Transcriptional Profiling of Tumorspheres Reveals TRPM4 as a Novel Stemness Regulator in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Pharmacological Agents
2.2. TRPM4 Knock-Down
2.3. Mammosphere Formation Assay
2.4. Flow Cytometry
2.5. Pharmacological Treatment
2.6. Immunoblot Analysis
2.7. Immunofluorescence Analysis
2.8. RNA Extraction, cDNA Synthesis and q-RT-PCR
2.9. RNA-Sequencing (RNA-seq) and Bioinformatic Analysis
2.10. Gene Ontology and Gene Set Enrichment Analysis of DEGs
2.11. Statistical Analysis
3. Results
3.1. RNA Sequencing Data Analysis
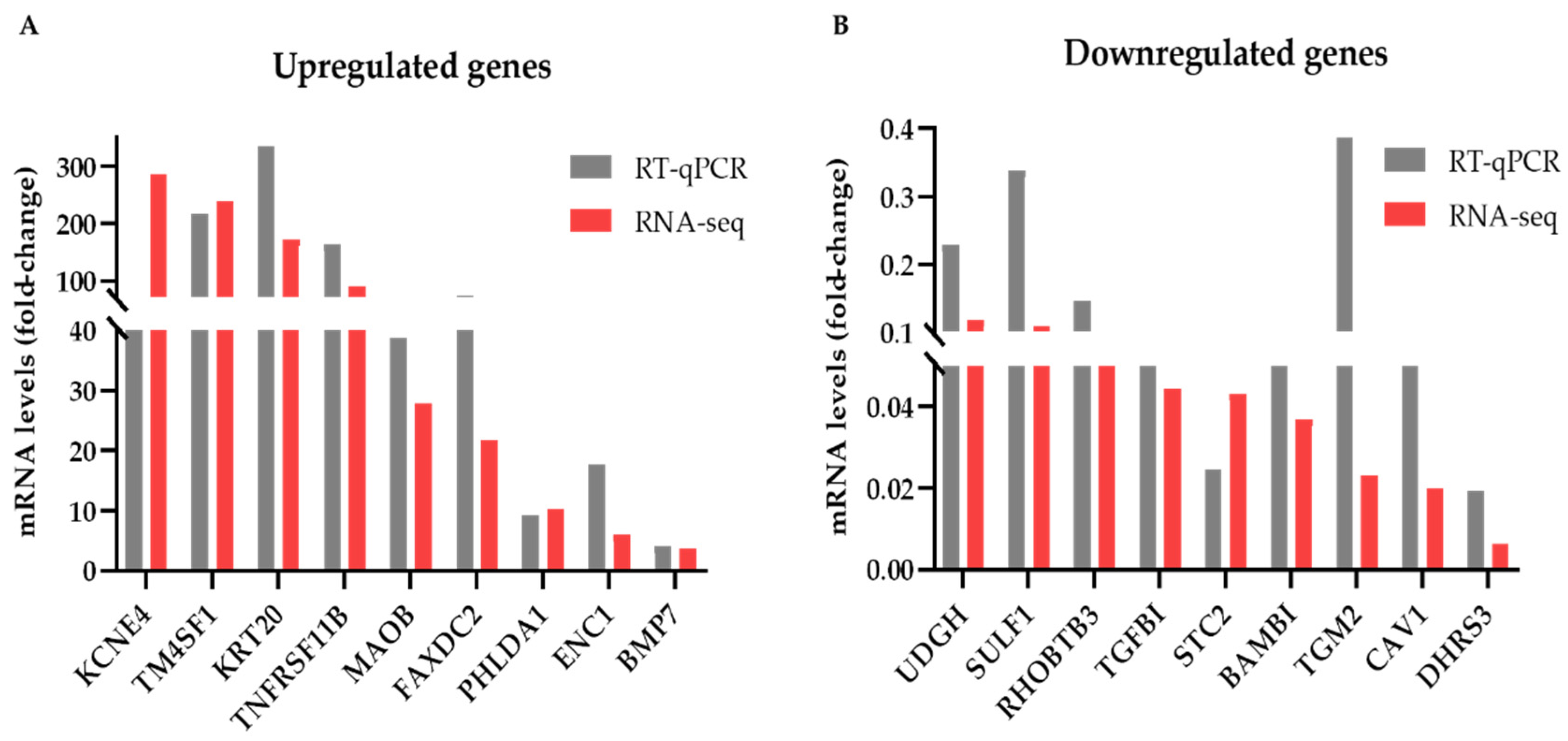
3.2. Identification and Validation of Differentially Expressed Genes
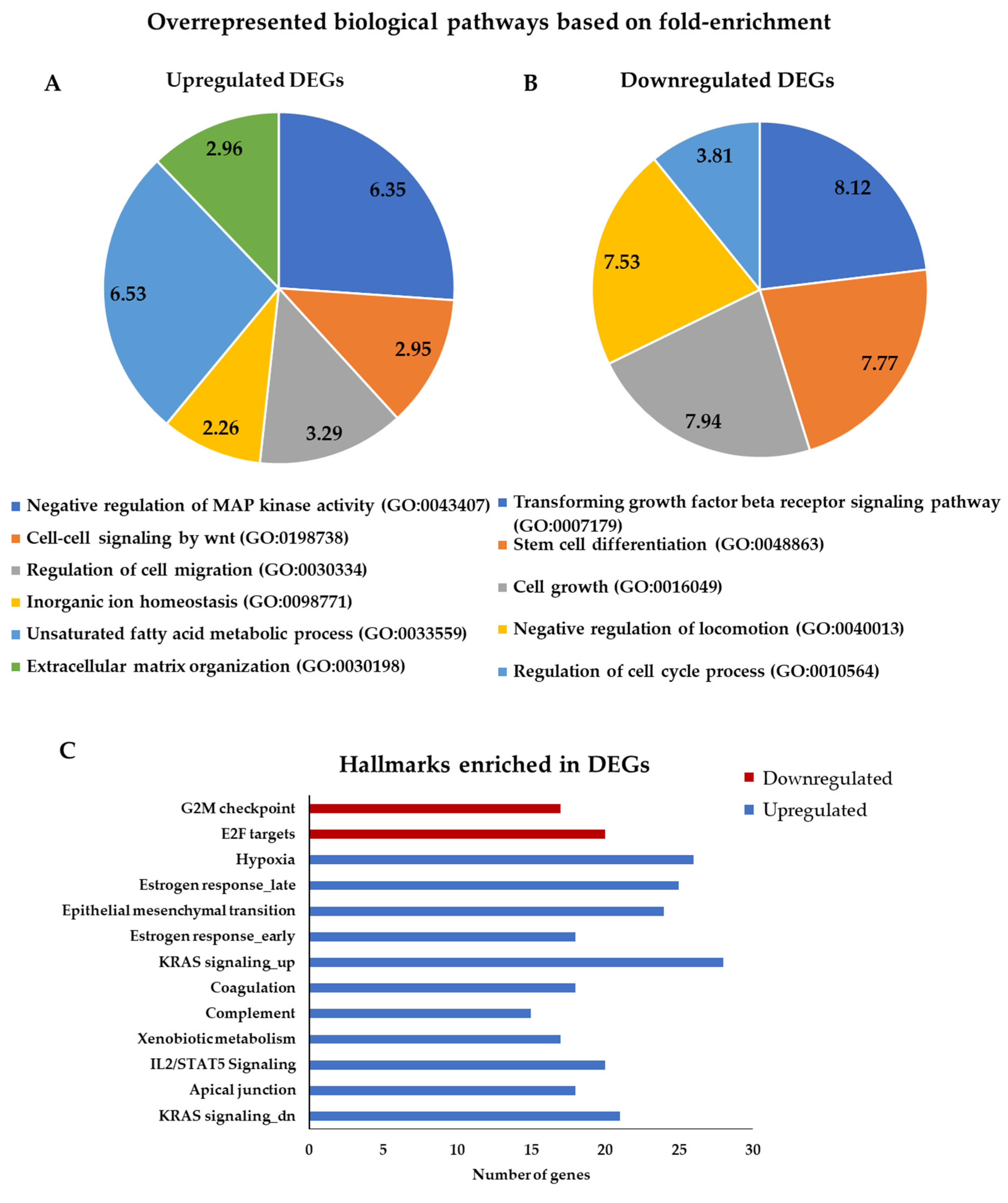
3.3. Gene Ontology and Gene Set Enrichment Analysis for DEGs
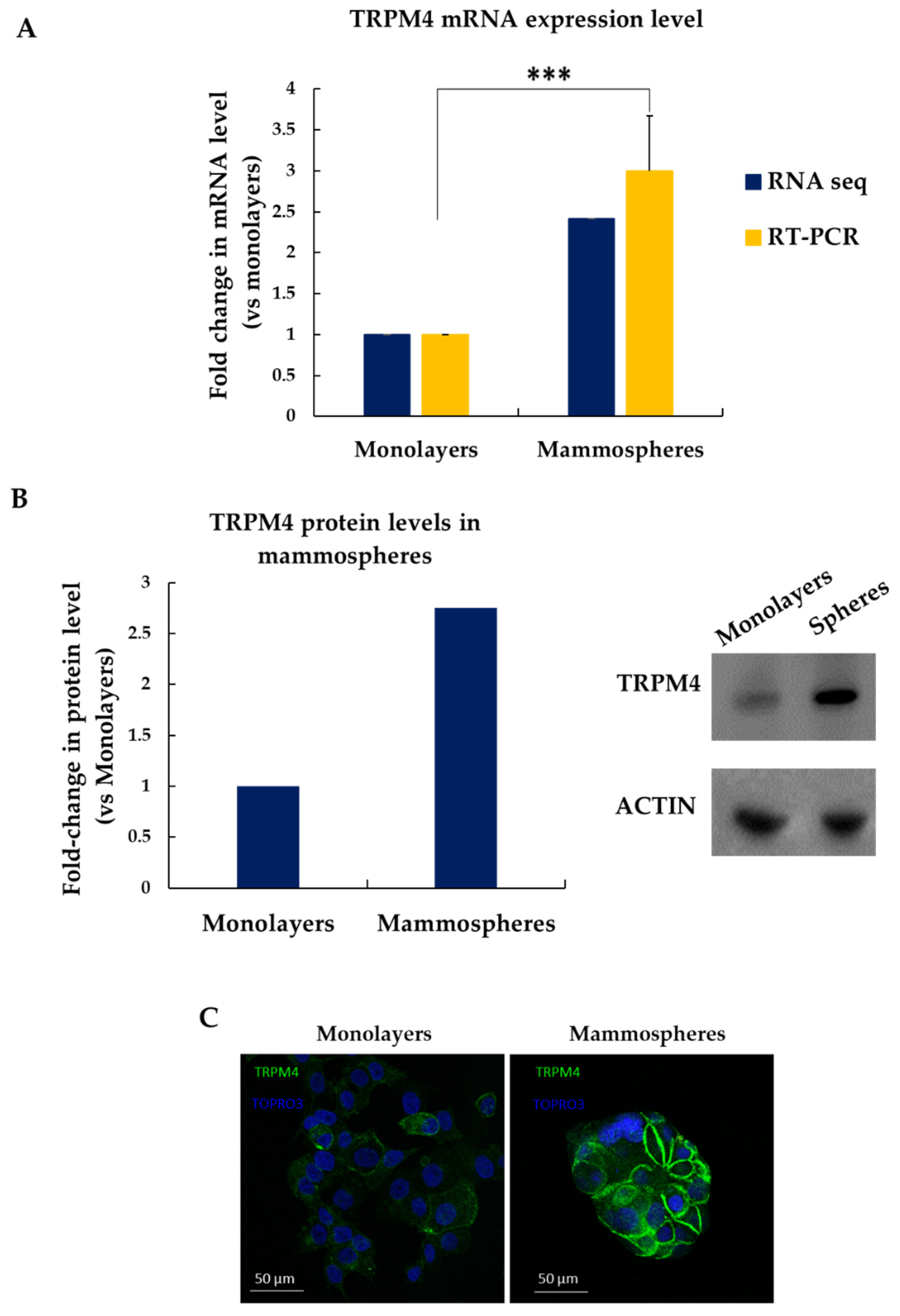
3.4. TRPM4 Is Overexpressed in MCF-7 Mammospheres
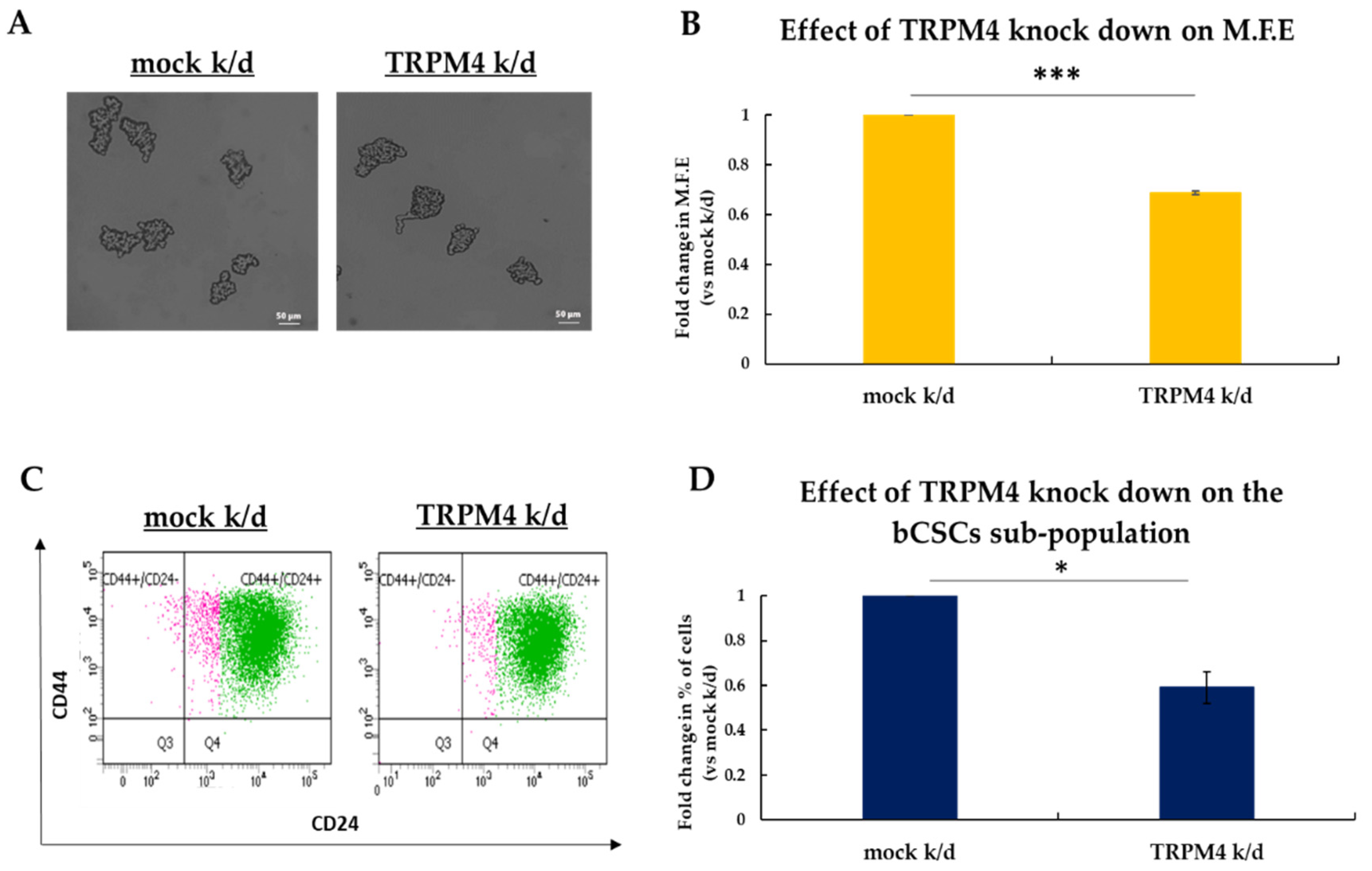
3.5. TRPM4 Regulates the Stemness Properties of bCSCs In Vitro
3.6. TRPM4 Is a Potential Druggable Target in bCSCs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 1 August 2021).
- Ji, X.; Lu, Y.; Tian, H.; Meng, X.; Wei, M.; Cho, W.C. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed. Pharm. 2019, 114, 108800. [Google Scholar] [CrossRef]
- Ignatov, A.; Eggemann, H.; Burger, E.; Ignatov, T. Patterns of breast cancer relapse in accordance to biological subtype. J. Cancer Res. Clin. Oncol. 2018, 144, 1347–1355. [Google Scholar] [CrossRef]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Fillmore, C.M.; Kuperwasser, C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. BCR 2008, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Ablett, M.P.; Singh, J.K.; Clarke, R.B. Stem cells in breast tumours: Are they ready for the clinic? Eur. J. Cancer 2012, 48, 2104–2116. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.Q.; Alexandrou, A.T.; Li, J.J. Breast cancer stem cells: Multiple capacities in tumor metastasis. Cancer Lett. 2014, 349, 1–7. [Google Scholar] [CrossRef]
- Saygin, C.; Matei, D.; Majeti, R.; Reizes, O.; Lathia, J.D. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell 2019, 24, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, M.; Maroufi, N.F.; Tazehkand, A.P.; Akbarzadeh, M.; Bastani, S.; Safdari, R.; Farzane, A.; Fattahi, A.; Nejabati, H.R.; Nouri, M.; et al. Current approaches in identification and isolation of cancer stem cells. J. Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [PubMed]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar] [CrossRef]
- Shaw, F.L.; Harrison, H.; Spence, K.; Ablett, M.P.; Simoes, B.M.; Farnie, G.; Clarke, R.B. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J. Mammary Gland. Biol. Neoplasia 2012, 17, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lv, Q.; Meng, W.; Tan, Q.; Zhang, S.; Mo, X.; Yang, X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J. Thorac. Dis. 2014, 6, 829–837. [Google Scholar] [CrossRef]
- Hantute-Ghesquier, A.; Haustrate, A.; Prevarskaya, N.; Lehen’kyi, V. TRPM Family Channels in Cancer. Pharmaceuticals 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.K.; Hussain, F.A. TRPM4 is overexpressed in breast cancer associated with estrogen response and epithelial-mesenchymal transition gene sets. PLoS ONE 2020, 15, e0233884. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, C.; Kappel, S.; Kilch, T.; Jochum, M.M.; Urban, S.K.; Jung, V.; Stockle, M.; Rother, K.; Greiner, M.; Peinelt, C. Transient receptor potential melastatin 4 channel contributes to migration of androgen-insensitive prostate cancer cells. Oncotarget 2015, 6, 41783–41793. [Google Scholar] [CrossRef]
