Can AI Be Useful in the Early Detection of Pancreatic Cancer in Patients with New-Onset Diabetes?
Abstract
1. Introduction
1.1. Pancreatic Cancer and New-Onset Diabetes
1.2. AI in Cancer Diagnosis
2. Former Perspectives on Pancreatic Cancer Screening in Diabetic Patients
3. The Role of AI in the Identification of High-Risk Pancreatic Cancer Group
| Study | AI | Non-AI | ||
|---|---|---|---|---|
| Models | Results | Models | Results | |
| Hsieh et al. (2018) [84] | ANN | AUROC: 0.642 Precision: 0.995 Recall: 0.873 F1: 0.930 | Logistic regression | AUROC: 0.707 Precision: 0.995 Recall: 0.998 F1: 0.996 |
| Chen et al. (2023) [86] | SVM | AUROC: 0.7721 Precision: 0.0001 Recall: 0.7500 F1: 0.0003 Accuracy: 0.7409 | Logistic regression | AUROC: 0.6669 Precision: 0.0001 Recall: 0.8889 F1: 0.0002 Accuracy: 0.3760 |
| LGBM | AUROC: 0.8632 Precision: 0.0002 Recall: 0.8333 F1: 0.0010 Accuracy: 0.7805 | |||
| XGB | AUROC: 0.8772 Precision: 0.0002 Recall: 0.8611 F1: 0.0009 Accuracy: 0.8375 | |||
| RF | AUROC: 0.8860 Precision: 0.0002 Recall: 0.8611 F1: 0.0015 Accuracy: 0.8336 | |||
| GBM | AUROC: 0.9000 Precision: 0.0002 Recall: 0.8889 F1: 0.0008 Accuracy: 0.8102 | |||
| Voting | AUROC: 0.9049 Precision: 0.0002 Recall: 0.8889 F1: 0.0009 Accuracy: 0.8373 | |||
| LDA | AUROC: 0.9073 Precision: 0.0002 Recall: 0.8611 F1: 0.0012 Accuracy: 0.8403 | |||
| Cichosz et al. (2024) [87] | RF | AUROC: 0.78 | - | - |
| Clift et al. (2024) [93] | ANN | Harrell’s C index: 0.650 Calibration slope: 1.855 CITL: 0.855 | Cox proportional hazard modeling | Harrell’s’ C index: 0.802 Calibration slope: 0.980 CITL: −0.020 |
| XGB | Harrell’s C index: 0.723 Calibration slope: 1.180 CITL: 0.180 | |||
| Khan et al. (2023) [95] | XGB | AUROC: 0.800 Precision: 0.012 Recall: 0.750 Accuracy: 0.700 | ENDPAC * | AUROC: 0.630 Precision: 0.008 Recall: 0.510 Accuracy: 0.700 |
| Boursi model * | AUROC: 0.680 Precision: 0.011 Recall: 0.540 Accuracy: 0.770 | |||
| Chen et al. (2023) [98] | RF | AUROC: 0.808–0.822 | - | - |
| Sun et al. (2024) [99] | RF | AUROC: 0.776 | Logistic regression | AUROC: 0.897 |
| XGB | AUROC: 0.824 | |||
| SVC | AUROC: 0.837 | |||
| MLP | AUROC: 0.884 | |||
4. Practical Challenges in Implementing AI-Based Technologies
5. Gaps and Future Directions in PCD Screening
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| NOD | New-onset diabetes |
| PDAC | Pancreatic ductal adenocarcinoma |
| AJCC | American Joint Committee on Cancer |
| SV | Splenic vein |
| ACG | American College of Gastroenterology |
| AGA | American Gastroenterology Associations |
| DM | Diabetes mellitus |
| T2DM | Type 2 diabetes mellitus |
| NOH | New-onset hyperglycemia |
| PCD | Pancreatic cancer-associated diabetes |
| IGF | Insulin-like growth factor |
| IGFBP-2 | Insulin-like growth factor binding protein-2 |
| PAI-1 | Plasminogen activator inhibitor-1 |
| ML | Machine learning |
| DL | Deep learning |
| GB | Gradient boosting |
| SVM | Support vector machine |
| KNNs | K-nearest neighbors |
| NB | Naïve Bayes |
| ANNs | Artificial neural networks |
| ENDPAC | Enriching New-Onset Diabetes for Pancreatic Cancer |
| QALYs | Quality-adjusted life years |
| BMI | Body Mass Index |
| PPV | Positive predictive value |
| LDL | Low-density lipoprotein |
| CA19-9 | Carbohydrate antigen 19-9 |
| FDA | Food and Drug Administration |
| PPIs | Proton pump inhibitors |
| ALT | Alanine aminotransferase |
| LDA | Linear discriminant analysis |
| GBM | Gradient boosting machine |
| XGB | Extreme gradient boosting |
| LGBM | Light gradient boosting machine |
| RF | Random forest |
| RR | Relative risk |
| EV | Ensemble voting |
| ICD-10 | International Classification of Diseases Version 10 |
| ATC | Anatomical Therapeutic Chemical |
| MPL | Multi-perceptron classifier |
| SNP | Single nucleotide polymorphism |
| NPV | Negative predictive value |
| MMTT | Mixed meal tolerance test |
| OGTT | Oral glucose tolerance test |
| GDPR | General Data Protection Regulation |
| MHRA | Medicine and Healthcare Products Regulatory Agency |
| GMLP | Good Machine Learning Practice |
| XAI | Explainable AI |
| FL | Federated learning |
| SSL | Self-supervised learning |
| CT | Computed Tomography |
| EUS | Endoscopic ultrasound |
| CONSORT-AI | Consolidated Standards of Reporting Trials AI Extension |
| TRIPOD | Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis |
| NFL | “No Free Lunch” (theorem) |
References
- Stoffel, E.M.; Brand, R.E.; Goggins, M. Pancreatic Cancer: Changing Epidemiology and New Approaches to Risk Assessment, Early Detection, and Prevention. Gastroenterology 2023, 164, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Cascinu, S.; Falconi, M.; Valentini, V.; Jelic, S. Pancreatic Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2010, 21, v55–v58. [Google Scholar] [CrossRef]
- SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Surveillance Research Program, National Cancer Institute; 17 April 2024. Data Source(s): SEER Incidence Data, November 2023 Submission (1975-2021), SEER 22 Registries (Excluding Illinois and Massachusetts). Expected Survival Life Tables by Socio-Economic Standards. Available online: https://seer.cancer.gov/statistics-network/explorer/application.html (accessed on 26 July 2024).
