Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Cell Culture
2.3. Lentiviral Transduction
2.4. Flow Cytometry Analysis
2.4.1. Transfection Efficiency
2.4.2. Flow Cytometric Analysis of Cell Size and p21 Expression
2.4.3. Cell Counting and Growth Rate Determination
2.4.4. Cell Cycle Analysis
2.5. Real-Time PCR
2.6. Fluorescence Microscopy
2.7. Colony Formation Assay
2.8. Sulforhodamine B (SRB) Assay
2.9. Western Immunoblotting
2.10. Sphere-Formation Assay
2.11. Scratch Assay
2.12. Statistical Analysis
3. Results
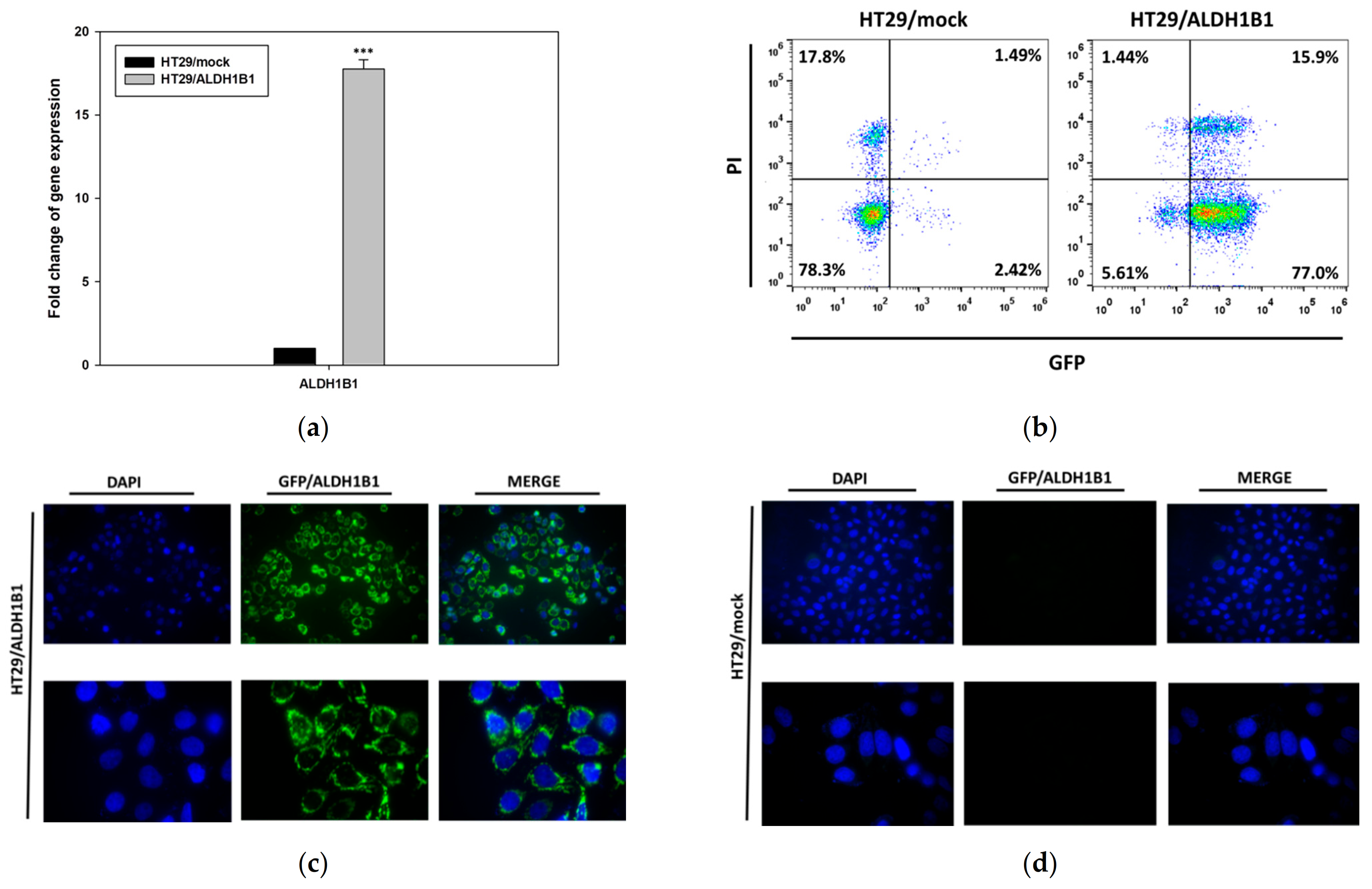
3.1. Generation and Characterization of the HT29 Isogenic Cell Line Pair
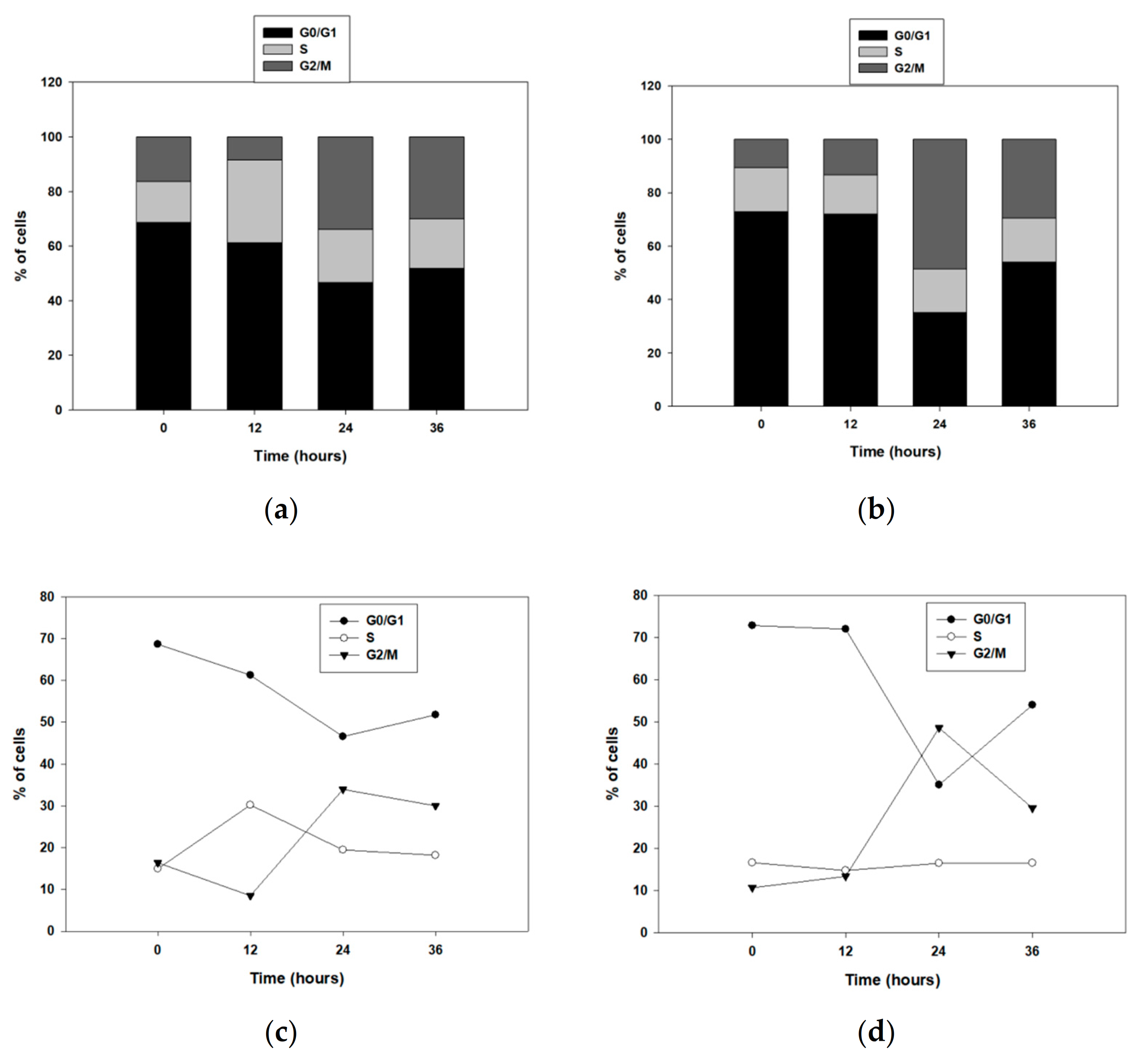
3.2. ALDH1B1 Is Related to Decreased Cell Proliferation and Clonogenicity and Induces G2/M Arrest through p53 Upregulation in HT29 Cells
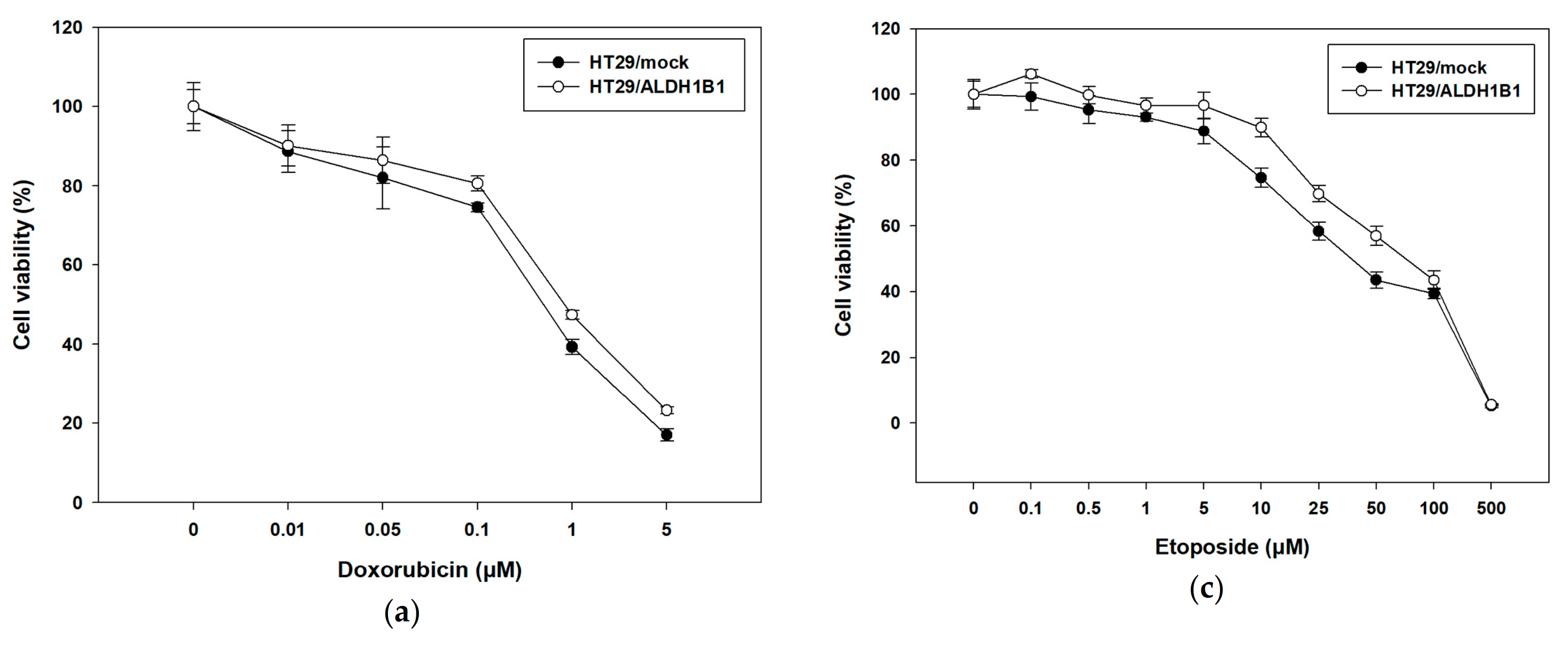
3.3. Expression of ALDH1B1 Confers Resistance to Various Chemotherapeutic Agents in HT29 Cells
3.4. ALDH1B1 Promotes Migration and EMT through ZEB1 and Vimentin Upregulation in HT29 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 2008, 4, 697–720. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Mou, Y.P.; Chen, K.; Cai, J.Q.; Zhou, Y.C.; Pan, Y.; Xu, X.W.; Zhou, W.; Gao, J.Q.; Chen, D.W.; et al. Aldehyde dehydrogenase 3A1 is robustly upregulated in gastric cancer stem-like cells and associated with tumorigenesis. Int. J. Oncol. 2016, 49, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Voulgaridou, G.P.; Kiziridou, M.; Mantso, T.; Chlichlia, K.; Galanis, A.; Koukourakis, M.I.; Franco, R.; Panayiotidis, M.I.; Pappa, A. Aldehyde dehydrogenase 3A1 promotes multi-modality resistance and alters gene expression profile in human breast adenocarcinoma MCF-7 cells. Int. J. Biochem. Cell Biol. 2016, 77, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, Y.; He, Y.; Cai, Q.; Gao, S.; Yao, W.; Liu, Z.; Tian, Z.; Han, Q.; Wang, W.; et al. Upregulation of ALDH1B1 promotes tumor progression in osteosarcoma. Oncotarget 2018, 9, 2502–2514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; De Melo, J.; Cutz, J.C.; Aziz, T.; Tang, D. Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br. J. Cancer 2014, 110, 2593–2603. [Google Scholar] [CrossRef] [PubMed]