- Verigos, J.; Karakaidos, P.; Kordias, D.; Papoudou-Bai, A.; Evangelou, Z.; Harissis, H.V.; Klinakis, A.; Magklara, A. The Histone Demethylase LSD1/KappaDM1A Mediates Chemoresistance in Breast Cancer via Regulation of a Stem Cell Program. Cancers 2019, 11, 1585. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Grimshaw, M.J.; Cooper, L.; Papazisis, K.; Coleman, J.A.; Bohnenkamp, H.R.; Chiapero-Stanke, L.; Taylor-Papadimitriou, J.; Burchell, J.M. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. BCR 2008, 10, R52. [Google Scholar] [CrossRef]
- Abd El-Rehim, D.M.; Pinder, S.E.; Paish, C.E.; Bell, J.; Blamey, R.W.; Robertson, J.F.; Nicholson, R.I.; Ellis, I.O. Expression of luminal and basal cytokeratins in human breast carcinoma. J. Pathol. 2004, 203, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Yue, X.; Li, X.; Taylor, J.S. Identification and characterization of high affinity antisense PNAs for the human unr (upstream of N-ras) mRNA which is uniquely overexpressed in MCF-7 breast cancer cells. Nucleic Acids Res. 2005, 33, 6700–6711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meric-Bernstam, F.; Chen, H.; Akcakanat, A.; Do, K.A.; Lluch, A.; Hennessy, B.T.; Hortobagyi, G.N.; Mills, G.B.; Gonzalez-Angulo, A. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res. BCR 2012, 14, R138. [Google Scholar] [CrossRef]