- Nikšić, M.; Minicozzi, P.; Weir, H.K.; Zimmerman, H.; Schymura, M.J.; Rees, J.R.; Coleman, M.P.; Allemani, C. Pancreatic Cancer Survival Trends in the US from 2001 to 2014: A CONCORD-3 Study. Cancer Commun. 2023, 43, 87–99. [Google Scholar] [CrossRef]
- Szymoński, K.; Milian-Ciesielska, K.; Lipiec, E.; Adamek, D. Current Pathology Model of Pancreatic Cancer. Cancers 2022, 14, 2321. [Google Scholar] [CrossRef] [PubMed]
- Keane, M.G.; Horsfall, L.; Rait, G.; Pereira, S.P. A Case-Control Study Comparing the Incidence of Early Symptoms in Pancreatic and Biliary Tract Cancer. BMJ Open 2014, 4, e005720. [Google Scholar] [CrossRef]
- Caban, M.; Małecka-Wojciesko, E. Gaps and Opportunities in the Diagnosis and Treatment of Pancreatic Cancer. Cancers 2023, 15, 5577. [Google Scholar] [CrossRef]
- Evans, J.; Chapple, A.; Salisbury, H.; Corrie, P.; Ziebland, S. “It Can’t Be Very Important Because It Comes and Goes”—Patients’ Accounts of Intermittent Symptoms Preceding a Pancreatic Cancer Diagnosis: A Qualitative Study. BMJ Open 2014, 4, e004215. [Google Scholar] [CrossRef]
- Wei, K.; Hackert, T. Surgical Treatment of Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 1971. [Google Scholar] [CrossRef]
- Edge, S.B.; Byrd, D.R.; Carducci, M.A.; Compton, C.C.; Fritz, A.G.; Greene, F.L. AJCC Cancer Staging Manual; Springer: New York, NY, USA, 2010; Volume 649. [Google Scholar]
- Robatel, S.; Schenk, M. Current Limitations and Novel Perspectives in Pancreatic Cancer Treatment. Cancers 2022, 14, 985. [Google Scholar] [CrossRef]
- Tamburrino, D. Selection Criteria in Resectable Pancreatic Cancer: A Biological and Morphological Approach. World J. Gastroenterol. 2014, 20, 11210. [Google Scholar] [CrossRef]
- Kang, J.S.; Choi, Y.J.; Byun, Y.; Han, Y.; Kim, J.H.; Lee, J.M.; Sohn, H.J.; Kim, H.; Kwon, W.; Jang, J.Y. Radiological Tumour Invasion of Splenic Artery or Vein in Patients with Pancreatic Body or Tail Adenocarcinoma and Effect on Recurrence and Survival. Br. J. Surg. 2022, 109, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Amemiya, H.; Izumo, W.; Nakata, Y.; Maruyama, S.; Takiguchi, K.; Shoda, K.; Shiraishi, K.; Furuya, S.; Kawaguchi, Y.; et al. Prognostic Significance of Splenic Vein Invasion for Pancreatic Cancer Patients with Pancreatectomy: A Retrospective Study. Anticancer. Res. 2025, 45, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Blackford, A.L.; Dal Molin, M.; Wolfgang, C.L.; Goggins, M. Time to Progression of Pancreatic Ductal Adenocarcinoma from Low-to-High Tumour Stages. Gut 2015, 64, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Wood, L.D.; Canto, M.I.; Jaffee, E.M.; Simeone, D.M. Pancreatic Cancer: Pathogenesis, Screening, Diagnosis, and Treatment. Gastroenterology 2022, 163, 386–402. [Google Scholar] [CrossRef]
- Pandharipande, P.V.; Heberle, C.; Dowling, E.C.; Kong, C.Y.; Tramontano, A.; Perzan, K.E.; Brugge, W.; Hur, C. Targeted Screening of Individuals at High Risk for Pancreatic Cancer: Results of a Simulation Model. Radiology 2015, 275, 177–187. [Google Scholar] [CrossRef]
- Frampas, E.; Morla, O.; Regenet, N.; Eugène, T.; Dupas, B.; Meurette, G. A Solid Pancreatic Mass: Tumour or Inflammation? Diagn. Interv. Imaging 2013, 94, 741–755. [Google Scholar] [CrossRef]
- Al-Hawary, M.M.; Kaza, R.K.; Azar, S.F.; Ruma, J.A.; Francis, I.R. Mimics of Pancreatic Ductal Adenocarcinoma. Cancer Imaging 2013, 13, 342–349. [Google Scholar] [CrossRef]
- Aslanian, H.R.; Lee, J.H.; Canto, M.I. AGA Clinical Practice Update on Pancreas Cancer Screening in High-Risk Individuals: Expert Review. Gastroenterology 2020, 159, 358–362. [Google Scholar] [CrossRef]
- Syngal, S.; E Brand, R.; Church, J.M.; Giardiello, F.M.; Hampel, H.L.; Burt, R.W. ACG Clinical Guideline: Genetic Testing and Management of Hereditary Gastrointestinal Cancer Syndromes. Am. J. Gastroenterol. 2015, 110, 223–262. [Google Scholar] [CrossRef]
- Koopmann, B.D.M.; Omidvari, A.H.; Lansdorp-Vogelaar, I.; Cahen, D.L.; Bruno, M.J.; de Kok, I.M.C.M. The Impact of Pancreatic Cancer Screening on Life Expectancy: A Systematic Review of Modeling Studies. Int. J. Cancer 2023, 152, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.K.; Korc, M.; Petersen, G.M.; Eibl, G.; Li, D.; Rickels, M.R.; Chari, S.T.; Abbruzzese, J.L. Diabetes, Pancreatogenic Diabetes, and Pancreatic Cancer. Diabetes 2017, 66, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.A.; Bellin, M.D.; Andersen, D.K.; Bradley, D.; Cruz-Monserrate, Z.; Forsmark, C.E.; Goodarzi, M.O.; Habtezion, A.; Korc, M.; Kudva, Y.C.; et al. Type 3c (Pancreatogenic) Diabetes Mellitus Secondary to Chronic Pancreatitis and Pancreatic Cancer. Lancet Gastroenterol. Hepatol. 2016, 1, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, G.; Kamada, P.; Chari, S.T. Prevalence of Diabetes Mellitus in Pancreatic Cancer Compared to Common Cancers. Pancreas 2013, 42, 198–201. [Google Scholar] [CrossRef]
- Amri, F.; Belkhayat, C.; Yeznasni, A.; Koulali, H.; Jabi, R.; Zazour, A.; Abda, N.; Bouziane, M.; Ismaili, Z.; Kharrasse, G. Association between Pancreatic Cancer and Diabetes: Insights from a Retrospective Cohort Study. BMC Cancer 2023, 23, 856. [Google Scholar] [CrossRef]
- Chari, S.T.; Leibson, C.L.; Rabe, K.G.; Timmons, L.J.; Ransom, J.; de Andrade, M.; Petersen, G.M. Pancreatic Cancer-Associated Diabetes Mellitus: Prevalence and Temporal Association with Diagnosis of Cancer. Gastroenterology 2008, 134, 95–101. [Google Scholar] [CrossRef]
- Pannala, R.; Leirness, J.B.; Bamlet, W.R.; Basu, A.; Petersen, G.M.; Chari, S.T. Prevalence and Clinical Profile of Pancreatic Cancer-Associated Diabetes Mellitus. Gastroenterology 2008, 134, 981–987. [Google Scholar] [CrossRef]