- Voulgaridou, G.P.; Tsochantaridis, I.; Tolkas, C.; Franco, R.; Giatromanolaki, A.; Panayiotidis, M.I.; Pappa, A. Aldehyde dehydrogenase 3A1 confers oxidative stress resistance accompanied by altered DNA damage response in human corneal epithelial cells. Free Radic. Biol. Med. 2020, 150, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.W.; Palle, K. Aldehyde dehydrogenases in cancer stem cells: Potential as therapeutic targets. Ann. Transl. Med. 2016, 4, 518. [Google Scholar] [CrossRef] [PubMed]
- King, G.; Holmes, R. Human corneal and lens aldehyde dehydrogenases. Purification and properties of human lens ALDH1 and differential expression as major soluble proteins in human lens (ALDH1) and cornea (ALDH3). Adv. Exp. Med. Biol. 1997, 414, 19–27. [Google Scholar] [CrossRef]
- Hilton, J. Role of aldehyde dehydrogenase in cyclophosphamide-resistant L1210 leukemia. Cancer Res. 1984, 44, 5156–5160. [Google Scholar]
- Sladek, N.E.; Landkamer, G.J. Restoration of Sensitivity to Oxazaphosphorines by Inhibitors of Aldehyde Dehydrogenase Activity in Cultured Oxazaphosphorine-resistant L1210 and Cross-Linking Agent-resistant P388 Cell Lines. Cancer Res. 1985, 45, 1549–1555. [Google Scholar]
- Yoshida, A.; Dave, V.; Han, H.; Scanlon, K.J. Enhanced transcription of the cytosolic ALDH gene in cyclophosphamide resistant human carcinoma cells. Adv. Exp. Med. Biol. 1993, 328, 63–72. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer stem cells and radioresistance: DNA repair and beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol. Ther. 2016, 160, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [Green Version]