- Mehra, R.; Varambally, S.; Ding, L.; Shen, R.; Sabel, M.S.; Ghosh, D.; Chinnaiyan, A.M.; Kleer, C.G. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005, 65, 11259–11264. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Sharpe, R.; Pearson, A.; Campbell, J.; Natrajan, R.; Ashworth, A.; Turner, N.C. An siRNA screen identifies the GNAS locus as a driver in 20q amplified breast cancer. Oncogene 2014, 33, 2478–2486. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase regulates estrogen receptor-alpha signaling in breast cancer cells. Oncogenesis 2017, 6, e299. [Google Scholar] [CrossRef] [PubMed]
- Burandt, E.; Jens, G.; Holst, F.; Janicke, F.; Muller, V.; Quaas, A.; Choschzick, M.; Wilczak, W.; Terracciano, L.; Simon, R.; et al. Prognostic relevance of AIB1 (NCoA3) amplification and overexpression in breast cancer. Breast Cancer Res. Treat. 2013, 137, 745–753. [Google Scholar] [CrossRef]
- Dubrovska, A.; Hartung, A.; Bouchez, L.C.; Walker, J.R.; Reddy, V.A.; Cho, C.Y.; Schultz, P.G. CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling. Br. J. Cancer 2012, 107, 43–52. [Google Scholar] [CrossRef]
- Shi, Y.; Riese, D.J., 2nd; Shen, J. The Role of the CXCL12/CXCR4/CXCR7 Chemokine Axis in Cancer. Front. Pharmacol 2020, 11, 574667. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhfyan, A.; Alhoshani, A.; Korashy, H.M. Aryl hydrocarbon receptor/cytochrome P450 1A1 pathway mediates breast cancer stem cells expansion through PTEN inhibition and beta-Catenin and Akt activation. Mol. Cancer 2017, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Chakraborty, G.; Zhang, Z.; Akalay, I.; Gadiya, M.; Gao, Y.; Sinha, S.; Hu, J.; Jiang, C.; Akram, M.; et al. Multi-organ Site Metastatic Reactivation Mediated by Non-canonical Discoidin Domain Receptor 1 Signaling. Cell 2016, 166, 47–62. [Google Scholar] [CrossRef]
