 419,807 | European | 23,861,814 | ǂ |
| TG | UKB | 400,639 | European | 23,861,718 | |
| HDL | UKB | 367,021 | European | 23,861,539 | |
| SBP | UKB | 396,663 | European | 23,861,710 | |
| DBP | UKB | 396,667 | European | 23,861,710 | |
| Glucose | UKB | 366,759 | European | 23,861,541 | |
| Dataset 2 | |||||
| 25(OH)D level | UKB + European GWAS | 443,734 (UKB: 401,460) | European | 16,668,957 | PMID: [41] 32059762 |
| Metabolic syndrome | (a) Multiple cohorts | 1,384,348 * | European | 2,265,555 | PMID: [42] 39349817 |
| Waist circumference | GIANT Consortium 2015 | 232,101 | European | 2,565,407 | PMID: [43] 25673412 |
| BMI | GIANT Consortium 2015 | 322,154 | European | 2,554,637 | PMID: [44] 25673413 |
| TG | GLGC Consortium | 864,240 | European | 37,005,452 | PMID: [45] 37237109 |
| HDL | GLGC Consortium | 888,227 | European | 36,588,494 | PMID: [45] 37237109 |
| Hypertension | FinnGen release 12 | 500,264 | European | 21,327,062 | † |
| Glucose | MAGIC Consortium | 200,622 | European | 34,064,006 | PMID: [46] 34059833 |
| Confounders, for reverse direction | |||||
| Skin color | UKB | 415,018 | European | 9,463,307 | ǂ |
| Physical activity (walking) | UKB | 418,278 | European | 23,088,387 | |
| Confounders, for forward direction | |||||
| Physical activity (strenuous sports) | UKB | 418,278 | European | 22,111,708 | ǂ |
| Alcohol consumption | UKB | 419,936 | European | 21,143,063 | |
| Smoking | UKB | 418,817 | European | 22,122,417 | |
| Exposure | Outcome | Heterogeneity | Horizontal Pleiotropy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cochran’s Q | Rücker’s Q’ | MR-PRESSO Global Test | MR–Egger | MR–Egger (SIMEX) | |||||||
| N | F | I2 (%) | p-Value | p-Value | p-Value | Intercept, β (SE) | p-Value | Intercept, β (SE) | p-Value | ||
| Dataset 1 (reverse direction) | |||||||||||
| Metabolic syndrome | 25(OH)D level | 71 | 65.21 | 89.27 | 0.044 | 0.049 | 0.042 | −0.0010 (0.0008) | 0.240 | −0.0010 (0.0009) | 0.252 |
| WC | 318 | 55.98 | 86.18 | 0.190 | 0.198 | 0.185 | −0.0005 (0.0004) | 0.198 | −0.0006 (0.0005) | 0.228 | |
| TG | 277 | 128.43 | 98.11 | <0.001 | <0.001 | <0.001 | −0.0002 (0.0003) | 0.415 | −0.0003 (0.0003) | 0.406 | |
| HDL | 327 | 127.80 | 97.72 | <0.001 | <0.001 | <0.001 | 0.0005 (0.0003) | 0.067 | 0.0005 (0.0003) | 0.072 | |
| SBP | 244 | 57.37 | 90.79 | 0.070 | 0.079 | 0.072 | −0.0008 (0.0005) | 0.128 | −0.0010 (0.0006) | 0.111 | |
| DBP | 229 | 56.92 | 82.45 | 0.039 | 0.036 | 0.040 | −0.0003 (0.0006) | 0.646 | −0.0003 (0.0007) | 0.647 | |
| Glucose | 114 | 115.39 | 97.99 | <0.001 | <0.001 | 0.001 | −0.0002 (0.0005) | 0.647 | −0.0002 (0.0005) | 0.645 | |
| Dataset 2 (forward direction) | |||||||||||
| 25(OH)D level | Metabolic syndrome | 89 | 114.98 | 97.72 | <0.001 | <0.001 | <0.001 | −0.0027 (0.0011) | 0.012 | −0.0027 (0.0011) | 0.013 |
| WC | 92 | 117.20 | 96.47 | <0.001 | <0.001 | <0.001 | −0.0020 (0.0015) | 0.199 | −0.0020 (0.0016) | 0.192 | |
| BMI | 93 | 116.40 | 96.72 | <0.001 | <0.001 | <0.001 | −0.0018 (0.0014) | 0.208 | −0.0018 (0.0014) | 0.208 | |
| TG | 104 | 118.64 | 97.46 | <0.001 | <0.001 | <0.001 | −0.0072 (0.0032) | 0.028 | −0.0072 (0.0033) | 0.030 | |
| HDL | 104 | 118.64 | 97.46 | <0.001 | <0.001 | <0.001 | −0.0023 (0.0051) | 0.650 | −0.0023 (0.0052) | 0.659 | |
| HTN | 101 | 124.84 | 97.79 | <0.001 | <0.001 | <0.001 | 0.0008 (0.0022) | 0.703 | 0.0009 (0.0022) | 0.697 | |
| Glucose | 104 | 118.64 | 98.17 | <0.001 | <0.001 | <0.001 | 0.0002 (0.0008) | 0.805 | 0.0002 (0.0009) | 0.813 | |
| Dataset 2 (reverse direction) | |||||||||||