- Ginestier, C.; Korkaya, H.; Dontu, G.; Birnbaum, D.; Wicha, M.S.; Charafe-Jauffret, E. The cancer stem cell: The breast cancer driver. Med. Sci. 2007, 23, 1133–1139. [Google Scholar] [CrossRef] [Green Version]
- Deng, S.; Yang, X.; Lassus, H.; Liang, S.; Kaur, S.; Ye, Q.; Li, C.; Wang, L.P.; Roby, K.F.; Orsulic, S.; et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS ONE 2010, 5, e10277. [Google Scholar] [CrossRef] [PubMed]
- van Niekerk, G.; Davids, L.M.; Hattingh, S.M.; Engelbrecht, A.M. Cancer stem cells: A product of clonal evolution? Int. J. Cancer 2017, 140, 993–999. [Google Scholar] [CrossRef]
- Bu, P.; Chen, K.Y.; Lipkin, S.M.; Shen, X. Asymmetric division: A marker for cancer stem cells? Oncotarget 2013, 4, 950–951. [Google Scholar] [CrossRef] [Green Version]
- Valent, P.; Bonnet, D.; De Maria, R.; Lapidot, T.; Copland, M.; Melo, J.V.; Chomienne, C.; Ishikawa, F.; Schuringa, J.J.; Stassi, G.; et al. Cancer stem cell definitions and terminology: The devil is in the details. Nat. Rev. Cancer 2012, 12, 767–775. [Google Scholar] [CrossRef]
- Sládek, N.E.; Kollander, R.; Sreerama, L.; Kiang, D.T. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: A retrospective study. Cancer Chemother. Pharmacol. 2002, 49, 309–321. [Google Scholar] [CrossRef]
- Honoki, K.; Fujii, H.; Kubo, A.; Kido, A.; Mori, T.; Tanaka, Y.; Tsujiuchi, T. Possible involvement of stem-like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol. Rep. 2010, 24, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Kawasoe, M.; Yamamoto, Y.; Okawa, K.; Funato, T.; Takeda, M.; Hara, T.; Tsurumi, H.; Moriwaki, H.; Arioka, Y.; Takemura, M.; et al. Acquired resistance of leukemic cells to AraC is associated with the upregulation of aldehyde dehydrogenase 1 family member A2. Exp. Hematol. 2013, 41, 597–603.