- Wang, S.; Wang, N.; Zheng, Y.; Yang, B.; Liu, P.; Zhang, F.; Li, M.; Song, J.; Chang, X.; Wang, Z. Caveolin-1 inhibits breast cancer stem cells via c-Myc-mediated metabolic reprogramming. Cell Death Dis. 2020, 11, 450. [Google Scholar] [CrossRef]
- Kang, Y.P.; Yoon, J.H.; Long, N.P.; Koo, G.B.; Noh, H.J.; Oh, S.J.; Lee, S.B.; Kim, H.M.; Hong, J.Y.; Lee, W.J.; et al. Spheroid-Induced Epithelial-Mesenchymal Transition Provokes Global Alterations of Breast Cancer Lipidome: A Multi-Layered Omics Analysis. Front. Oncol. 2019, 9, 145. [Google Scholar] [CrossRef]
- Mukherjee, A.; Kenny, H.A.; Lengyel, E. Unsaturated Fatty Acids Maintain Cancer Cell Stemness. Cell Stem Cell 2017, 20, 291–292. [Google Scholar] [CrossRef]
- de Sousa, E.M.F.; Vermeulen, L. Wnt Signaling in Cancer Stem Cell Biology. Cancers 2016, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lazaro, M. The migration ability of stem cells can explain the existence of cancer of unknown primary site. Rethinking metastasis. Oncoscience 2015, 2, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Biol. 2019, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Stelling, M.P.; Motta, J.M.; Mashid, M.; Johnson, W.E.; Pavao, M.S.; Farrell, N.P. Metal ions and the extracellular matrix in tumor migration. FEBS J. 2019, 286, 2950–2964. [Google Scholar] [CrossRef]
- Bellomo, C.; Caja, L.; Moustakas, A. Transforming growth factor beta as regulator of cancer stemness and metastasis. Br. J. Cancer 2016, 115, 761–769. [Google Scholar] [CrossRef]
- Borgstrom, A.; Peinelt, C.; Stoklosa, P. TRPM4 in Cancer—A New Potential Drug Target. Biomolecules 2021, 11, 229. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Song, K.; Farzaneh, M. Signaling pathways governing breast cancer stem cells behavior. Stem Cell Res. Ther. 2021, 12, 245. [Google Scholar] [CrossRef]