- Permert, J.; Ihse, I.; Jorfeldt, L.; von Schenck, H.; Arnqvist, H.J.; Larsson, J. Pancreatic Cancer Is Associated with Impaired Glucose Metabolism. Eur. J. Surg. 1993, 159, 101–107. [Google Scholar]
- Roy, A.; Sahoo, J.; Kamalanathan, S.; Naik, D.; Mohan, P.; Kalayarasan, R. Diabetes and Pancreatic Cancer: Exploring the Two-Way Traffic. World J. Gastroenterol. 2021, 27, 4939. [Google Scholar] [CrossRef]
- Sharma, A.; Smyrk, T.C.; Levy, M.J.; Topazian, M.A.; Chari, S.T. Fasting Blood Glucose Levels Provide Estimate of Duration and Progression of Pancreatic Cancer Before Diagnosis. Gastroenterology 2018, 155, 490–500. [Google Scholar] [CrossRef]
- Aggarwal, G.; Rabe, K.G.; Petersen, G.M.; Chari, S.T. New-Onset Diabetes in Pancreatic Cancer: A Study in the Primary Care Setting. Pancreatology 2012, 12, 156–161. [Google Scholar] [CrossRef]
- Pannala, R.; Basu, A.; Petersen, G.M.; Chari, S.T. New-Onset Diabetes: A Potential Clue to the Early Diagnosis of Pancreatic Cancer. Lancet Oncol. 2009, 10, 88–95. [Google Scholar] [CrossRef]
- Jensen, M.H.; Cichosz, S.L.; Hejlesen, O.; Henriksen, S.D.; Drewes, A.M.; Olesen, S.S. Risk of Pancreatic Cancer in People with New-Onset Diabetes: A Danish Nationwide Population-Based Cohort Study. Pancreatology 2023, 23, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Chari, S.T. Detecting Early Pancreatic Cancer: Problems and Prospects. Semin. Oncol. 2007, 34, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Chari, S.T. Pancreatic Cancer and Diabetes Mellitus. Curr. Treat. Options Gastroenterol. 2018, 16, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Eibl, G.; Cruz-Monserrate, Z.; Korc, M.; Petrov, M.S.; Goodarzi, M.O.; Fisher, W.E.; Habtezion, A.; Lugea, A.; Pandol, S.J.; Hart, P.A.; et al. Diabetes Mellitus and Obesity as Risk Factors for Pancreatic Cancer. J. Acad. Nutr. Diet. 2018, 118, 555–567. [Google Scholar] [CrossRef]
- Pollak, M. Insulin and Insulin-like Growth Factor Signalling in Neoplasia. Nat. Rev. Cancer 2008, 8, 915–928. [Google Scholar]
- Mutgan, A.C.; Besikcioglu, H.E.; Wang, S.; Friess, H.; Ceyhan, G.O.; Demir, I.E. Insulin/IGF-Driven Cancer Cell-Stroma Crosstalk as a Novel Therapeutic Target in Pancreatic Cancer. Mol. Cancer 2018, 17, 66. [Google Scholar]
- Włodarczyk, B.; Borkowska, A.; Włodarczyk, P.; Małecka-Panas, E.; Gąsiorowska, A. Insulin-like Growth Factor 1 and Insulin-like Growth Factor Binding Protein 2 Serum Levels as Potential Biomarkers in Differential Diagnosis between Chronic Pancreatitis and Pancreatic Adenocarcinoma in Reference to Pancreatic Diabetes. Prz. Gastroenterol. 2021, 16, 36–42. [Google Scholar] [CrossRef]
- Wang, R.; Liu, Y.; Liang, Y.; Zhou, L.; Chen, M.-J.; Liu, X.-B.; Tan, C.-L.; Chen, Y.-H. Regional Differences in Islet Amyloid Deposition in the Residual Pancreas with New-Onset Diabetes Secondary to Pancreatic Ductal Adenocarcinoma. World J. Gastrointest. Surg. 2023, 15, 1703–1711. [Google Scholar] [CrossRef]
- Bures, J.; Kohoutova, D.; Skrha, J.; Bunganic, B.; Ngo, O.; Suchanek, S.; Skrha, P.; Zavoral, M. Diabetes Mellitus in Pancreatic Cancer: A Distinct Approach to Older Subjects with New-Onset Diabetes Mellitus. Cancers 2023, 15, 3669. [Google Scholar] [CrossRef]
- Chowdhary, K.R. Fundamentals of Artificial Intelligence; Springer: New Delhi, India, 2020; pp. 603–649. [Google Scholar]
- Huang, B.; Huang, H.; Zhang, S.; Zhang, D.; Shi, Q.; Liu, J.; Guo, J. Artificial Intelligence in Pancreatic Cancer. Theranostics 2022, 12, 6931. [Google Scholar] [CrossRef]
- Roscher, R.; Bohn, B.; Duarte, M.F.; Garcke, J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2020, 8, 42200–42216. [Google Scholar] [CrossRef]
- Kenner, B.; Chari, S.T.; Kelsen, D.; Klimstra, D.S.; Pandol, S.J.; Rosenthal, M.; Rustgi, A.K.; Taylor, J.A.; Yala, A.; Abul-Husn, N.; et al. Artificial Intelligence and Early Detection of Pancreatic Cancer: 2020 Summative Review. Pancreas 2021, 50, 251–279. [Google Scholar] [CrossRef] [PubMed]
- Mobarak, M.H.; Mimona, M.A.; Islam, M.A.; Hossain, N.; Zohura, F.T.; Imtiaz, I.; Rimon, M.I.H. Scope of Machine Learning in Materials Research—A Review. Appl. Surf. Sci. Adv. 2023, 18, 100523. [Google Scholar] [CrossRef]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Santos, C.S.; Amorim-Lopes, M. Externally Validated and Clinically Useful Machine Learning Algorithms to Support Patient-Related Decision-Making in Oncology: A Scoping Review. BMC Med. Res. Methodol. 2025, 25, 45. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, K.P.; Papaloukas, C.; Sakaloglou, P.; Exarchos, T.; Fotiadis, D.I. Applied Machine Learning in Cancer Research: A Systematic Review for Patient Diagnosis, Classification and Prognosis. Comput. Struct. Biotechnol. J. 2021, 19, 5546–5555. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, H.; Wang, H. Machine Learning and AI in Cancer Prognosis, Prediction, and Treatment Selection: A Critical Approach. J. Multidiscip. Healthc. 2023, 16, 1779–1791. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Feature Engineering and Selection: A Practical Approach for Predictive Models; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019. [Google Scholar]
- Majda-Zdancewicz, E.; Potulska-Chromik, A.; Jakubowski, J.; Nojszewska, M.; Kostera-Pruszczyk, A. Deep Learning vs. Feature Engineering in the Assessment of Voice Signals for Diagnosis in Parkinson’s Disease. Bull. Pol. Acad. Sci. 2021, 69, e137347. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and Deep Learning Methods for Radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar]