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cojoc, M.; Peitzsch, C.; Kurth, I.; Trautmann, F.; Kunz-Schughart, L.A.; Telegeev, G.D.; Stakhovsky, E.A.; Walker, J.R.; Simin, K.; Lyle, S.; et al. Aldehyde dehydrogenase is regulated by β-Catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res. 2015, 75, 1482–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/ electrophilicstress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Kim, R.J.; Park, J.R.; Roh, K.J.; Choi, A.R.; Kim, S.R.; Kim, P.H.; Yu, J.H.; Lee, J.W.; Ahn, S.H.; Gong, G.; et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2α. Cancer Lett. 2013, 333, 18–31. [Google Scholar] [CrossRef]
- Feng, H.; Liu, Y.; Bian, X.; Zhou, F.; Liu, Y. ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status. Br. J. Cancer 2018, 118, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Muzio, G.; Maggiora, M.; Paiuzzi, E.; Oraldi, M.; Canuto, R.A. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012, 52, 735–746. [Google Scholar] [CrossRef]
- Stagos, D.; Chen, Y.; Brocker, C.; Donald, E.; Jackson, B.C.; Orlicky, D.J.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenase 1B1: Molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab. Dispos. 2010, 38, 1679–1687. [Google Scholar] [CrossRef] [Green Version]
- Jackson, B.C.; Reigan, P.; Miller, B.; Thompson, D.C.; Vasiliou, V. Human ALDH1B1 Polymorphisms may Affect the Metabolism of Acetaldehyde and All-trans retinaldehyde—In Vitro Studies and Computational Modeling. Pharm. Res. 2015, 32, 1648–1662. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Arcaroli, J.; Chen, Y.; Thompson, D.C.; Messersmith, W.; Jimeno, A.; Vasiliou, V. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/β-catenin, Notch and PI3K/Akt signaling pathways. PLoS ONE 2015, 10, e0121648. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Arcaroli, J.; Chen, Y.; Gasparetto, M.; Neumeister, V.; Thompson, D.C.; Singh, S.; Smith, C.; Messersmith, W.; Vasiliou, V. Aldehyde dehydrogenase 1B1: A novel immunohistological marker for colorectal cancer. Br. J. Cancer 2017, 117, 1537–1543. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Orlicky, D.J.; Matsumoto, A.; Singh, S.