- Kotiyal, S.; Bhattacharya, S. Breast cancer stem cells, EMT and therapeutic targets. Biochem. Biophys. Res. Commun. 2014, 453, 112–116. [Google Scholar] [CrossRef]
- Daulat, A.M.; Bertucci, F.; Audebert, S.; Serge, A.; Finetti, P.; Josselin, E.; Castellano, R.; Birnbaum, D.; Angers, S.; Borg, J.P. PRICKLE1 Contributes to Cancer Cell Dissemination through Its Interaction with mTORC2. Dev. Cell 2016, 37, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Kappel, S.; Stoklosa, P.; Hauert, B.; Ross-Kaschitza, D.; Borgstrom, A.; Baur, R.; Galvan, J.A.; Zlobec, I.; Peinelt, C. TRPM4 is highly expressed in human colorectal tumor buds and contributes to proliferation, cell cycle, and invasion of colorectal cancer cells. Mol. Oncol. 2019, 13, 2393–2405. [Google Scholar] [CrossRef]
- Loo, S.K.; Ch’ng, E.S.; Md Salleh, M.S.; Banham, A.H.; Pedersen, L.M.; Moller, M.B.; Green, T.M.; Wong, K.K. TRPM4 expression is associated with activated B cell subtype and poor survival in diffuse large B cell lymphoma. Histopathology 2017, 71, 98–111. [Google Scholar] [CrossRef]
- Sagredo, A.I.; Sagredo, E.A.; Pola, V.; Echeverria, C.; Andaur, R.; Michea, L.; Stutzin, A.; Simon, F.; Marcelain, K.; Armisen, R. TRPM4 channel is involved in regulating epithelial to mesenchymal transition, migration, and invasion of prostate cancer cell lines. J. Cell Physiol. 2019, 234, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Rivas, J.; Diaz, N.; Silva, I.; Morales, D.; Lavanderos, B.; Alvarez, A.; Saldias, M.P.; Pulgar, E.; Cruz, P.; Maureira, D.; et al. KCTD5, a novel TRPM4-regulatory protein required for cell migration as a new predictor for breast cancer prognosis. FASEB J. 2020, 34, 7847–7865. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, M.; Zhou, F.; Zhang, L.; Meng, X. The Breast Cancer Stem Cells Traits and Drug Resistance. Front. Pharmacol. 2020, 11, 599965. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wicha, M.S. Targeting breast cancer stem cells. J. Clin. Oncol. 2010, 28, 4006–4012. [Google Scholar] [CrossRef]
- Yan, W.; Chen, Y.; Yao, Y.; Zhang, H.; Wang, T. Increased invasion and tumorigenicity capacity of CD44+/CD24- breast cancer MCF7 cells in vitro and in nude mice. Cancer Cell Int. 2013, 13, 62. [Google Scholar] [CrossRef] [PubMed]
- Biasiotta, A.; D’Arcangelo, D.; Passarelli, F.; Nicodemi, E.M.; Facchiano, A. Ion channels expression and function are strongly modified in solid tumors and vascular malformations. J. Transl. Med. 2016, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, M.A.; Baskin, D.S. Monoamine oxidase B levels are highly expressed in human gliomas and are correlated with the expression of HiF-1alpha and with transcription factors Sp1 and Sp3. Oncotarget 2016, 7, 3379–3393. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Feng, Z.; He, M.L. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics 2020, 10, 7053–7069. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Chen, A.; Du, Q.; Liao, Q.; Shuai, Z.; Chen, C.; Yang, X.; Hu, Y.; Zhao, J.; Liu, S.; et al. Novel insights into ion channels in cancer stem cells (Review). Int. J. Oncol. 2018, 53, 1435–1441. [Google Scholar] [CrossRef]