- Boumaraf, S.; Liu, X.; Wan, Y.; Zheng, Z.; Ferkous, C.; Ma, X.; Li, Z.; Bardou, D. Conventional Machine Learning versus Deep Learning for Magnification Dependent Histopathological Breast Cancer Image Classification: A Comparative Study with Visual Explanation. Diagnostics 2021, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Yadavendra; Chand, S. A Comparative Study of Breast Cancer Tumor Classification by Classical Machine Learning Methods and Deep Learning Method. Mach. Vis. Appl. 2020, 31, 46. [Google Scholar] [CrossRef]
- Wang, X.; Yang, W.; Weinreb, J.; Han, J.; Li, Q.; Kong, X.; Yan, Y.; Ke, Z.; Luo, B.; Liu, T.; et al. Searching for Prostate Cancer by Fully Automated Magnetic Resonance Imaging Classification: Deep Learning versus Non-Deep Learning. Sci. Rep. 2017, 7, 15415. [Google Scholar] [CrossRef]
- Brehar, R.; Mitrea, D.A.; Vancea, F.; Marita, T.; Nedevschi, S.; Lupsor-Platon, M.; Rotaru, M.; Badea, R.I. Comparison of Deep-Learning and Conventional Machine-Learning Methods for the Automatic Recognition of the Hepatocellular Carcinoma Areas from Ultrasound Images. Sensors 2020, 20, 3085. [Google Scholar] [CrossRef]
- Bouamrane, A.; Derdour, M. Enhancing Lung Cancer Detection and Classification Using Machine Learning and Deep Learning Techniques: A Comparative Study. In Proceedings of the 6th International Conference on Networking and Advanced Systems, ICNAS 2023, Algiers, Algeria, 21–23 October 2023. [Google Scholar]
- Painuli, D.; Bhardwaj, S.; Köse, U. Recent Advancement in Cancer Diagnosis Using Machine Learning and Deep Learning Techniques: A Comprehensive Review. Comput. Biol. Med. 2022, 146, 105580. [Google Scholar]
- Claridge, H.; Price, C.A.; Ali, R.; Cooke, E.A.; De Lusignan, S.; Harvey-Sullivan, A.; Hodges, C.; Khalaf, N.; O’callaghan, D.; Stunt, A.; et al. Determining the Feasibility of Calculating Pancreatic Cancer Risk Scores for People with New-Onset Diabetes in Primary Care (DEFEND PRIME): Study Protocol. BMJ Open 2024, 14, e079863. [Google Scholar] [CrossRef]
- Sharma, A.; Kandlakunta, H.; Nagpal, S.J.S.; Feng, Z.; Hoos, W.; Petersen, G.M.; Chari, S.T. Model to Determine Risk of Pancreatic Cancer in Patients with New-Onset Diabetes. Gastroenterology 2018, 155, 730–739.e3. [Google Scholar] [CrossRef]
- Khan, S.; Safarudin, R.F.; Kupec, J.T. Validation of the ENDPAC Model: Identifying New-Onset Diabetics at Risk of Pancreatic Cancer. Pancreatology 2021, 21, 550–555. [Google Scholar] [CrossRef]
- Chen, W.; Butler, R.K.; Lustigova, E.; Chari, S.T.; Wu, B.U. Validation of the Enriching New-Onset Diabetes for Pancreatic Cancer Model in a Diverse and Integrated Healthcare Setting. Dig. Dis. Sci. 2020, 66, 78–87. [Google Scholar] [CrossRef]
- Wang, L.; Levinson, R.; Mezzacappa, C.; Katona, B.W. Review of the Cost-Effectiveness of Surveillance for Hereditary Pancreatic Cancer. Fam. Cancer 2024, 23, 351–360. [Google Scholar] [PubMed]
- Schwartz, N.R.M.; Matrisian, L.M.; Shrader, E.E.; Feng, Z.; Chari, S.; Roth, J.A. Potential Cost-Effectiveness of Risk-Based Pancreatic Cancer Screening in Patients with New-Onset Diabetes. JNCCN J. Natl. Compr. Cancer Netw. 2022, 20, 451–459. [Google Scholar] [CrossRef]
- Bertram, M.Y.; Lauer, J.A.; De Joncheere, K.; Edejer, T.; Hutubessy, R.; Kieny, M.-P.; Hill, S.R. Cost-Effectiveness Thresholds: Pros and Cons. Bull. World Health Organ. 2016, 94, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Klatte, D.C.F.; Clift, K.E.; Mantia, S.K.; Millares, L.; Hoogenboom, S.A.M.; Presutti, R.J.; Wallace, M.B. Identification of Individuals at High-Risk for Pancreatic Cancer Using a Digital Patient-Input Tool Combining Family Cancer History Screening and New-Onset Diabetes. Prev. Med. Rep. 2023, 31, 102110. [Google Scholar] [CrossRef] [PubMed]
- Boursi, B.; Finkelman, B.; Giantonio, B.J.; Haynes, K.; Rustgi, A.K.; Rhim, A.D.; Mamtani, R.; Yang, Y.-X. A Clinical Prediction Model to Assess Risk for Pancreatic Cancer Among Patients with New-Onset Diabetes. Gastroenterology 2017, 152, 840–850.e3. [Google Scholar] [CrossRef]
- Ali, S.; Coory, M.; Donovan, P.; Na, R.; Pandeya, N.; Pearson, S.A.; Spilsbury, K.; Tuesley, K.; Jordan, S.J.; Neale, R.E. Predicting the Risk of Pancreatic Cancer in Women with New-Onset Diabetes Mellitus. J. Gastroenterol. Hepatol. 2024, 39, 1057–1064. [Google Scholar] [CrossRef]
- Wilson, J.A.P. Colon Cancer Screening in the Elderly: When Do We Stop? Trans. Am. Clin. Climatol. Assoc. 2010, 121, 94–103. [Google Scholar]
- Higuera, O.; Ghanem, I.; Nasimi, R.; Prieto, I.; Koren, L.; Feliu, J. Management of Pancreatic Cancer in the Elderly. World J. Gastroenterol. 2016, 22, 764–775. [Google Scholar] [CrossRef]
- Smith, L.M.; Mahoney, D.W.; Bamlet, W.R.; Yu, F.; Liu, S.; Goggins, M.G.; Darabi, S.; Majumder, S.; Wang, Q.-L.; Coté, G.A.; et al. Early Detection of Pancreatic Cancer: Study Design and Analytical Considerations in Biomarker Discovery and Early Phase Validation Studies. Pancreatology 2024, 24, 1265–1279. [Google Scholar] [CrossRef]
- O’Neill, R.S.; Stoita, A. Biomarkers in the Diagnosis of Pancreatic Cancer: Are We Closer to Finding the Golden Ticket? World J. Gastroenterol. 2021, 27, 4045–4087. [Google Scholar] [CrossRef]
- Gong, J.; Li, X.; Feng, Z.; Lou, J.; Pu, K.; Sun, Y.; Hu, S.; Zhou, Y.; Song, T.; Shangguan, M.; et al. Sorcin Can Trigger Pancreatic Cancer-Associated New-Onset Diabetes through the Secretion of Inflammatory Cytokines Such as Serpin E1 and CCL5. Exp. Mol. Med. 2024, 56, 2535–2547. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. National Diabetes Statistics Report Website. Available online: https://Www.Cdc.Gov/Diabetes/Php/Data-Research/Index.Html (accessed on 27 July 2024).
- Elsayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. American Diabetes Association. 2.Classification and Diagnosis of Diabetes: Standards of Care in Diabetes. Diabetes Care 2023, 46, S19. [Google Scholar] [PubMed]
- Wu, B.U.; Butler, R.K.; Lustigova, E.; Lawrence, J.M.; Chen, W. Association of Glycated Hemoglobin Levels with Risk of Pancreatic Cancer. JAMA Netw. Open 2020, 3, e204945. [Google Scholar] [CrossRef]
- Boursi, B.; Finkelman, B.; Giantonio, B.J.; Haynes, K.; Rustgi, A.K.; Rhim, A.D.; Mamtani, R.; Yang, Y.-X. A Clinical Prediction Model to Assess Risk for Pancreatic Cancer among Patients with Prediabetes. Eur. J. Gastroenterol. Hepatol. 2022, 34, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Kearns, M.D.; Boursi, B.; Yang, Y.-X. Proton Pump Inhibitors on Pancreatic Cancer Risk and Survival. Cancer Epidemiol. 2017, 46, 80–84. [Google Scholar] [CrossRef]
- Hong, H.E.; Kim, A.S.; Kim, M.R.; Ko, H.J.; Jung, M.K. Does the Use of Proton Pump Inhibitors Increase the Risk of Pancreatic Cancer? A Systematic Review and Meta-Analysis of Epidemiologic Studies. Cancers 2020, 12, 2220. [Google Scholar] [CrossRef]
- Berrington de Gonzalez, A.; Yun, J.E.; Lee, S.-Y.; Klein, A.P.; Jee, S.H. Pancreatic Cancer and Factors Associated with the Insulin Resistance Syndrome in the Korean Cancer Prevention Study. Cancer Epidemiol. Biomark. Prev. 2008, 17, 359–364. [Google Scholar] [CrossRef]
- Hsieh, M.H.; Sun, L.M.; Lin, C.L.; Hsieh, M.J.; Hsu, C.Y.; Kao, C.H. Development of a Prediction Model for Pancreatic Cancer in Patients with Type 2 Diabetes Using Logistic Regression and Artificial Neural Network Models. Cancer Manag. Res. 2018, 10, 6317–6324. [Google Scholar] [CrossRef]
- Klein, A.P. Pancreatic Cancer Epidemiology: Understanding the Role of Lifestyle and Inherited Risk Factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar]
- Chen, S.M.; Phuc, P.T.; Nguyen, P.A.; Burton, W.; Lin, S.J.; Lin, W.C.; Lu, C.Y.; Hsu, M.H.; Cheng, C.T.; Hsu, J.C. A Novel Prediction Model of the Risk of Pancreatic Cancer among Diabetes Patients Using Multiple Clinical Data and Machine Learning. Cancer Med. 2023, 12, 19987–19999. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Jensen, M.H.; Hejlesen, O.; Henriksen, S.D.; Drewes, A.M.; Olesen, S.S. Prediction of Pancreatic Cancer Risk in Patients with New-Onset Diabetes Using a Machine Learning Approach Based on Routine Biochemical Parameters; Prediction of Pancreatic Cancer Risk in New Onset Diabetes. Comput. Methods Programs Biomed. 2024, 244, 107965. [Google Scholar] [CrossRef]
- White, M.J.; Sheka, A.C.; Larocca, C.J.; Irey, R.L.; Ma, S.; Wirth, K.M.; Benner, A.; Denbo, J.W.; Jensen, E.H.; Ankeny, J.S.; et al. The Association of New-Onset Diabetes with Subsequent Diagnosis of Pancreatic Cancer—Novel Use of a Large Administrative Database. J. Public Health 2023, 45, e266–e274. [Google Scholar] [CrossRef]
- Mellenthin, C.; Balaban, V.D.; Dugic, A.; Cullati, S. Risk Factors for Pancreatic Cancer in Patients with New-Onset Diabetes: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 4684. [Google Scholar] [CrossRef] [PubMed]
- Ozsay, O.; Karabacak, U.; Cetin, S.; Majidova, N. Is Diabetes Onset at Advanced Age a Sign of Pancreatic Cancer? Ann. Ital. Chir. 2022, 93, 476–480. [Google Scholar]
- Sharma, S.; Tapper, W.J.; Collins, A.; Hamady, Z.Z.R. Predicting Pancreatic Cancer in the UK Biobank Cohort Using Polygenic Risk Scores and Diabetes Mellitus. Gastroenterology 2022, 162, 1665–1674. [Google Scholar] [CrossRef]
- Sah, R.P.; Sharma, A.; Nagpal, S.; Patlolla, S.H.; Sharma, A.; Kandlakunta, H.; Anani, V.; Angom, R.S.; Kamboj, A.K.; Ahmed, N.; et al. Phases of Metabolic and Soft Tissue Changes in Months Preceding a Diagnosis of Pancreatic Ductal Adenocarcinoma. Gastroenterology 2019, 156, 1742–1752. [Google Scholar] [CrossRef]
- Clift, A.K.; Tan, P.S.; Patone, M.; Liao, W.; Coupland, C.; Bashford-Rogers, R.; Sivakumar, S.; Hippisley-Cox, J. Predicting the Risk of Pancreatic Cancer in Adults with New-Onset Diabetes: Development and Internal–External Validation of a Clinical Risk Prediction Model. Br. J. Cancer 2024, 130, 1969–1978. [Google Scholar]
- NICE Overview|Suspected Cancer: Recognition and Referral|Guidance|NICE. Available online: https://www.nice.org.uk/guidance/ng12/chapter/Recommendations-organised-by-site-of-cancer (accessed on 10 January 2025).