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem. Biophys. Res. Commun. 2011, 405, 173–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langan, R.C.; Mullinax, J.E.; Ray, S.; Raiji, M.T.; Schaub, N.; Xin, H.W.; Koizumi, T.; Steinberg, S.M.; Anderson, A.; Wiegand, G.; et al. A pilot study assessing the potential role of non-CD133 colorectal cancer stem cells as biomarkers. J. Cancer 2012, 3, 231–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Arcaroli, J.J.; Orlicky, D.J.; Chen, Y.; Messersmith, W.A.; Bagby, S.; Purkey, A.; Quackenbush, K.S.; Thompson, D.C.; Vasiliou, V. Aldehyde Dehydrogenase 1B1 as a Modulator of Pancreatic Adenocarcinoma. Pancreas 2016, 45, 117–122. [Google Scholar] [CrossRef] [Green Version]
- Mameishvili, E.; Serafimidis, I.; Iwaszkiewicz, S.; Lesche, M.; Reinhardt, S.; Bölicke, N.; Büttner, M.; Stellas, D.; Papadimitropoulou, A.; Szabolcs, M.; et al. Aldh1b1 expression defines progenitor cells in the adult pancreas and is required for Kras-induced pancreatic cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 20679–20688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.K.; Wang, X.K.; Liao, X.W.; Han, C.Y.; Yu, T.D.; Qin, W.; Zhu, G.Z.; Su, H.; Yu, L.; Liu, X.G.; et al. Aldehyde dehydrogenase 1 (ALDH1) isoform expression and potential clinical implications in hepatocellular carcinoma. PLoS ONE 2017, 12, e0182208. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Guo, X.; Wang, Z.; Li, X.; Bu, Y.; Bai, X.; Zheng, L.; Huang, Y. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco Targets Ther. 2016, 9, 3405–3414. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Song, X.; Yu, L.; Li, J.; Qiao, Z.; Jiu, R.; Yu, B.; Liu, X. Increased expression of acetaldehyde dehydrogenase in cisplatin-resistant human lung adenocarcinoma A549/DDP cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2015, 31, 625–629. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25940289 (accessed on 12 April 2020).