| Metabolic syndrome | 25(OH)D level | 543 | 74.23 | 88.79 | <0.001 | <0.001 | <0.001 | −0.0004 (0.0005) | 0.385 | −0.0003 (0.0005) | 0.537 |
| WC | 41 | 59.80 | 87.11 | <0.001 | <0.001 | <0.001 | −0.0030 (0.0017) | 0.087 | −0.0029 (0.0018) | 0.119 | |
| BMI | 67 | 67.60 | 90.07 | <0.001 | <0.001 | <0.001 | −0.0016 (0.0011) | 0.162 | −0.0015 (0.0012) | 0.211 | |
| TG | 369 | 138.96 | 97.82 | <0.001 | <0.001 | <0.001 | −0.0004 (0.0005) | 0.402 | −0.0003 (0.0005) | 0.446 | |
| HDL | 414 | 147.02 | 98.17 | <0.001 | <0.001 | <0.001 | 0.0008 (0.0004) | 0.075 | 0.0008 (0.0004) | 0.077 | |
| HTN | 268 | 55.19 | 91.24 | <0.001 | <0.001 | <0.001 | −0.0009 (0.0007) | 0.178 | −0.0011 (0.0008) | 0.181 | |
| Glucose | 71 | 121.30 | 96.85 | <0.001 | <0.001 | <0.001 | −0.0035 (0.0012) | 0.003 | −0.0035 (0.0012) | 0.004 | |
| Datasets | Exposure | Outcome | Model 1 β (95% CI) | Model 2 β (95% CI) |
|---|---|---|---|---|
| Dataset 1 (reverse direction) | Metabolic syndrome | 25(OH)D level | −0.002 (−0.018, 0.013) | −0.006 (−0.020, 0.008) |
| Waist circumference | −0.032 (−0.070, 0.006) | −0.036 (−0.065, −0.006) * | ||
| TG | −0.009 (−0.068, 0.050) | −0.010 (−0.068, 0.047) | ||
| HDL | 0.013 (−0.017, 0.042) | 0.014 (−0.011, 0.040) | ||
| SBP | −0.024 (−0.089, 0.041) | 0.0001 (−0.026, 0.026) | ||
| DBP | 0.047 (−0.107, 0.201) | −0.015 (−0.040, 0.011) | ||
| Glucose | −0.017 (−0.05, 0.015) | −0.022 (−0.057, 0.014) | ||
| Dataset 2 (forward direction) | 25(OH)D level | Metabolic syndrome | −0.030 (−0.145, 0.085) | −0.060 (−0.159, 0.040) |
| Waist circumference | 0.145 (−0.0004, 0.291) | 0.106 (−0.028, 0.239) | ||
| BMI | 0.095 (−0.062, 0.251) | 0.034 (−0.094, 0.163) | ||
| TG | −0.118 (−0.388, 0.151) | −0.121 (−0.362, 0.120) | ||
| HDL | −0.134 (−1.342, 1.074) | −0.111 (−0.329, 0.107) | ||
| Hypertension | −0.107 (−0.286, 0.072) | −0.115 (−0.241, 0.011) | ||
| Glucose | −0.010 (−0.034, 0.014) | −0.012 (−0.048, 0.024) | ||
| Dataset 2 (reverse direction) | Metabolic syndrome | 25(OH)D level | −0.135 (−0.175, −0.095) * | −0.141 (−0.175, −0.107) * |
| Waist circumference | −0.087 (−0.135, −0.039) * | −0.097 (−0.143, −0.051) * | ||
| BMI | −0.055 (−0.096, −0.015) * | −0.059 (−0.095, −0.024) * | ||
| TG | −0.120 (−0.164, −0.076) * | −0.120 (−0.164, −0.077) * | ||
| HDL | −0.024 (−0.084, 0.035) | −0.024 (−0.083, 0.035) | ||
| Hypertension | 0.006 (−0.015, 0.027) | 0.007 (−0.014, 0.028) | ||
| Glucose | −0.058 (−0.116, 0.001) | −0.058 (−0.118, 0.002) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.; Seo, J.H.; Lee, J.; Kim, H.S. Causal Effects of 25-Hydroxyvitamin D on Metabolic Syndrome and Metabolic Risk Traits: A Bidirectional Two-Sample Mendelian Randomization Study. Biomedicines 2025, 13, 723. https://doi.org/10.3390/biomedicines13030723
Lee Y, Seo JH, Lee J, Kim HS. Causal Effects of 25-Hydroxyvitamin D on Metabolic Syndrome and Metabolic Risk Traits: A Bidirectional Two-Sample Mendelian Randomization Study. Biomedicines. 2025; 13(3):723. https://doi.org/10.3390/biomedicines13030723
Chicago/Turabian StyleLee, Young, Je Hyun Seo, Junyong Lee, and Hwa Sun Kim. 2025. "Causal Effects of 25-Hydroxyvitamin D on Metabolic Syndrome and Metabolic Risk Traits: A Bidirectional Two-Sample Mendelian Randomization Study" Biomedicines 13, no. 3: 723. https://doi.org/10.3390/biomedicines13030723
APA StyleLee, Y., Seo, J. H., Lee, J., & Kim, H. S. (2025). Causal Effects of 25-Hydroxyvitamin D on Metabolic Syndrome and Metabolic Risk Traits: A Bidirectional Two-Sample Mendelian Randomization Study. Biomedicines, 13(3), 723. https://doi.org/10.3390/biomedicines13030723