- Khan, S.; Bhushan, B. Machine Learning Predicts Patients with New-Onset Diabetes at Risk of Pancreatic Cancer. J. Clin. Gastroenterol. 2023, 58, 681–691. [Google Scholar] [CrossRef]
- Khan, S.; Al Heraki, S.; Kupec, J.T. Noninvasive Models Screen New-Onset Diabetics at Low Risk of Early-Onset Pancreatic Cancer. Pancreas 2021, 50, 1326–1330. [Google Scholar] [CrossRef]
- Hajibandeh, S.; Intrator, C.; Carrington-Windo, E.; James, R.; Hughes, I.; Hajibandeh, S.; Satyadas, T. Accuracy of the END-PAC Model in Predicting the Risk of Developing Pancreatic Cancer in Patients with New-Onset Diabetes: A Systematic Review and Meta-Analysis. Biomedicines 2023, 11, 3040. [Google Scholar] [CrossRef]
- Chen, W.; Butler, R.K.; Lustigova, E.; Chari, S.T.; Maitra, A.; Rinaudo, J.A.; Wu, B.U. Risk Prediction of Pancreatic Cancer in Patients with Recent-Onset Hyperglycemia: A Machine-Learning Approach. J. Clin. Gastroenterol. 2023, 57, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, C.; Hu, S.; Xu, H.; Gong, J.; Wu, Y.; Fan, Y.; Lv, C.; Song, T.; Lou, J.; et al. Predicting Pancreatic Cancer in New-Onset Diabetes Cohort Using a Novel Model with Integrated Clinical and Genetic Indicators: A Large-Scale Prospective Cohort Study. Cancer Med. 2024, 13, e70388. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Li, L.; Sun, C.; Qi, L.; Tuason, J.P.W.; Kim, N.H.; Bhan, C.; Abdul, H.M. S88 Pancreatic Hormones Response-Generated Machine Learning Model Help Distinguish Sporadic Pancreatic Cancer from New-Onset Diabetes Cohort. Am. J. Gastroenterol. 2021, 116, S38. [Google Scholar] [CrossRef]
- Health Organization, W. Definition and Diagnosis of Diabetes Mellitus and Intermediate Hyperglycemia: Report of a WHO/IDF Consultation; World Health Organization: Geneva, Switzerland, 2006; p. 50. [Google Scholar]
- Lages, M.; Barros, R.; Moreira, P.; Guarino, M.P. Metabolic Effects of an Oral Glucose Tolerance Test Compared to the Mixed Meal Tolerance Tests: A Narrative Review. Nutrients 2022, 14, 2032. [Google Scholar] [CrossRef]
- Śliwińska-Mossoń, M.; Marek, G.; Milnerowicz, H. The Role of Pancreatic Polypeptide in Pancreatic Diseases. Adv. Clin. Exp. Med. 2017, 26, 1447. [Google Scholar] [CrossRef]
- Xu, Q.; Xie, W.; Liao, B.; Hu, C.; Qin, L.; Yang, Z.; Xiong, H.; Lyu, Y.; Zhou, Y.; Luo, A. Interpretability of Clinical Decision Support Systems Based on Artificial Intelligence from Technological and Medical Perspective: A Systematic Review. J. Healthc. Eng. 2023, 2023, 9919269. [Google Scholar] [CrossRef]
- Gryz, J.; Rojszczak, M. Black Box Algorithms and the Rights of Individuals: No Easy Solution to the “Explainability” Problem. Internet Policy Rev. 2021, 10, 1–24. [Google Scholar] [CrossRef]
- Kesa, A.; Kerikmäe, T. Artificial Intelligence and the GDPR: Inevitable Nemeses. TalTech J. Eur. Stud. 2020, 10, 68–90. [Google Scholar] [CrossRef]
- Verdicchio, M.; Perin, A. When Doctors and AI Interact: On Human Responsibility for Artificial Risks. Philos. Technol. 2022, 35, 11. [Google Scholar] [CrossRef]
- Topol, E.J. High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- US FDA. Good Machine Learning Practice for Medical Device Development: Guiding Principles. 2021. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles (accessed on 5 March 2025).
- European Comission AI Act. Available online: https://digital-strategy.ec.europa.eu/en/policies/regulatory-framework-ai (accessed on 6 March 2025).
- Gilbert, S. The EU Passes the AI Act and Its Implications for Digital Medicine Are Unclear. NPJ Digit. Med. 2024, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E. Are Medical Oncologists Ready for the Artificial Intelligence Revolution? Evaluation of the Opinions, Knowledge, and Experiences of Medical Oncologists about Artificial Intelligence Technologies. Med. Oncol. 2023, 40, 327. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, X.M.; Xu, B.; Dickson, C. Concerns on Integrating Artificial Intelligence in Clinical Practice: Cross-Sectional Survey Study. JMIR Form. Res. 2024, 8, e53918. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Habli, I.; Lawton, T.; Porter, Z. Artificial Intelligence in Health Care: Accountability and Safety. Bull. World Health Organ. 2020, 98, 251. [Google Scholar] [CrossRef]