- Jaroszeski, M.J.; Radcliff, G. Fundamentals of flow cytometry. Appl. Biochem. Biotechnol. Part B Mol. Biotechnol. 1999, 11, 37–53. [Google Scholar] [CrossRef]
- Karapetsas, A.; Voulgaridou, G.P.; Iliadi, D.; Tsochantaridis, I.; Michail, P.; Kynigopoulos, S.; Lambropoulou, M.; Stavropoulou, M.I.; Stathopoulou, K.; Karabournioti, S.; et al. Honey extracts exhibit cytoprotective properties against UVB-induced photodamage in human experimental skin models. Antioxidants 2020, 9, 566. [Google Scholar] [CrossRef]
- Anestopoulos, I.; Sfakianos, A.P.; Franco, R.; Chlichlia, K.; Panayiotidis, M.I.; Kroll, D.J.; Pappa, A. A novel role of silibinin as a putative epigenetic modulator in human prostate carcinoma. Molecules 2017, 22, 62. [Google Scholar] [CrossRef] [PubMed]
- Rybak, A.P.; He, L.; Kapoor, A.; Cutz, J.C.; Tang, D. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 683–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olejniczak, A.; Szaryńska, M.; Kmieć, Z. In vitro characterization of spheres derived from colorectal cancer cell lines. Int. J. Oncol. 2018, 52, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [PubMed]
- Floor, S.; Van Staveren, W.C.G.; Larsimont, D.; Dumont, J.E.; Maenhaut, C. Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: Distinct, overlapping or same populations. Oncogene 2011, 30, 4609–4621. [Google Scholar] [CrossRef] [Green Version]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Anastasiou, V.; Ninou, E.; Alexopoulou, D.; Stertmann, J.; Müller, A.; Dahl, A.; Solimena, M.; Speier, S.; Serafimidis, I.; Gavalas, A. Aldehyde dehydrogenase activity is necessary for beta cell development and functionality in mice. Diabetologia 2016, 59, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Gibb, Z.; Lambourne, S.R.; Curry, B.J.; Hall, S.E.; Aitken, R.J. Aldehyde Dehydrogenase Plays a Pivotal Role in the Maintenance of Stallion Sperm Motility. Biol. Reprod. 2016, 94, 133. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chen, Y.; Matsumoto, A.; Orlicky, D.J.; Dong, H.; Thompson, D.C.; Vasiliou, V. ALDH1B1 links alcohol consumption and diabetes. Biochem. Biophys. Res. Commun. 2015, 463, 768–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Dinavahi, S.S.; Bazewicz, C.G.; Gowda, R.; Robertson, G.P. Aldehyde Dehydrogenase Inhibitors for Cancer Therapeutics. Trends Pharmacol. Sci. 2019, 40, 774–789. [Google Scholar] [CrossRef]
- Ginestier, C.; Wicinski, J.; Cervera, N.; Monville, F.; Finetti, P.; Bertucci, F.; Wicha, M.S.; Birnbaum, D.; Charafe-Jauffret1, E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle 2009, 8, 3297–3302. [Google Scholar] [CrossRef]
- Ying, M.; Wang, S.; Sang, Y.; Sun, P.; Lal, B.; Goodwin, C.R.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Laterra, J.; Xia, S. Regulation of glioblastoma stem cells by retinoic acid: Role for Notch pathway inhibition. Oncogene 2011, 30, 3454–3467. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.M.; Khauv, D.; Bissell, M.J.; Radisky, D.C. Change in cell shape is required for matrix metalloproteinase-induced epithelial-mesenchymal transition of mammary epithelial cells. J. Cell Biochem. 2008, 105, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Leggett, S.E.; Sim, J.Y.; Rubins, J.E.; Neronha, Z.J.; Williams, E.K.; Wong, I.Y. Morphological single cell profiling of the epithelial-mesenchymal transition. Integr. Biol. 2016, 8, 1133–1144. [Google Scholar] [CrossRef] [Green Version]