- Burris, S.K.; Wang, Q.; Bulley, S.; Neeb, Z.P.; Jaggar, J.H. 9-Phenanthrol inhibits recombinant and arterial myocyte TMEM16A channels. Br. J. Pharmacol. 2015, 172, 2459–2468. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- VanderVorst, K.; Dreyer, C.A.; Konopelski, S.E.; Lee, H.; Ho, H.H.; Carraway, K.L., 3rd. Wnt/PCP Signaling Contribution to Carcinoma Collective Cell Migration and Metastasis. Cancer Res. 2019, 79, 1719–1729. [Google Scholar] [CrossRef]
- Arullampalam, P.; Preti, B.; Ross-Kaschitza, D.; Lochner, M.; Rougier, J.S.; Abriel, H. Species-Specific Effects of Cation Channel TRPM4 Small-Molecule Inhibitors. Front. Pharmacol. 2021, 12, 712354. [Google Scholar] [CrossRef]
- Low, S.W.; Gao, Y.; Wei, S.; Chen, B.; Nilius, B.; Liao, P. Development and characterization of a monoclonal antibody blocking human TRPM4 channel. Sci. Rep. 2021, 11, 10411. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verigos, J.; Kordias, D.; Papadaki, S.; Magklara, A. Transcriptional Profiling of Tumorspheres Reveals TRPM4 as a Novel Stemness Regulator in Breast Cancer. Biomedicines 2021, 9, 1368. https://doi.org/10.3390/biomedicines9101368
Verigos J, Kordias D, Papadaki S, Magklara A. Transcriptional Profiling of Tumorspheres Reveals TRPM4 as a Novel Stemness Regulator in Breast Cancer. Biomedicines. 2021; 9(10):1368. https://doi.org/10.3390/biomedicines9101368
Chicago/Turabian StyleVerigos, John, Dimitris Kordias, Styliani Papadaki, and Angeliki Magklara. 2021. "Transcriptional Profiling of Tumorspheres Reveals TRPM4 as a Novel Stemness Regulator in Breast Cancer" Biomedicines 9, no. 10: 1368. https://doi.org/10.3390/biomedicines9101368
APA StyleVerigos, J., Kordias, D., Papadaki, S., & Magklara, A. (2021). Transcriptional Profiling of Tumorspheres Reveals TRPM4 as a Novel Stemness Regulator in Breast Cancer. Biomedicines, 9(10), 1368. https://doi.org/10.3390/biomedicines9101368