- Tamori, H.; Yamashina, H.; Mukai, M.; Morii, Y.; Suzuki, T.; Ogasawara, K. Acceptance of the Use of Artificial Intelligence in Medicine among Japan’s Doctors and the Public: A Questionnaire Survey. JMIR Hum. Factors 2022, 9, e24680. [Google Scholar] [CrossRef]
- Daniyal, M.; Qureshi, M.; Marzo, R.R.; Aljuaid, M.; Shahid, D. Exploring Clinical Specialists’ Perspectives on the Future Role of AI: Evaluating Replacement Perceptions, Benefits, and Drawbacks. BMC Health Serv. Res. 2024, 24, 587. [Google Scholar] [CrossRef]
- Toussaint, P.A.; Leiser, F.; Thiebes, S.; Schlesner, M.; Brors, B.; Sunyaev, A. Explainable Artificial Intelligence for Omics Data: A Systematic Mapping Study. Brief. Bioinform. 2024, 25, bbad453. [Google Scholar]
- Nazir, S.; Dickson, D.M.; Akram, M.U. Survey of Explainable Artificial Intelligence Techniques for Biomedical Imaging with Deep Neural Networks. Comput. Biol. Med. 2023, 156, 106668. [Google Scholar]
- Murray, K.; Oldfield, L.; Stefanova, I.; Gentiluomo, M.; Aretini, P.; O’Sullivan, R.; Greenhalf, W.; Paiella, S.; Aoki, M.N.; Pastore, A.; et al. Biomarkers, Omics and Artificial Intelligence for Early Detection of Pancreatic Cancer. Semin. Cancer Biol. 2025, 111, 76–88. [Google Scholar] [CrossRef]
- Phillips, P.J.; Hahn, C.A.; Fontana, P.C.; Broniatowski, D.A.; Przybocki, M.A. Four Principles of Explainable Artificial Mark; NIST Interagency/Internal Report (NISTIR); National Institute of Standards and Technology: Gaithersburg, MD, USA, 2020. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Raab, R.; Küderle, A.; Zakreuskaya, A.; Stern, A.D.; Klucken, J.; Kaissis, G.; Rueckert, D.; Boll, S.; Eils, R.; Wagener, H.; et al. Federated Electronic Health Records for the European Health Data Space. Lancet Digit. Health 2023, 5, e840–e847. [Google Scholar] [CrossRef]
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-Supervised Learning in Medicine and Healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar]
- Chowdhury, A.; Rosenthal, J.; Waring, J.; Umeton, R. Applying Self-Supervised Learning to Medicine: Review of the State of the Art and Medical Implementations. Informatics 2021, 8, 1346–1352. [Google Scholar] [CrossRef]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable Artificial Intelligence (XAI) in Deep Learning-Based Medical Image Analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [PubMed]
- Korfiatis, P.; Suman, G.; Patnam, N.G.; Trivedi, K.H.; Karbhari, A.; Mukherjee, S.; Cook, C.; Klug, J.R.; Patra, A.; Khasawneh, H.; et al. Automated Artificial Intelligence Model Trained on a Large Data Set Can Detect Pancreas Cancer on Diagnostic Computed Tomography Scans As Well As Visually Occult Preinvasive Cancer on Prediagnostic Computed Tomography Scans. Gastroenterology 2023, 165, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yang, X.; Sun, L.; Pan, P.; Peng, L.; Li, K.; Zhang, D.; Cui, F.; Xia, C.; Huang, H.; et al. The Value of Artificial Intelligence Techniques in Predicting Pancreatic Ductal Adenocarcinoma with EUS Images: A Meta-Analysis and Systematic Review. Endosc. Ultrasound 2023, 12, 50–58. [Google Scholar]
- Tallam, H.; Elton, D.C.; Lee, S.; Wakim, P.; Pickhardt, P.J.; Summers, R.M. Fully Automated Abdominal CT Biomarkers for Type 2 Diabetes Using Deep Learning. Radiology 2022, 304, 85–95. [Google Scholar] [CrossRef]
- Wright, D.E.; Mukherjee, S.; Patra, A.; Khasawneh, H.; Korfiatis, P.; Suman, G.; Chari, S.T.; Kudva, Y.C.; Kline, T.L.; Goenka, A.H. Radiomics-Based Machine Learning (ML) Classifier for Detection of Type 2 Diabetes on Standard-of-Care Abdomen CTs: A Proof-of-Concept Study. Abdom. Radiol. 2022, 47, 3806–3816. [Google Scholar] [CrossRef]