- Rao, Q.X.; Yao, T.T.; Zhang, B.Z.; Lin, R.C.; Chen, Z.L.; Zhou, H.; Wang, L.J.; Lu, H.W.; Chen, Q.; Di, N.; et al. Expression and functional role of ALDH1 in cervical carcinoma cells. Asian Pacific J. Cancer Prev. 2012, 13, 1325–1331. [Google Scholar] [CrossRef] [Green Version]
- Moreb, J.S.; Ucar, D.; Han, S.; Amory, J.K.; Goldstein, A.S.; Ostmark, B.; Chang, L.J. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem. Biol. Interact. 2012, 195, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Awad, O.; Yustein, J.T.; Shah, P.; Gul, N.; Katuri, V.; O’Neill, A.; Kong, Y.; Brown, M.L.; Toretsky, J.A.; Loeb, D.M. High ALDH activity identifies chemotherapy-resistant Ewing’s sarcoma stem cells that retain sensitivity to EWS-Fli1 inhibition. PLoS ONE 2010, 5, e13943. [Google Scholar] [CrossRef] [Green Version]
- Ajani, J.A.; Wang, X.; Song, S.; Suzuki, A.; Taketa, T.; Sudo, K.; Wadhwa, R.; Hofstetter, W.L.; Komaki, R.; Maru, D.M.; et al. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol. Oncol. 2014, 8, 142–149. [Google Scholar] [CrossRef]
- Atashzar, M.R.; Baharlou, R.; Karami, J.; Abdollahi, H.; Rezaei, R.; Pourramezan, F.; Moghaddam, S.H.Z. Cancer stem cells: A review from origin to therapeutic implications. J. Cell Physiol. 2020, 235, 790–803. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Brueckmann, I.; Scheel, C.; Kaestli, A.J.; Wiggins, P.A.; Rodrigues, L.O.; Brooks, M.; Reinhardt, F.; Su, Y.; Polyak, K.; et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA 2011, 108, 7950–7955. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Tilló, E.; Lázaro, A.; Torrent, R.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Engel, P.; Postigo, A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010, 29, 3490–3500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Li, B.; Liu, F.; Zhang, M.; Wang, Q.; Liu, Y.; Yao, Y.; Li, D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Mol. Cancer 2017, 16, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Wolf, K. Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Moreb, J.S.; Baker, H.V.; Chang, L.J.; Amaya, M.; Lopez, M.C.; Ostmark, B.; Chou, W. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol. Cancer 2008, 7, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croker, A.K.; Rodriguez-Torres, M.; Xia, Y.; Pardhan, S.; Leong, H.S.; Lewis, J.D.; Allan, A.L. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. Int. J. Mol. Sci. 2017, 18, 2039. [Google Scholar] [CrossRef] [PubMed]
- Evdokimova, V.; Tognon, C.; Ng, T.; Sorensen, P.H.B. Reduced proliferation and enhanced migration: Two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 2009, 8, 2901–2906. [Google Scholar] [CrossRef]
- Collins, N.L.; Reginato, M.J.; Paulus, J.K.; Sgroi, D.C.; LaBaer, J.; Brugge, J.S. G1/S Cell Cycle Arrest Provides Anoikis Resistance through Erk-Mediated Bim Suppression. Mol. Cell Biol. 2005, 25, 5282–5291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell 2014, 14, 306–321. [Google Scholar] [CrossRef] [Green Version]