- Siontis, G.C.M.; Sweda, R.; Noseworthy, P.A.; Friedman, P.A.; Siontis, K.C.; Patel, C.J. Development and Validation Pathways of Artificial Intelligence Tools Evaluated in Randomised Clinical Trials. BMJ Health Care Inform. 2021, 28, e100466. [Google Scholar] [CrossRef]
- Douville, C.; Lahouel, K.; Kuo, A.; Grant, H.; Avigdor, B.E.; Curtis, S.D.; Summers, M.; Cohen, J.D.; Wang, Y.; Mattox, A.; et al. Machine Learning to Detect the SINEs of Cancer. Sci. Transl. Med. 2024, 16, eadi3883. [Google Scholar] [CrossRef]
- Liu, X.; Cruz Rivera, S.; Moher, D.; Calvert, M.J.; Denniston, A.K.; Chan, A.W.; Darzi, A.; Holmes, C.; Yau, C.; Ashrafian, H.; et al. Reporting Guidelines for Clinical Trial Reports for Interventions Involving Artificial Intelligence: The CONSORT-AI Extension. Nat. Med. 2020, 26, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. BMJ 2015, 350, g7594. [Google Scholar] [CrossRef] [PubMed]
- Wolpert, D.H. The Lack of a Priori Distinctions between Learning Algorithms. Neural Comput. 1996, 8, 1341–1390. [Google Scholar] [CrossRef]
- Oikonomou, E.K.; Khera, R. Machine Learning in Precision Diabetes Care and Cardiovascular Risk Prediction. Cardiovasc. Diabetol. 2023, 22, 259. [Google Scholar]
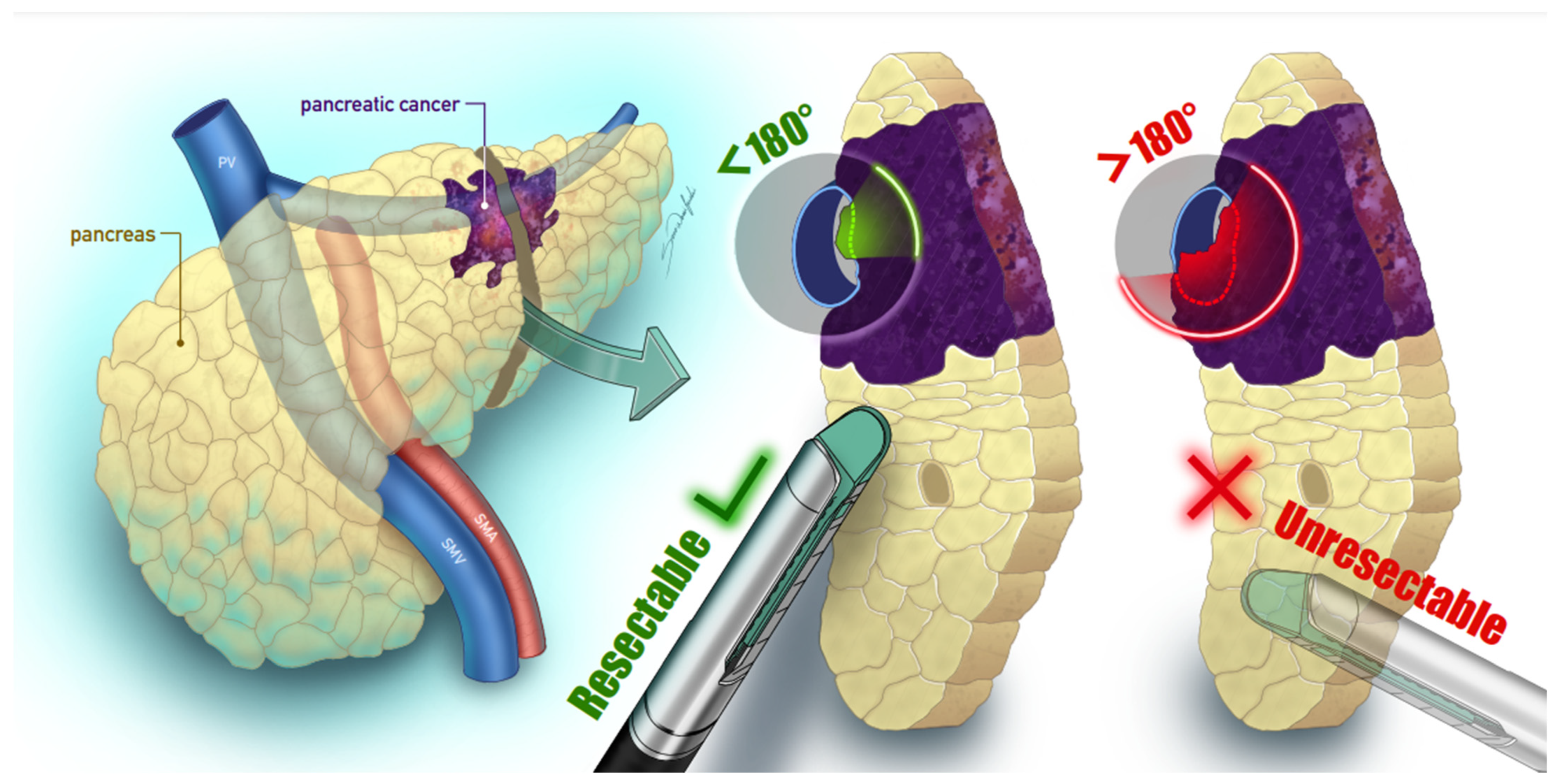
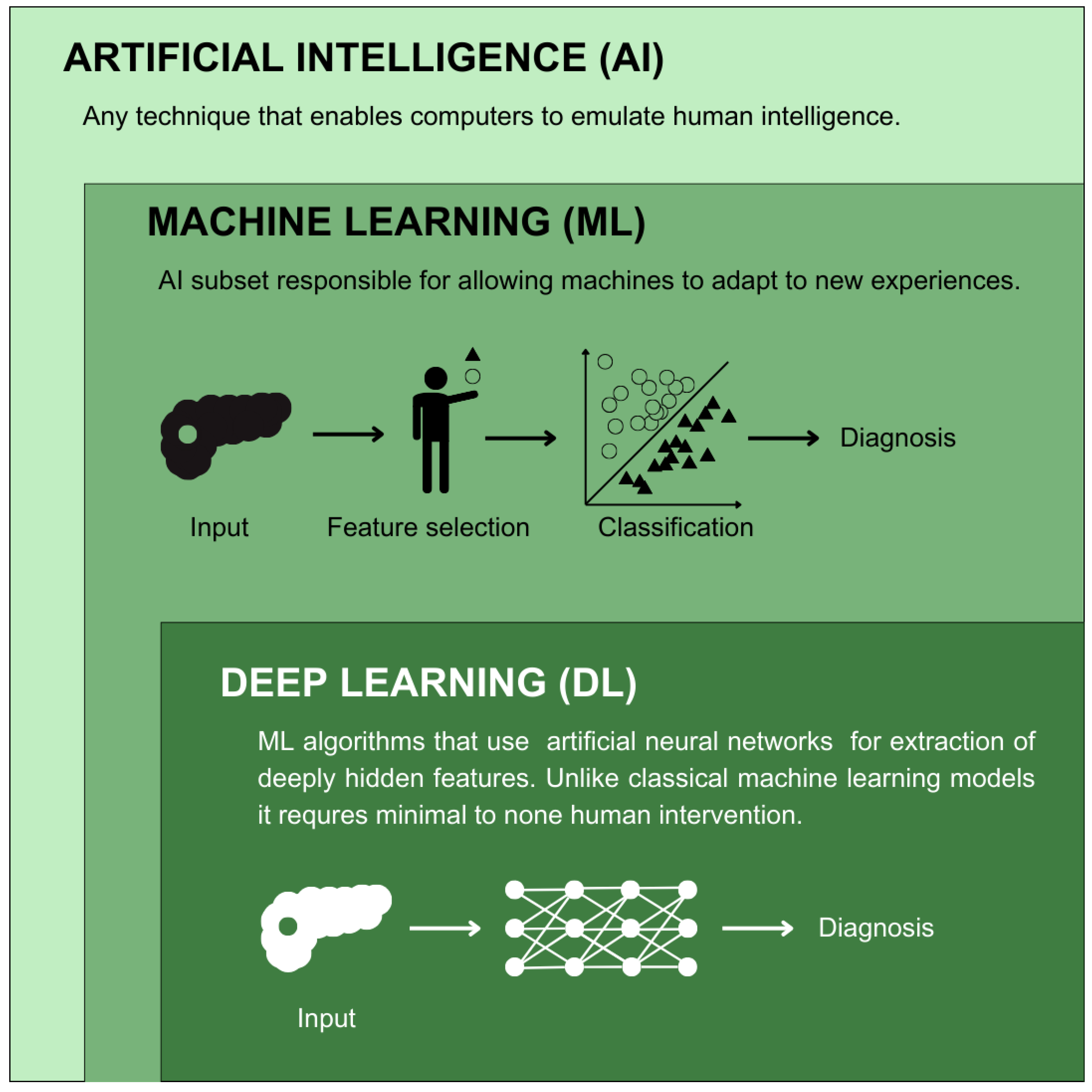
| Study | Data Source | Objective | Population Characteristics | Data Needed | Performance |
|---|---|---|---|---|---|
| Sharma et al. (2018) [63] | The Rochester Epidemiology Project | Determining risk of pancreatic cancer in NOD patients | ≥50 years old who met the glycemic criteria of NOD | Age at onset of diabetes, weight alterations from onset, change in blood glucose over 1 year before NOD | Sensitivity: 78% Specificity: 82% (initial validation cohort) AUROC 0.72–0.75 (validation studies [64,65]) |
| Boursi et al. (2016) [70] | THIN database | Determining risk of pancreatic cancer in NOD patients | ≥35 years old at the time of NOD diagnosis | Age, BMI, change in BMI, smoking, use of proton pump inhibitors and anti-diabetic medication, HbA1c, cholesterol, hemoglobin, creatinine and alkaline phosphatase levels | Sensitivity: 44.7% Specificity: 94% AUROC: 0.82 |
| Ali et al. (2024) [71] | IMPROVE data set | Determining risk of pancreatic cancer in women with NOD | ≥50-year-old women with diagnosed NOD | Age at NOD diagnosis, severity of diabetes, use of prescription medication | Sensitivity: 69% Specificity: 69% AUROC: 0.73 |
| Boursi et al. (2022) [80] | THIN database | Determining risk of pancreatic cancer in patients with prediabetes | ≥35 years old at the time of impaired fasting glucose diagnosis (100–125 mg/dL) | Age, BMI, use of proton pump inhibitors, total cholesterol, LDL (low-density lipoprotein), alkaline phosphatase, ALT (alanine aminotransferase) | Sensitivity: 66.53% Specificity: 54.91% AUROC: 0.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mejza, M.; Bajer, A.; Wanibuchi, S.; Małecka-Wojciesko, E. Can AI Be Useful in the Early Detection of Pancreatic Cancer in Patients with New-Onset Diabetes? Biomedicines 2025, 13, 836. https://doi.org/10.3390/biomedicines13040836
Mejza M, Bajer A, Wanibuchi S, Małecka-Wojciesko E. Can AI Be Useful in the Early Detection of Pancreatic Cancer in Patients with New-Onset Diabetes? Biomedicines. 2025; 13(4):836. https://doi.org/10.3390/biomedicines13040836
Chicago/Turabian StyleMejza, Maja, Anna Bajer, Sora Wanibuchi, and Ewa Małecka-Wojciesko. 2025. "Can AI Be Useful in the Early Detection of Pancreatic Cancer in Patients with New-Onset Diabetes?" Biomedicines 13, no. 4: 836. https://doi.org/10.3390/biomedicines13040836
APA StyleMejza, M., Bajer, A., Wanibuchi, S., & Małecka-Wojciesko, E. (2025). Can AI Be Useful in the Early Detection of Pancreatic Cancer in Patients with New-Onset Diabetes? Biomedicines, 13(4), 836. https://doi.org/10.3390/biomedicines13040836