- Lim, W.; Kim, H.E.; Kim, Y.; Na, R.; Li, X.; Jeon, S.; Choi, H.; Kim, O. Association between cancer stem cell-like properties and epithelial-to-mesenchymal transition in primary and secondary cancer cells. Int. J. Oncol. 2016, 49, 991–1000. [Google Scholar] [CrossRef]
- Anjomshoaa, A.; Nasri, S.; Humar, B.; McCall, J.L.; Chatterjee, A.; Yoon, H.S.; McNoe, L.; Black, M.A.; Reeve, A.E. Slow proliferation as a biological feature of colorectal cancer metastasis. Br. J. Cancer 2009, 101, 822–828. [Google Scholar] [CrossRef] [PubMed]






| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| β-actin | GCGCGGCTACAGCTTCA | CTTAATGTCACGCACGATTTCC |
| ALDH1B1 | AGCCTCTGTTCAAGTTCAAG | CCTTAAAGCCTCCGAATGG |
| E-cadherin | TACACTGCCCAGGAGCCAGA | TGGCACCAGTGTCCGGATTA |
| SNAI1 | ACTATGCCGCGCTCTTTCCT | GGTGGGGTTGAGGATCTCCG |
| SNAI2 | CTACAGCGAACTGGACACAC | TGTGGTATGACAGGCATGGAG |
| vimentin | TGAGTACCGGAGACAGGTGCAG | TAGCAGCTTCAACGGCAAAGTTC |
| ZEB1 | CGAGTCAGATGCAGAAAATGAGCAA | ACCCAGACTGCGTCACATGTCTT |
| ZEB2 | ACTATGGGGCCAGAAGCCAC | CTGCATGACCATCGCGTTCC |
| TWIST1 | AGCTACGCCTTCTCGGTCTG | TGGGAATCACTGTCCACGGG |
| N-cadherin | CCTACTGGACGGTTCGCCAT | GTTGCAGTTGACTGAGGCGG |
| Parameter | HT29/mock | HT29/ALDH1B1 | Statistical Significance | |
|---|---|---|---|---|
| Median Fluorescence Intensity | FSC | 247,552 ± 13,408.88 † | 292,973.2 ± 37,072.24 † | * |
| SSC | 208,992 ± 20,580.28 † | 167,270.4 ± 19,655.07 † | * |
| Sample | Doubling Time (tD) (h) | Growth Rates (GR) |
|---|---|---|
| HT29/mock | 24.16 ± 0.05 | 0.029 ± 0.0004 |
| HT29/ALDH1B1 | 26.75 ± 0.02 | 0.026 ± 0.0007 |
| Time (h) | HT29/mock | HT29/ALDH1B1 | Statistical Significance | |
|---|---|---|---|---|
| Median Fluorescence Intensity | 0 | 90 ± 3.68 † | 142 ± 5.66 † | * |
| 24 | 92.93 ± 2.08 † | 168.67 ± 10.01 † | ** |
| Sample | Doxorubicin (μM) (72 h) | 5-FU (μM) (72 h) | Etoposide (μM) (48 h) |
|---|---|---|---|
| HT29/mock | 0.59 ± 0.079 † | 0.92 ± 0.15 † | 67.85 ± 1.67 † |
| HT29/ALDH1B1 | 1.073 ± 0.13 † | 1.79 ± 0.056 † | 92.47 ± 0.65 † |
| Statistical significance | ** | *** | ** |
| Sample | Untreated | Etoposide 20 μM | Etoposide 25 μM | |
|---|---|---|---|---|
| HT29/mock | PI− | 77 ± 1.91 † | 71.2 ± 1.37 † | 63.3 ± 2.33 † |
| PI+ | 23 ± 2.16 † | 28.8 ± 0.95 † | 36.7 ± 1.44 † | |
| HT29/ALDH1B1 | PI− | 80.5 ± 1.66 † | 76.6 ± 1.14 † | 71.1 ± 1.77 † |
| PI+ | 19.5 ± 1.31 † | 23.4 ± 2.03 † | 28.9 ± 1.82 † | |
| Statistical significance | PI− vs. PI− | - | * | *** |
| PI+ vs. PI+ | - | * | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsochantaridis, I.; Roupas, A.; Voulgaridou, G.-P.; Giatromanolaki, A.; Koukourakis, M.I.; Panayiotidis, M.I.; Pappa, A. Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells. Biomedicines 2021, 9, 44. https://doi.org/10.3390/biomedicines9010044
Tsochantaridis I, Roupas A, Voulgaridou G-P, Giatromanolaki A, Koukourakis MI, Panayiotidis MI, Pappa A. Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells. Biomedicines. 2021; 9(1):44. https://doi.org/10.3390/biomedicines9010044
Chicago/Turabian StyleTsochantaridis, Ilias, Angelos Roupas, Georgia-Persephoni Voulgaridou, Alexandra Giatromanolaki, Michael I. Koukourakis, Mihalis I. Panayiotidis, and Aglaia Pappa. 2021. "Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells" Biomedicines 9, no. 1: 44. https://doi.org/10.3390/biomedicines9010044







